Abstract
In vitro studies have shown that enteroviruses employ strategies that may impair the ability of DCs to trigger T cell immunity, but it is unclear how these viruses affect DCs in vivo. Here, we evaluate the effects of wild-type (wt) coxsackievirus B3 on DCs in vitro and in a murine model in vivo. Although CVB3 does not productively infect the vast majority of DCs, virus infection profoundly reduces splenic conventional DCs numbers and diminishes their capacity to prime naïve CD8+ T cells in vitro. In contrast to recombinant CVB3, highly pathogenic wt virus infection significantly diminishes the host’s capacity to mount T cell responses, which is temporally associated with the loss of CD8α+ DCs. Our findings demonstrate that enterovirus infection substantially alters the number, heterogeneity, and stimulatory capacity of DCs in vivo, and these dramatic immunomodulatory effects may weaken the host’s capacity to mount antiviral T cell responses.
Keywords: coxsackievirus, lymphocytic choriomeningitis virus, dendritic cell, T cell, mouse, spleen, infection, immunity
Introduction
Dendritic cells (DCs) are specialized antigen presenting cells that link the immediate innate immune response to viral infection with the subsequent generation of an adaptive immune response (Lee and Iwasaki, 2007). DCs comprise 2 major groups, plasmacytoid DCs (pDC) and conventional DCs (cDC). pDCs play an important role in shaping the innate and adaptive antiviral immune responses by their ability to secrete large amounts of type I interferon (IFN) in response to viral nucleic acid recognition (Gilliet et al., 2008; Swiecki and Colonna, 2010b). Although pDCs are capable of antigen presentation (Villadangos and Young, 2008), it is the cDCs that play a major role in priming virus-specific T cell responses (Heath and Carbone, 2009; Lopez-Bravo and Ardavin, 2008). cDCs in the spleen can be divided into 3 major subsets: CD8α+ DCs, CD4+ DCs, and CD8−CD4− “double negative” DCs. These subsets also are found in peripheral lymph nodes, along with other subsets of migratory conventional DCs (Heath and Carbone, 2009; Lopez-Bravo and Ardavin, 2008). In addition, the inflammatory environment generated by virus infection can trigger the differentiation of monocyte-derived DCs that contribute to antiviral immunity and stimulate virus-specific T cell responses (Dominguez and Ardavin, 2010; Leon and Ardavin, 2008). Studies have shown that cDC subsets exert distinct functions in activating antiviral T cell responses. CD8α+ DCs are particularly efficient at capturing apoptotic cells and priming virus-specific CD8+ T cell responses. In contrast, CD8α− DCs are specialized for processing and presentation of antigens on MHC class II and preferentially trigger CD4+ T cell responses (Lopez-Bravo and Ardavin, 2008; Segura and Villadangos, 2009; Shortman and Heath, 2010; Villadangos and Schnorrer, 2007). The functional specialization of CD8α+ and CD8α− subsets may be related to differences in their expression of viral RNA sensors and/or machinery involved in antigen processing and presentation (Dudziak et al., 2007; Luber et al., 2010).
Type B Coxsackieviruses (CVB) are important human pathogens that belong to the Picornavirus family and human enterovirus B genus. A considerable proportion of CVB infections trigger severe acute and chronic diseases and cause morbidity and mortality, particularly in infants, young children, and immunocompromised individuals (Modlin and Rotbart, 1997; Romero, 2008; Whitton, 2002). CVB are the most common cause of infectious myocarditis, a serious disease that can lead to dilated cardiomyopathy and cardiac failure (O'Connell, 1987; Sole and Liu, 1993; Tam, 2006). These viruses also can induce other diseases, including pancreatitis, aseptic meningitis, and encephalitis (Daley et al., 1998; Feuer et al., 2005; Huber and Ramsingh, 2004; Mena et al., 2000; Rhoades et al., 2011; Tracy and Chapman, 2010). Control of CVB infection depends on both humoral and cell mediated-immunity (Kemball et al., 2010a; Kemball et al., 2010b). CD8+ T cell responses directed at native epitopes in the CVB polyprotein are remarkably weak (Jakel et al., 2009; Varela-Calvino et al., 2004; Voigt et al., 2010; Weinzierl et al., 2008a) and, in order to enhance our ability to detect virus-specific T cells, our laboratory generated recombinant CVB3 (rCVB3) that encode well characterized CD8+ and/or CD4+ T cell epitopes from lymphocytic choriomeningitis virus (LCMV). Despite their ability to replicate to high titers in vivo, wild-type (wt) CVB3 and rCVB3 induce minimal T cell activation and weak endogenous virus-specific T cell responses (Crocker et al., 2007; Kemball et al., 2008; Slifka et al., 2001). Using epitope-specific CD8+ transgenic T cells as sensors to evaluate in vivo antigen presentation by rCVB3, we demonstrated that this virus almost completely evades presentation through the MHC class I pathway (Kemball et al., 2009). This strategy is extremely effective, as naïve virus-specific CD8+ transgenic T cells fail to expand in rCVB3-infected mice. Memory CD8+ T cells, however, can respond to a lower level of antigen presentation than their naïve counterparts (Slifka and Whitton, 2001), and we found that epitope-specific memory CD8+ T cells were activated by rCVB3 infection in vivo (Kemball et al., 2008). Therefore, we have suggested that the quantity of MHC class I/viral peptide complex that is expressed on a CVB3-infected cell is too low to trigger strong responses by naïve CD8+ T cells, but is sufficient to activate memory CD8+ T cells.
Activation of naïve CD8+ T cells is profoundly dependent upon antigen presentation by cDCs. However, the effects of coxsackievirus infection on dendritic cell populations in vivo are not well understood. In vitro studies have shown that other enteroviruses can infect DCs and employ tactics that may impair the ability of DCs to prime robust T cell responses. Poliovirus, a close relative of CVB, can inhibit protein synthesis in DCs and dampens MHC class I antigen presentation (Choe et al., 2005; Deitz et al., 2000; Wahid et al., 2005); the virus also inhibits proinflammatory cytokine secretion and the expression of antiviral cytokine receptors (Dodd et al., 2001; Neznanov et al., 2001). Furthermore, echovirus infection limits the ability of DCs to up-regulate costimulatory molecules and to produce cytokines following TLR stimulation (Kramer et al., 2007). However, it is not known whether picornaviruses exert similar effects on DCs in vivo. A recent study reported that positive and negative strand CVB3 RNA was present in DCs following infection in vitro and in vivo but, in both cases, viral replication was incomplete and the infection was non-productive (Weinzierl et al., 2008b). It is unclear what frequency of DCs is infected by CVB3 in vivo, and whether the virus can modulate presentation of viral epitopes at the surface of these professional antigen presenting cells. Furthermore, infection of the host may indirectly affect DCs, quantitatively and/or qualitatively, potentially limiting their ability to trigger T cell responses. We have previously shown that rCVB3 did not globally suppress the host’s capacity to mount CD8+ T cell responses (Kemball et al., 2009); however, rCVB3 are attenuated in vivo, and we considered it vital to determine if highly-pathogenic wild-type (wt) virus profoundly affects one or more DC subsets in vivo, and whether such changes are associated with a reduced ability of the host to trigger strong antiviral T cell responses.
In this study, we have used the murine model of CVB3 infection to evaluate the effects of the wt virus on dendritic cells in vivo. We report that, for the vast majority of DCs in vitro and in vivo, we observe neither productive infection nor viral protein synthesis. Nevertheless, wtCVB3 infection rapidly induces phenotypic changes in conventional DCs in vivo, and also dramatically reduces their abundance. In vitro analyses show that splenocytes taken from wtCVB3-infected mice have a diminished capacity to stimulate naïve T cells; however, on a per-cell basis, DCs from wtCVB3-infected mice remain capable of triggering CD8+ T cell division. In contrast to rCVB3, wt virus infection significantly diminished the host’s capacity to mount T cell responses, which was temporally associated with the loss of CD8α+ DCs. To our knowledge, our study is the first to precisely enumerate cDCs during picornavirus infection, and to show that their abundance is substantially altered in vivo.
Results
CVB3 infection substantially alters conventional and plasmacytoid dendritic cell populations in vivo
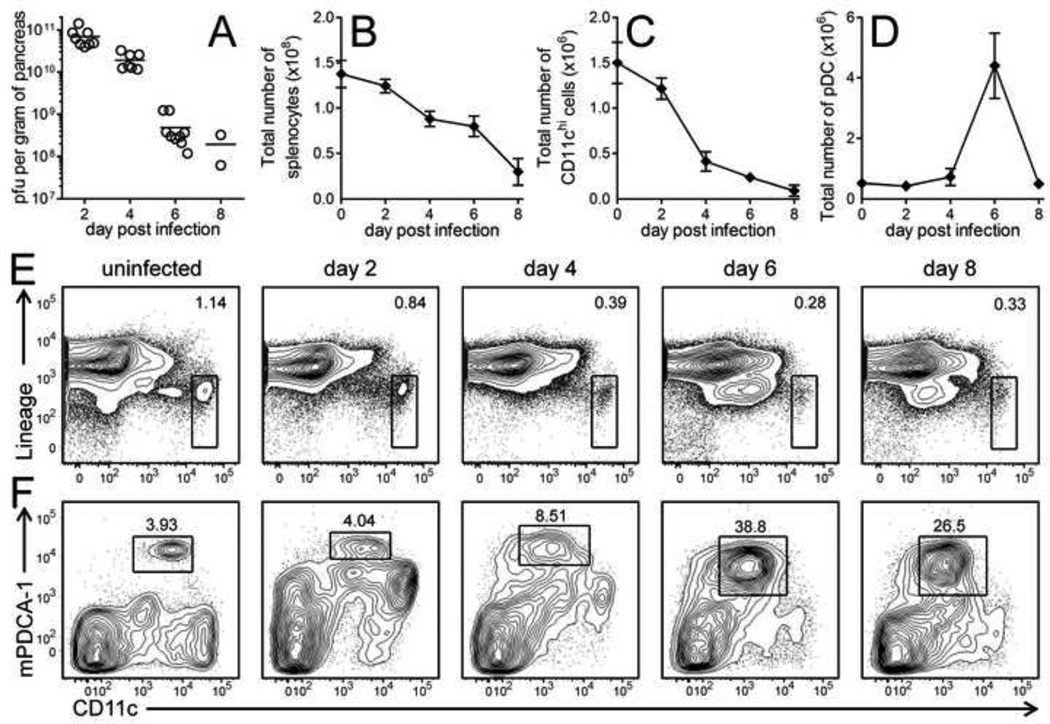
We first examined whether CVB infection had any quantitative or qualitative effect on conventional and plasmacytoid DCs in vivo. Mice were inoculated with 103 PFU of wild-type CVB3 (wtCVB3). Virus replicated rapidly and reached very high titers in the pancreas on day 2–4 p.i. (>1010 PFU/g), then declined by day 6 (Figure 1A). In contrast to many acute virus infections, which induce mild or moderate splenomegaly together with strong virus-specific CD8+ T cell responses, acute infection with wtCVB3 was associated with a gradual and substantial reduction in the total number of splenocytes, which declined ~4-fold by day 8 compared to uninfected mice (Figure 1B). Because primary virus-specific T cell responses are usually initiated within a few days of infection, we analyzed conventional and plasmacytoid DCs numbers on days 2, 4, 6, and 8 p.i. cDCs were classified as CD11chi and negative for expression of lineage markers for T cells, B cells, and NK cells (CD3−CD19−CD49b−). pDCs were defined by being CD11c intermediate (CD11cint), lineage negative, and expressing high levels of bone marrow stromal cell antigen 2 (BST-2, also known as mPDCA-1). cDCs were readily detected in uninfected mice, but declined in CVB3-infected mice after day 2, and very few of these cells remained by day 8 (Figure 1C & E). pDCs also were detected in uninfected mice, and their total number increased ~8-fold in virus-infected mice by day 6 and then declined by day 8 (Figure 1D); their frequency, too, increased markedly between 4 and 6 days p.i. (Figure 1F). Thus, CVB3 infection substantially alters the frequency and number of both conventional and plasmacytoid DCs.
Figure 1. CVB3 infection substantially alters the numbers of conventional and plasmacytoid dendritic cells in vivo.
Mice were infected with wtCVB3, and the virus titers in the pancreas (A, titers shown for individual mice, with mean) and total number of mononuclear cells in the spleen (B) were determined at the indicated time points p.i. Data show the mean ± SEM of 2–12 mice per time point, combined from 4 independent experiments. Conventional and plasmacytoid DCs in the spleen were analyzed by flow cytometry on days 2, 4, 6, and 8 p.i. (C) Total number of cDCs in the spleen. (D) Total number of pDCs in the spleen. Data show the mean ± SEM of 2–9 mice per time point, combined from 3 independent experiments. Representative contour plots are shown, and the rectangular gates identify cDCs [CD11chi and lineage negative (CD3−CD19−CD49b−), panel E] and pDCs [CD11cint and mPDCA-1hi (plots are gated on lineage negative cells), panel F]; the numbers shown on the contour plots are the percentages of conventional or plasmacytoid DCs among all cells in the plot.
CVB3 infection dramatically reduces the subset of conventional dendritic cells that is thought to be the most potent inducer of antiviral CD8+ T cell responses
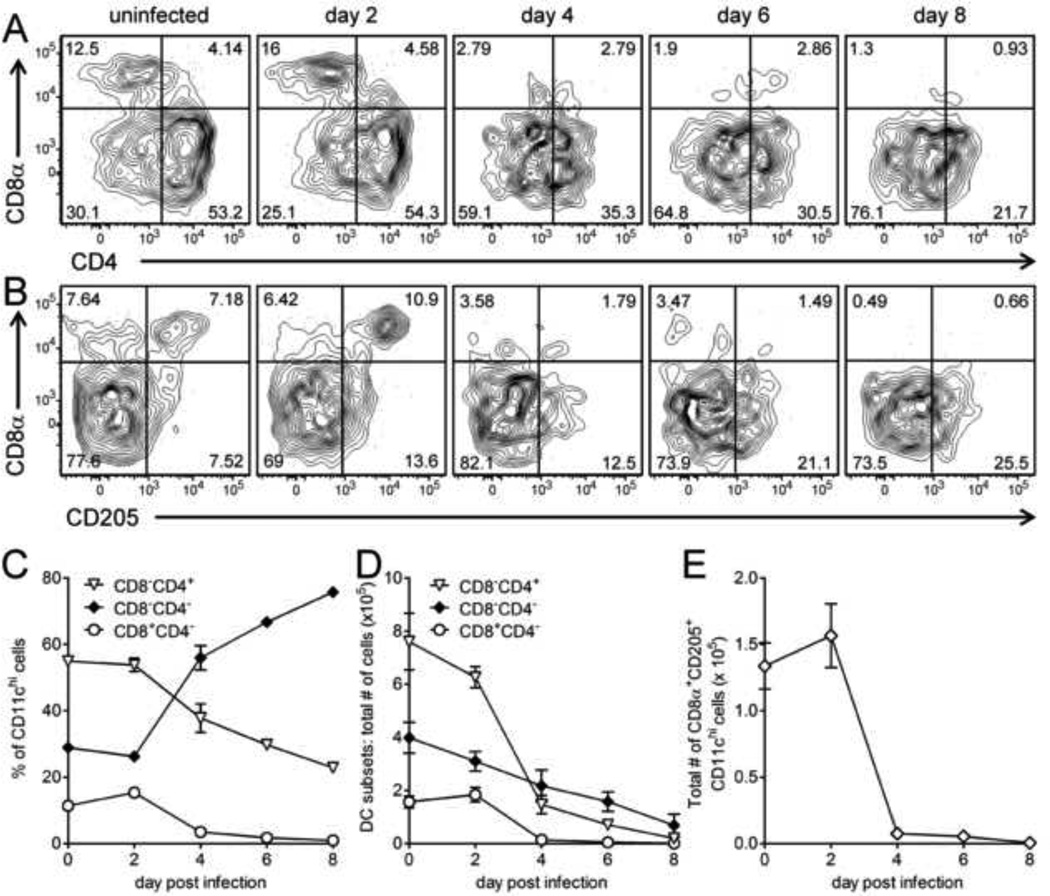
Conventional DCs, and in particular their CD8α+ subset, are known to be very efficient at priming virus-specific CD8+ T cell responses (Shortman and Heath, 2010). To evaluate the effects of wtCVB3 infection on these cells, splenocytes were harvested from infected mice on days 2, 4, 6, and 8, stained with antibodies to discriminate among the 3 major subsets of conventional DCs, and analyzed by flow cytometry. As expected, and in order of decreasing frequency, CD4+, double negative, and CD8α+ subsets were detected among CD11chi DCs in uninfected spleens (Figure 2A, top left panel). The frequency of CD8α+ DCs increased slightly on day 2, and then declined sharply thereafter; very few CD8α+ DCs remained in the spleen from day 4 onward (Figure 2A, C & D). The frequency of CD4+ DCs also declined dramatically following CVB3 infection (Figure 2A, & C) and, by 8 days p.i., the number of these cells had decreased almost 40-fold compared to uninfected animals (Figure 2D). Double negative DCs were proportionately less affected by infection, and became the dominant subset by day 4 (as a percentage of all DCs, Figure 2A & C). However, the absolute number of all three cDC subsets declined over the course of infection (Figure 2D). We next focused on the CD205+ subset of CD8α+ cDCs. These cells are potent inducers of antiviral CD8+ T cell responses in vivo and are thought to be specialized for the processing and presentation of exogenous antigens on MHC class I (cross-presentation) (Dudziak et al., 2007; Lopez-Bravo and Ardavin, 2008; Villadangos and Schnorrer, 2007). Previous findings from our laboratory have suggested that cross-priming is inefficient during CVB3 infection (Kemball et al., 2009; Kemball et al., 2008), and led us to question whether the virus modifies the CD8α+CD205+ cDC subset in vivo. The frequency and number of CD8α+CD205+ cDCs shared similar kinetics with the total CD8α+ cDC population, and virus infection led to a dramatic reduction in number of CD8α+CD205+ cDCs by day 4 (Figure 2B & E). Taken together, these data show that CVB3 infection substantially modifies the relative proportions of cDC subsets, and is associated with a profound reduction in the number of all cDC subsets, including the cells that are optimally suited to prime virus-specific CD8+ T cell responses.
Figure 2. CVB3 infection profoundly reduces the numbers of CD8α+, CD4+, and double negative conventional dendritic cells.
Mice were infected with wtCVB3, and splenocytes were harvested and analyzed by flow cytometry on days 2, 4, 6, and 8 p.i. Representative contour plots are shown (A & B), and were gated to include CD11chi lineageneg cells. The numbers indicate the proportion of the various subsets of cDCs (CD8α+, CD4+, double negative, and CD8α+CD205+) in each quadrant, as a percentage of all CD11chi lineageneg cells. (C) Frequency of CD8α+, CD4+, and double negative cDCs (as a percentage of CD11chi lineageneg cells) in the spleen. (D) Total number of CD8α+, CD4+, double negative cDCs in the spleen. (E) Total number of CD8α+CD205+ cDCs in the spleen. Data show the mean ± SEM of 2–9 mice per time point, combined from 3 independent experiments.
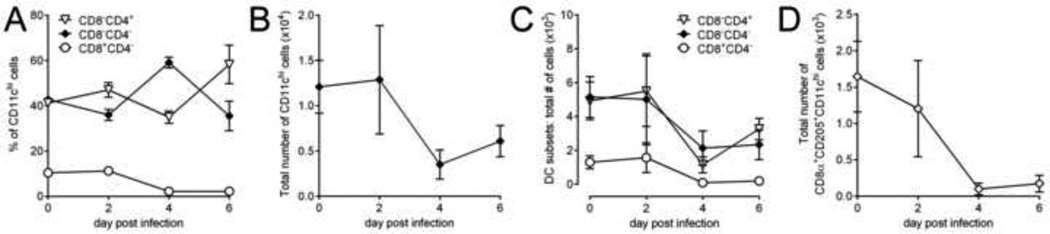
To assess whether this loss of DCs occurs in other secondary lymphoid organs, we evaluated the effects of wtCVB3 infection on DC populations in the pancreatic lymph nodes (pLNs). The frequency of CD8α+ DCs in the pLNs declined sharply after day 2 p.i. in a similar manner to CD8α+ DCs in the spleen (Figure 2C & Figure 3A). The total number of CD11chi DCs, and the number of all three DC subsets also declined after infection; few CD8α+ DCs remained in the pLNs from day 4 onward (Figure 3B–D). In addition, very few CD11chi cDCs were present in virus-infected pancreas; although other lymphocytes (T cells, macrophages, neutrophils) were readily detected in this organ following infection, DCs were present at a very low level and their numbers did not increase with infection (data not shown). Overall, CVB3 infection leads to a similar reduction in cDCs in the spleen and pLNs.
Figure 3. CVB3 infection diminishes the number of conventional dendritic cells in lymph nodes.
Mice were infected with wtCVB3, and lymphocytes were harvested from pancreatic lymph nodes (pLNs) and analyzed by flow cytometry on days 2, 4, and 6 p.i. (A) Frequency of CD8α+, CD4+, and double negative cDCs (as a percentage of CD11chi lineageneg cells) in the pLNs. (B) Total number of CD11chi cDCs in the pLNs. (C) Total number of CD8α+, CD4+, and double negative cDCs in the pLNs. (D) Total number of CD8α+CD205+cDCs in the pLNs. Data show the mean ± SEM of 3–4 mice per time point, combined from 2 independent experiments.
CVB3 infection does not globally inhibit the up-regulation of MHC and costimulatory molecules on conventional dendritic cells in vivo
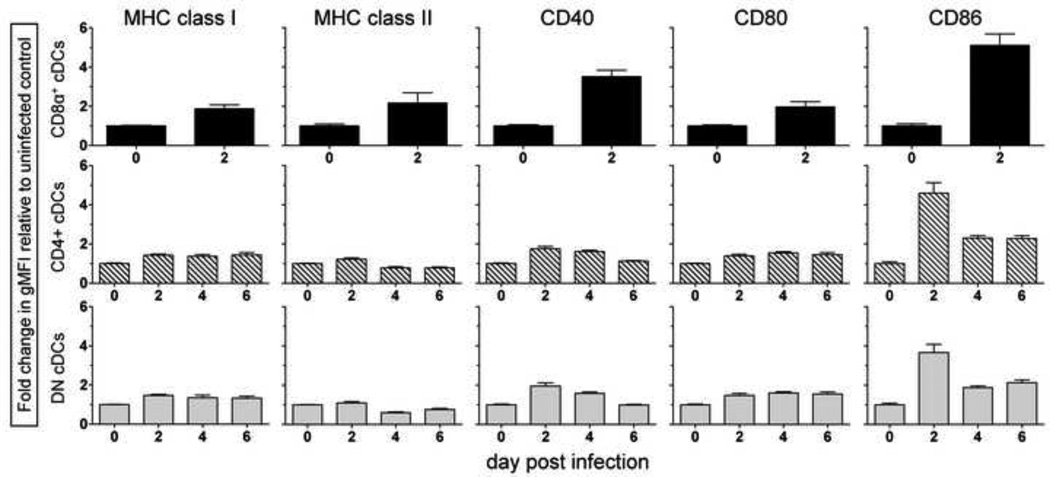
Besides reducing the number of DCs available for presenting viral antigen and/or priming virus-specific T cells in vivo, CVB3 also could avoid activating naïve T cells by limiting the expression of MHC and costimulatory molecules on the remaining DCs. To assess this, mice were infected with wtCVB3, and the cell surface phenotypes of the three major subsets of cDCs were analyzed. The levels of MHC class I, class II, and CD80 increased ~2-fold on CD8α+ DCs by day 2, while CD40 and CD86 were more strongly up-regulated (~3-fold and ~5-fold, respectively) (Figure 4, top row). Because of the profound reduction in the frequency and number of CD8α+ DCs by day 4 (Figure 2), we were unable to track the phenotype of this subset at later time points. The CD4+ and double negative subsets of DCs also expressed elevated levels of MHC class I, class II, CD40, and CD80 by day 2, although the increase was quite modest (≤ 2-fold); the level of these molecules was maintained, or decreased slightly, between 2 and 6 days post infection (Figure 4, middle and bottom rows). In contrast, CD86 expression was substantially up-regulated on CD4+ and double negative subsets on day 2 (~4-fold), and then declined by day 4 (middle and bottom rows). Overall, the 3 major subsets of cDCs in the spleen displayed an activated phenotype within 48 hrs of CVB3 infection, and strongly up-regulated the costimulatory molecule CD86. A recent study examined changes in CD11c+ DC phenotype in vivo at 36 hrs post CVB3 infection, but failed to detect a substantial change in MHC class I and II expression at this time point (Rahnefeld et al., 2011). In contrast, our detailed analysis of cDC subsets revealed that CD8α+ DCs up-regulate these MHC molecules by 48 hrs p.i. (Figure 3, top row). These data suggest that the virus does not prevent the activation of splenic cDCs; on the contrary, CD8α+ DCs display a mature phenotype and appear poised to prime CVB3-specific CD8+ T cells within the first 2 days of infection.
Figure 4. Conventional dendritic cells up-regulate the expression of MHC and costimulatory molecules following virus infection.
Mice were infected with wtCVB3, and the level of cell surface expression of MHC class I, MHC class II, CD40, CD80, and CD86 on cDCs in the spleen was measured by flow cytometry on day 2, 4, and 6. The expression kinetics for MHC and costimulatory molecules are shown for each of the 3 major cDC subsets (CD8α+, top row; CD4+, middle row; and double negative, bottom row). The fold change in the geometric mean fluorescence intensity of each protein over the course of infection was normalized and compared to the level of expression on cells from uninfected mice (day 0). Data show the mean + SEM of 4–9 mice per time point, combined from 3 independent experiments. The expression level of these proteins on the CD8α+ subset could not be determined after day 2 because these cells were too few in number.
Infection with a recombinant virus expressing DsRed suggests that very few DCs are infected in vivo
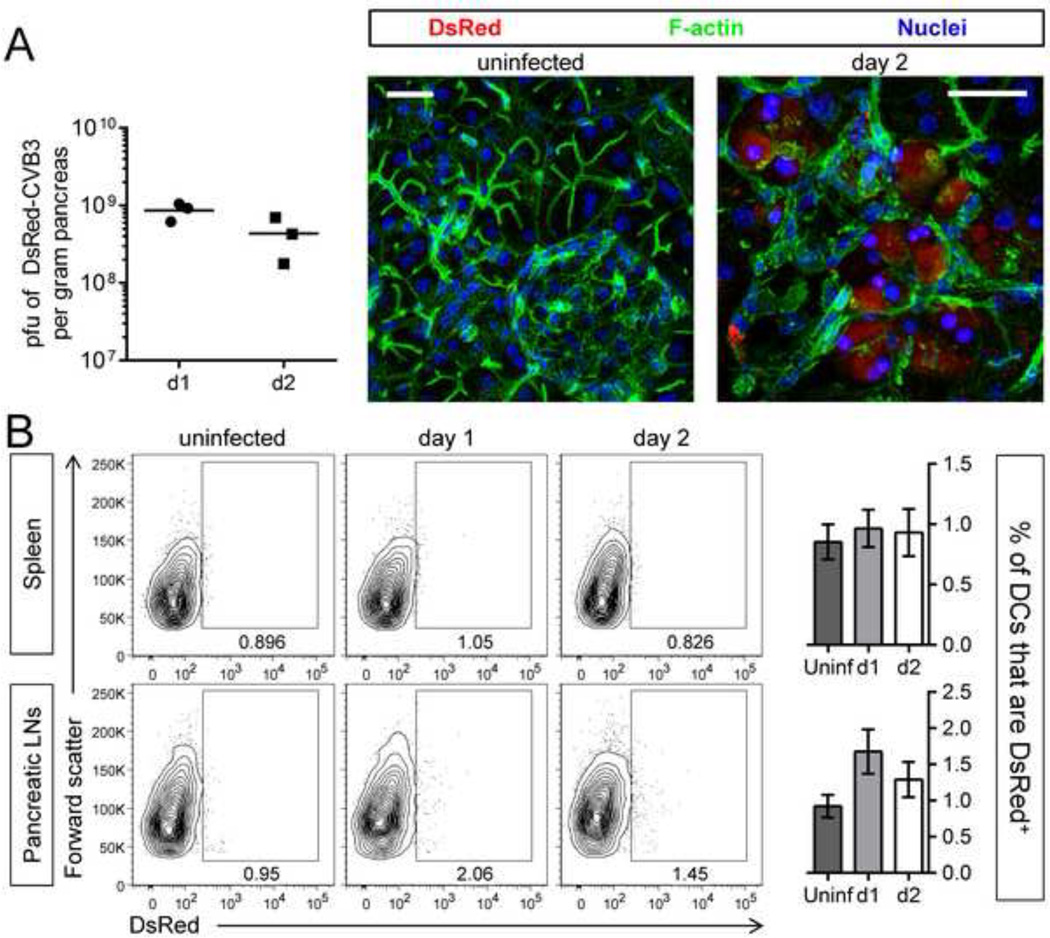
One obvious explanation for the dramatic reduction in DC numbers was that the virus directly infects DCs. In support of this idea, a recent study found that CVB3 could infect DCs, and both positive and negative strand viral RNA was detected in DCs in vitro and in vivo (Weinzierl et al., 2008b). However, the proportion of DCs that were infected was not reported, viral replication was incomplete and non-productive, and viral protein synthesis within the cells was not assessed. Thus, we have assessed these issues, in vivo and in vitro. To investigate whether CVB3 proteins were expressed in cDCs in vivo, we used a rCVB3 that expresses DsRed (DsRed-CVB3, ref Tabor-Godwin et al., 2010). Mice were infected with this virus and, to confirm infection and DsRed expression, we evaluated virus titers at days 1 & 2 post infection (p.i.), and used confocal microscopy to analyze tissue sections; the studies focused on the pancreas, a primary target organ of CVB3 in vivo. Viral titers were high (~109 pfu/g) at both time points, and DsRed expression was readily detected in many pancreatic acinar cells at 2 days p.i. (Figure 5A), indicative of successful infection and marker protein expression in vivo. Next we evaluated DsRed expression in DCs from the spleen and pancreatic lymph nodes (pLNs) at days 1 and 2 p.i., using flow cytometry. Several thousand DCs were analyzed in both tissues (~3–7×103 cells per sample), but the frequency of DsRed+ DCs was very low and similar to the background level seen in uninfected mice (Figure 5B). The differences in the percentage of DsRed+ DCs between uninfected and infected mice were not statistically significant by a t-test comparing uninfected versus day 1 (spleen p=0.6266, pLNs p=0.0939) and uninfected versus day 2 (spleen p= 0.7664, pLNs p=0.2746). Flow cytometric analysis of DsRed expression in CD8α+, CD4+, and double negative subsets of conventional DCs in the spleen yielded similar results (data not shown). Although very slightly more DsRed+ DCs were identified in infected mice, the intensity of the fluorescent signal was low, and comparable to the negative control fluorescence intensity observed in DCs from uninfected mice (Figure 5B). We conclude that very few, if any, DCs express this CVB3 protein following infection in vivo.
Figure 5. Few, if any, dendritic cells are infected by CVB3 in vivo.
Mice were infected with DsRed-CVB3, and were sacrificed 1 or 2 days later. (A) Viral titers in the pancreas were measured (graph) and DsRed protein was imaged by confocal microscopy of vibratome sections of pancreas; scale bar = 30 µm. (B) The frequency of DsRed+ DCs in the spleen and pancreatic lymph nodes was analyzed by flow cytometry on days 1 and 2. Contour plots are gated on DCs [CD11chi and “lineage” negative (CD3−CD19− CD49b−) cells for the spleen (top row), and CD11cint-hi lineage− cells for the pancreatic lymph nodes (bottom row)]. The large rectangular gates shown on representative contour plots identify DsRed+ cells, and the numbers shown are the percentage of DsRed+ cells among all cells in the plot. ~3–7.5×103 DCs were analyzed in these plots. Bar graphs display the mean ± SEM of 3 mice per group. No statistically significant differences were detected among these groups by a t-test.
CVB3 and dendritic cells in vitro: no productive infection, protein synthesis, or down-regulation of MHC expression
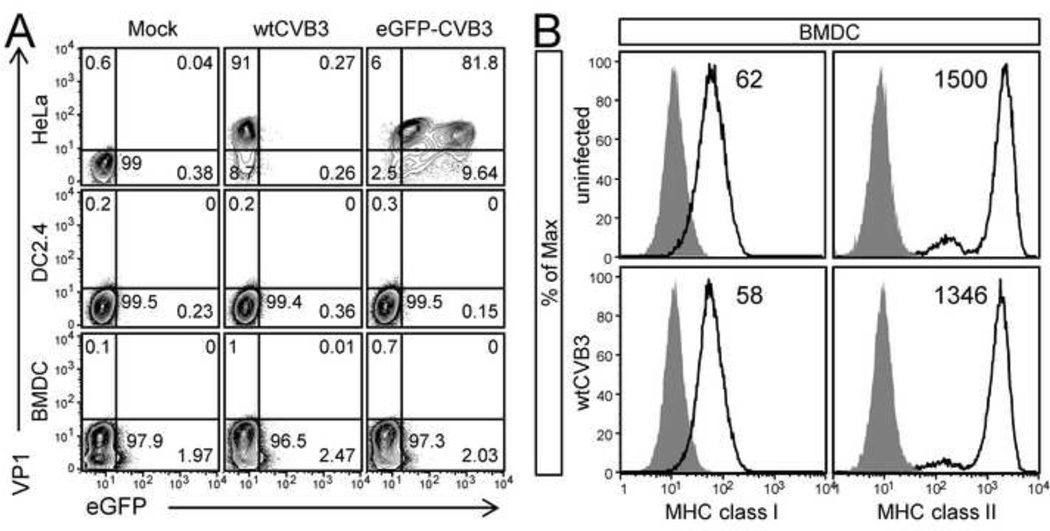
The above data suggest that DCs are not a primary target for CVB3 infection in vivo, but we considered it important to determine if DCs might be susceptible to in vitro infection. An in vitro approach allowed us to generate large numbers of DCs, expose them to CVB3 synchronously at high moi, and characterize the progress of infection at defined time points thereafter. Two different populations of DCs were analyzed: immature C57BL/6 bone marrow monocyte-derived DCs (cultured with GM-CSF); and DC2.4 cells, an immortalized cell line generated from C57BL/6 monocyte-derived DCs (Shen et al., 1997). DCs were exposed to wtCVB3, or to a recombinant CVB3 expressing eGFP (eGFP-CVB3), at a high multiplicity of infection (MOI=10), or were mock infected; 22–23 hrs later the cells were stained for intracellular expression of the viral capsid protein VP1 and analyzed by flow cytometry to detect VP1 ± eGFP. HeLa cells were infected as a positive control. As expected, viral proteins were readily detected in HeLa cells (6 hrs post infection); for both viruses, ~90% of HeLa cells were GFP+ and/or VP1+ (Figure 6A, top row). In contrast, we did not detect a significant number of DCs that were VP1+ or GFP+; 96.5–99.5% of DCs were negative for CVB protein expression (Figure 6A, middle & bottom rows). Furthermore, no increase in virus titer was detected in infected DC2.4 or BMDC cultures, nor did we observe virus-induced cytopathic effect (not shown). Thus, there is little evidence that the majority of DCs can be productively infected by CVB3; indeed, the absence of viral protein synthesis in DCs suggests that even non-productive infection is very unusual. Moreover, no substantial changes to MHC class I, or II was observed on BMDCs 18–24 hrs after exposure to CVB3 in vitro (Figure 6B); this was true, too, of CD86 (not shown). Taken together, the data in Figure 5 and Figure 6 suggest that the great majority of DCs are resistant to CVB3 infection. Therefore, the dramatic reduction in DC numbers that follows CVB3 infection in vivo does not appear to result from direct infection-mediated cell death of DCs (Figure 1 & Figure 2).
Figure 6. wtCVB3 has little effect on dendritic cells in vitro.
(A) Bone marrow monocyte-derived dendritic cells (BMDCs) and the DC2.4 dendritic cell line were evaluated for their susceptibility to CVB3 infection; HeLa cells were infected as a positive control. HeLa, DC2.4, and BMDCs were incubated with wtCVB3 or eGFP-CVB3 (MOI=10) for 1 hr, or mock infected. Inoculum was removed, and cells were harvested (HeLa cells, 6 hrs p.i.; BMDC & DC2.4, 24 hrs p.i.), stained for intracellular VP1, and the percentage of cells expressing VP1 ± eGFP was determined by flow cytometry. Representative contour plots are shown for each cell type, and the numbers indicate the proportion of cells in each quadrant, as a percentage of all cells in the plot (VP1+, upper left quadrant; eGFP+, lower right quadrant; VP1+eGFP+, upper right quadrant). (B) BMDCs (cultured with GM-CSF) were infected with wtCVB3 (MOI=100), or left uninfected, and the surface expression of MHC class I and II were analyzed by flow cytometry 18 hrs p.i. Histograms are gated on CD11c+CD11b+ cells, and show the expression of MHC class I or II (black lines); isotype control staining is shaded grey. The number on each histogram indicates the geometric mean fluorescence intensity of MHC class I or II. Data are representative of 2 independent experiments.
The loss of conventional dendritic cells in CVB3-infected mice is associated with weaker T cell responses to a secondary virus infection
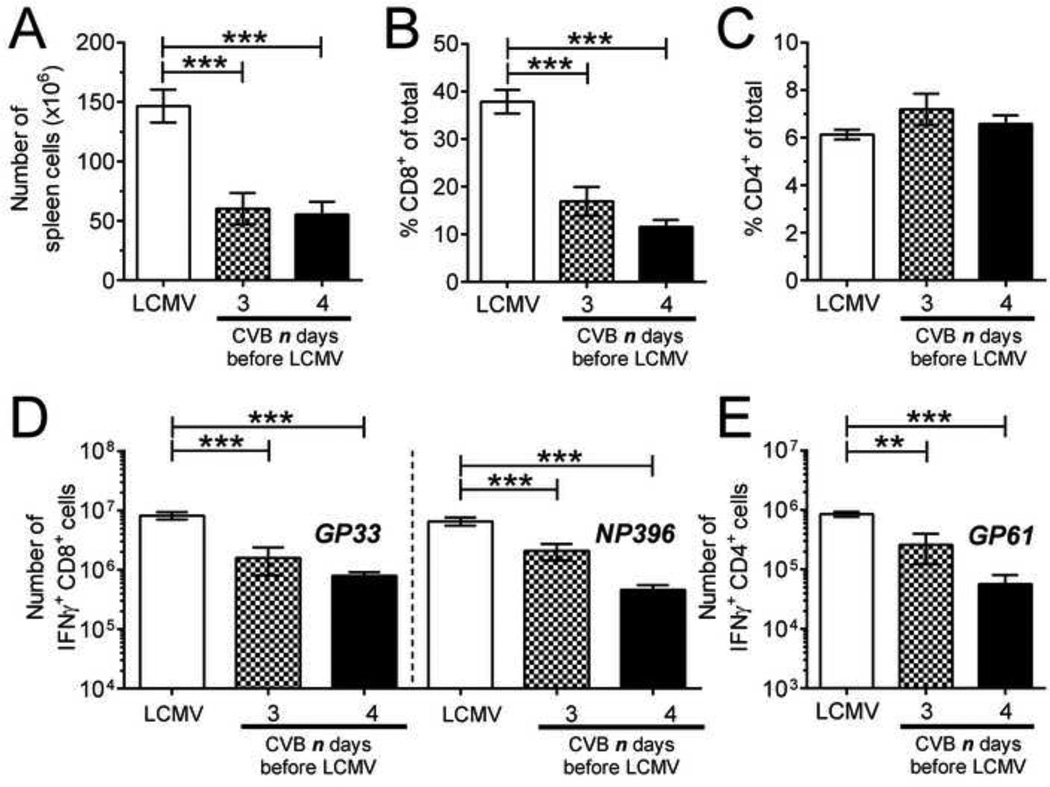
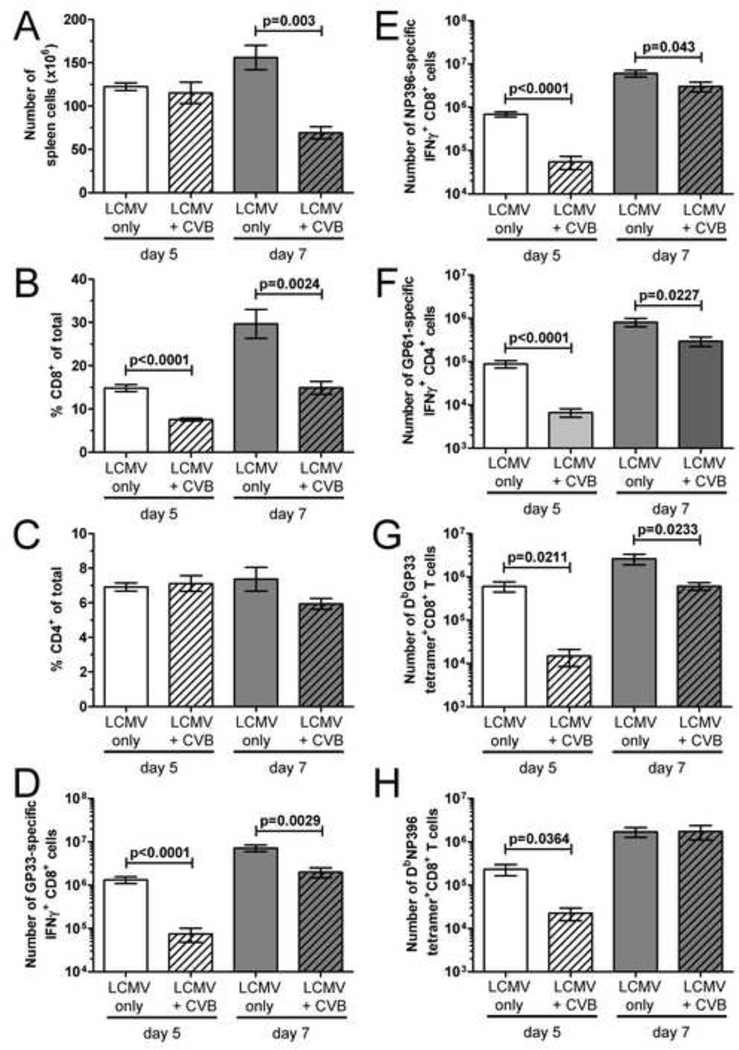
We have previously shown that infection of mice with rCVB3 does not weaken their ability to mount T cell responses against a subsequent infection with LCMV. Rather, mice that receive rCVB3 three days before LCMV mount larger LCMV-specific T cell responses than mice infected with LCMV alone (Kemball et al., 2009). However, rCVB3 are attenuated in vivo, and in light of the profound changes in CD8α+ DC numbers that accompany wtCVB3 infection, we considered it important to determine if the highly pathogenic wt virus would reduce the ability of the host to mount a subsequent antiviral T cell response to LCMV. Studies have shown that priming of LCMV-specific CD8+ T cells requires DCs, and that CD8α+ cDCs are the predominant subset that initiates this response (Belz et al., 2005; Probst and van den, 2005). We reasoned that the loss of CD8α+CD205+ DCs in the spleen of wtCVB3-infected mice between 2 and 4 days p.i. (Figure 2B & E) might be associated with diminished T cell responses to LCMV that was inoculated 3 or 4 days post-wtCVB3. Therefore, groups of mice were infected with wtCVB3, and then co-infected with LCMV 3 or 4 days later. The frequencies of total CD8+ and CD4+ T cells, and LCMV-specific CD8+ and CD4+ T cell responses, were analyzed in the spleen 7 days after LCMV infection. Our analyses of co-infected mice (presented in Figure 7–Figure 12) were carried out in animals that exhibited clear indications of a robust CVB3 infection, by meeting three criteria: the presence of infectious virus in the feces (determined by plaque assay, 2 days after CVB3 infection); the presence of infectious virus in the pancreas (assayed at the time point of animal sacrifice and T cell analysis); and the presence of calcium deposits in the peritoneal cavity, a classic phenotypic signature of CVB3 infection and pancreatic disease (Mena et al., 2000). The total number of mononuclear cells in the spleen (Figure 7A) and the frequency of splenic CD8+ T cells (Figure 7B) were significantly reduced (~2–3 fold) in mice that had received wtCVB3 3 or 4 days before LCMV, as compared to mice that received LCMV infection alone. In contrast, the frequency of CD4+ T cells was not significantly altered by CVB3 co-infection (Figure 7C). In mice that had received wtCVB3 3 or 4 days before LCMV, the total numbers of GP33- and NP396-specific CD8+ T cells were significantly lower than in mice infected only with LCMV; when 4 days elapsed between wtCVB and LCMV infections, the LCMV-specific T cell responses were 10- to 14-fold lower (Figure 7D). Mice that had received wtCVB3 4 days before LCMV did not have a lower frequency of total CD4+ T cells (Figure 7C), but the overall reduction in splenocyte numbers (Figure 7A), combined with a reduced frequency of GP61-specific cells among total CD4+ T cells (not shown), led to a strong and significant reduction in the total number of GP61-specific CD4+ T cells, to an extent very similar to that observed for CD8+ T cells (~14-fold; Figure 7E).
Figure 7. CVB3-infected mice mount weaker CD4+ and CD8+ T cell responses to a secondary virus infection.
Groups of mice were infected with wtCVB3, and then co-infected with LCMV (3 or 4 days after the primary infection with CVB3). An additional group of mice was infected only with LCMV. LCMV-specific CD8+ and CD4+ T cell responses were analyzed 7 days after LCMV infection by flow cytometry. (A) Total number of mononuclear cells in the spleen. (B & C) Frequency of CD8+ and CD4+ T cells in the spleen, as a percentage of total mononuclear cells. (D & E) Splenocytes were stimulated with peptide, and the total numbers of T cells producing IFNγ in response to GP33 & NP396 peptides (D), and GP61 peptide (E) were determined by flow cytometry. Data show the mean ± SEM of 6 or 8 mice per group, combined from 2 independent experiments (** p<0.01, *** p<0.001).
Figure 12. Purified dendritic cells from LCMV-infected or co-infected mice are similarly capable of stimulating T cell proliferation in vitro.
Mice were infected with wtCVB3 and, 4 days later, were co-infected with LCMV. Control mice received CVB3 alone, LCMV alone, or no virus. Splenocytes were harvested from infected mice 2 days after LCMV infection and/or 6 days post CVB3, and CD11chi DCs were purified by magnetic enrichment (splenocytes were pooled within mouse groups). Naïve CD8+ transgenic T cells were also purified by magnetic enrichment from uninfected P14 mice, and then labeled with CFSE. An equal number (6×104/well) of purified DCs from each group were incubated with purified P14 T cells (1.2×105/well) in vitro. An additional set of cultures included P14 cells incubated with GP33–41 peptide-pulsed purified DCs from co-infected mice. Culture wells were harvested on days 2, 3, or 4, and analyzed by flow cytometry. (A) Proliferation (loss of CFSE fluorescence) of P14 T cells. Histograms are gated on CD8+Thy1.1+ cells, and, for each histogram, the percentage of P14 cells that have divided is shown; the gate used to define P14 division is shown by dotted lines in the upper left panel. (B) Kinetics of P14 T cell division in vitro. Data show the percentage of total P14 cells in each well that have divided at least once, and are expressed as the mean ± SEM of triplicate wells. (C) Prior to the start of in vitro culture, the purified DC stimulator populations were analyzed by flow cytometry to determine the frequency of cDCs subsets. Representative contour plots are gated on CD11chi lineageneg cells. The numbers indicate the proportion of the cDC subset (CD8α+, CD4+, double negative) in each quadrant, as a percentage of all CD11chi lineageneg cells.
The effect of wtCVB3 on T cell responses to LCMV appears early, but its impact is time-dependent
The data in Figure 7 show that wtCVB3 infection limits the T cell response to LCMV administered 4 days later, a time point at which CD8α+CD205+ DCs in the spleen have reached their numerical nadir (Figure 2). In contrast to their status at day 4 following wtCVB3 infection, CD8α+CD205+ DCs are abundant in the spleen at day 2 p.i., and display and activated phenotype (Figure 2B & E, Figure 4). Therefore, we considered the possibility that these abundant and highly-activated CD8α+ DCs might enhance, rather than limit, T cell responses to LCMV that was administered at this time point. To test this hypothesis, mice were infected with wtCVB3, and co-infected with LCMV 2 days later. The frequencies of total CD8+ and CD4+ T cells, and LCMV-specific CD8+ and CD4+ T cell responses, were analyzed in the spleen on day 5 and 7 after LCMV infection. Compared to mice that received LCMV infection alone, the total number of mononuclear cells in the spleen (day 7, Figure 8A) and the frequency of splenic CD8+ T cells (day 5 and 7, Figure 8B) were significantly reduced (~2-fold) in co-infected mice. In contrast, the frequency of CD4+ T cells was not significantly altered by CVB3 co-infection (Figure 8C). The total numbers of GP33- and NP396-specific CD8+ T cells were significantly lower than in mice infected only with LCMV (Figure 8D & E). Co-infection with wtCVB3 had similar effects on the LCMV-specific CD4+ T cell response; the total number of GP61-specific CD4+ T cells was reduced at both days 5 and 7 after LCMV infection (Figure 8F). The above evaluation of the LCMV-specific T cell responses relied on a functional readout (IFNγ production). Thus, it was formally possible that the number of LCMV-induced T cells in CVB3-infected mice was normal, and that the majority of the cells were dysfunctional (IFNγ-deficient), rendering them undetectable by the above assay. Therefore, we enumerated the cells using appropriate MHC class I tetramers. Compared to mice infected with LCMV alone, the total number of DbGP33 and DbNP396 tetramer+CD8+ T cells was substantially reduced (>10-fold) in co-infected mice 5 days after LCMV (Figure 8G & H), which parallels the reduction in the total number LCMV-specific IFN-γ+ CD8+ T cells (Figure 8D & E). Taken together, these ICCS and tetramer data show that wtCVB3 infection is associated not with enhanced, but rather with significantly weaker CD8+ and CD4+ T cell responses to LCMV, even if LCMV is administered shortly after CVB3 (2 days later) when activated DCs are abundant in the spleen.
Figure 8. The effect of wtCVB3 infection is more profound at the early stage of LCMV-specific T cell expansion.
Mice were infected with wtCVB3 and co-infected with LCMV 2 days later. An additional group of mice was infected only with LCMV. LCMV-specific CD8+ and CD4+ T cell responses were analyzed 5 or 7 days after LCMV infection by flow cytometry. (A) Total number of mononuclear cells in the spleen. (B & C) Frequency of CD8+ and CD4+ T cells in the spleen, as a percentage of total mononuclear cells. (D, E, & F) Splenocytes were stimulated with peptide, and the total numbers of T cells producing IFNγ in response to GP33 (D), NP396 (E), and GP61 peptides (F) were determined by flow cytometry. Data show the mean ± SEM of 6 or 8 mice per group, combined from 3 independent experiments. (G & H) The total number of DbGP33 and DbNP396 tetramer+ CD8+ T cells in the spleen was determined by flow cytometry. Data show the mean ± SEM of 3 or 4 mice per group, combined from 2 independent experiments. Statistical significance was determined by an unpaired t-test.
The effect of wtCVB3 on the magnitude of the LCMV-specific CD8+ and CD4+ T cell responses varied with, and was dependent upon, two distinct time intervals. First, the time allowed to elapse between the CVB3 and LCMV infections. When CVB3 was administered only 2 days prior to LCMV, the responses to LCMV 7 days later were only modestly reduced (Figure 8D–H), while the suppressive effects of CVB3 were much more pronounced when 4 days had elapsed between wtCVB and LCMV infections (Figure 7D & E). Second, the time point post-LCMV at which the responses are measured. For CD8+ T cells (Figure 8D, E, G & H) the reduction in LCMV-specific responses was markedly greater at day 5 post-LCMV (~10–20-fold) compared to day 7 (~2–4-fold). A similar observation applies to CD4+ T cells: the total number of GP61-specific CD4+ T cells was strongly reduced 5 days after LCMV (13-fold), but this numerical deficit was partially restored by day 7, with only a ~3-fold difference between co-infected mice and mice infected with LCMV alone (Figure 8F). Although the effects of wtCVB3 on the magnitude of LCMV-specific T cell responses varies with these time intervals, in all cases the LCMV-specific effector cells appear to be functionally competent to produce IFNγ.
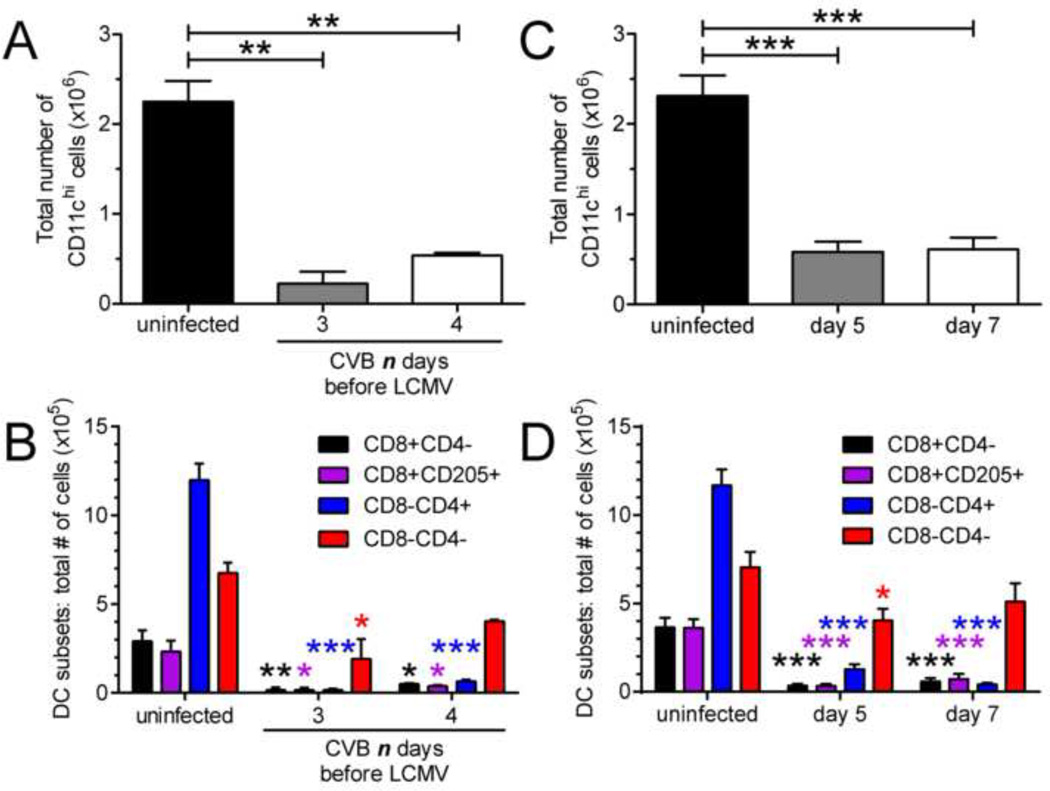
Conventional dendritic cell numbers are diminished in co-infected mice
To determine if co-infection with wtCVB3 and LCMV led to a similar reduction in cDC numbers as wtCVB3 infection alone, the number of splenic DCs in co-infected mice was assessed at the time points when LCMV-specific T cell responses were measured ex vivo (day 5 and 7 post LCMV, Figure 7 & Figure 8). In mice that were infected with wtCVB3 and co-infected with LCMV 3 or 4 days later, the total number of CD11chi cDCs was significantly diminished 7 days post LCMV infection (10 or 11 days post CVB3) (Figure 9A). Analysis of cDC subsets showed that the total number of CD8α+ and CD4+ cDCs were most profoundly reduced (Figure 9B). Similar results were obtained in mice that were infected with wtCVB3 and co-infected with LCMV 2 days later; DC numbers were significantly reduced at 5 and 7 days post LCMV infection (7 or 9 days post CVB3) (Figure 9C & D). Overall, the number of DCs did not vary substantially between co-infected mice and mice infected with wtCVB3 alone. Rather, cDC numbers were significantly diminished and to a similar extent in both groups of mice (Figure 1, Figure 2, Figure 9).
Figure 9. Conventional dendritic cell numbers are diminished in co-infected mice.
Groups of mice were infected with wtCVB3, and then co-infected with LCMV. (A & B) Mice received LCMV 3 or 4 days after the primary infection with CVB3, and cDCs were analyzed 7 days after LCMV infection by flow cytometry. (C & D) Mice received LCMV 2 days after CVB3, and cDCs were analyzed on day 5 and 7 after LCMV infection. (A & C) Total number of cDCs in the spleen. (B & D) Total number of CD8α+, CD8α+CD205+, CD4+, and double negative cDCs in the spleen. Data show the mean ± SEM of 2 or 3 mice per group (A & B), or 6–8 mice per group combined from 3 independent experiments (C & D). Statistically significant differences between uninfected and co-infected groups were detected by ANOVA (* p<0.05, ** p<0.01, *** p<0.001).
The reduced T cell responses to LCMV do not result from lack of replication, and LCMV antigen presentation by splenic APCs from co-infected mice can trigger naïve T cell proliferation
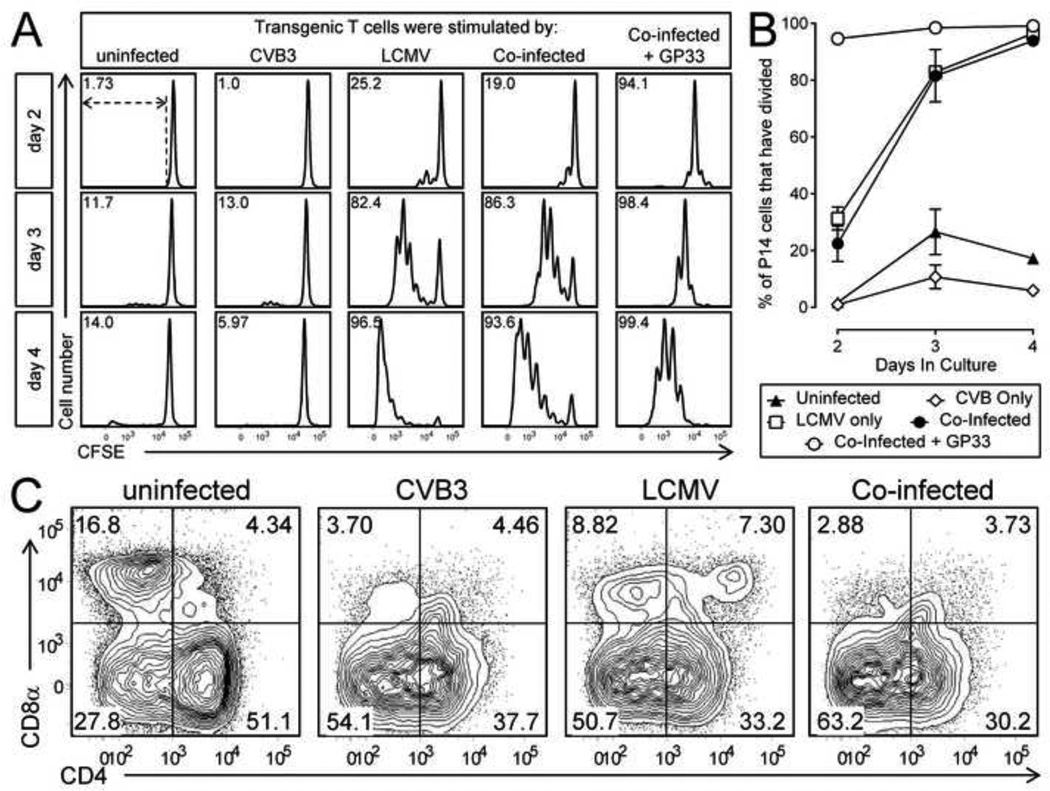
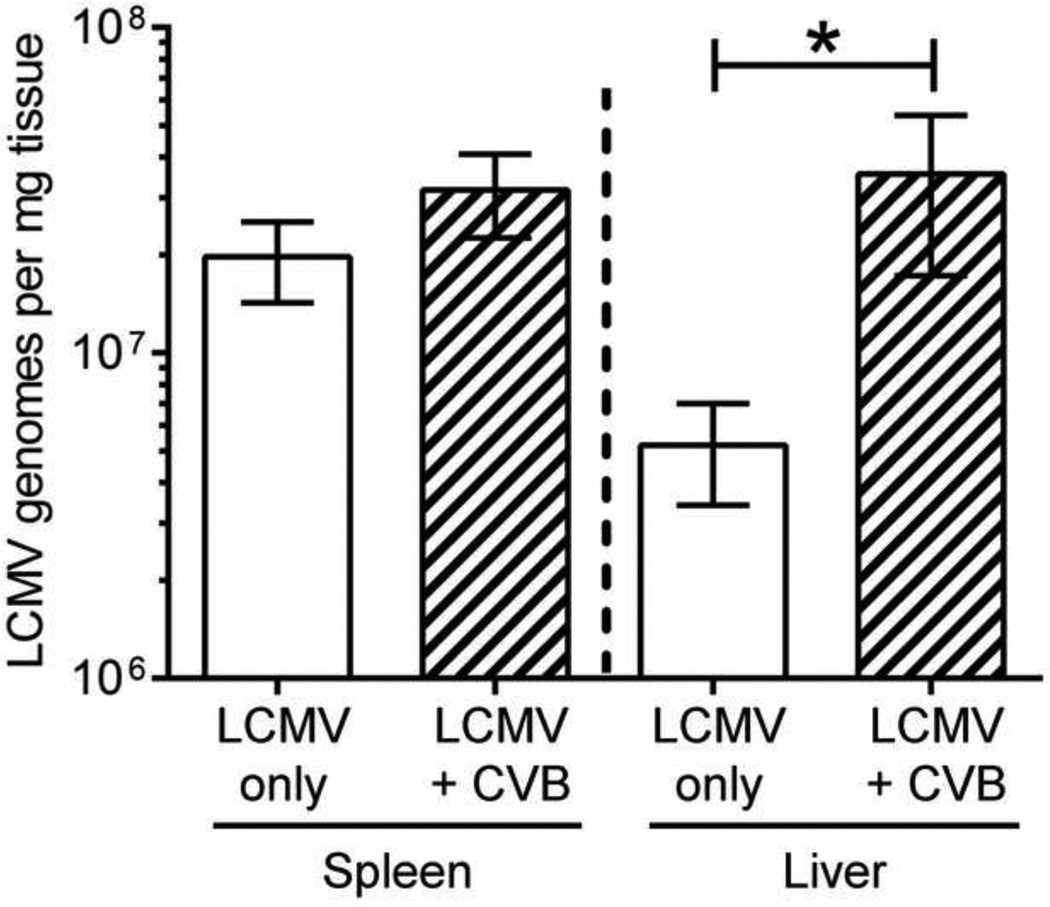
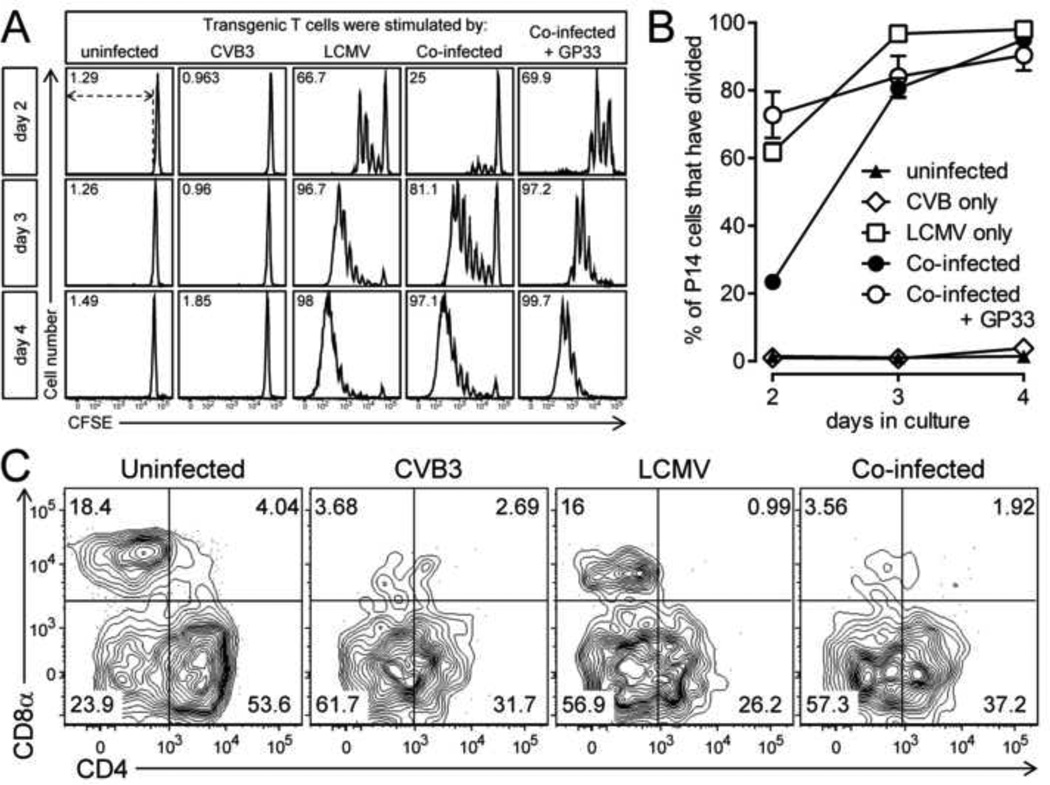
An alternative explanation for the reduced LCMV-specific T cell response in co-infected mice is that CVB3 infection establishes a microenvironment that suppresses LCMV replication, thereby reducing the amount of viral antigen available for priming naïve LCMV-specific T cells. This possibility was assessed in two ways. First, mice were infected with wtCVB3 and then infected with LCMV 2 days later. A control group of mice received only LCMV. Five days after LCMV infection, RNA was isolated from the spleen and liver and the copy number of LCMV genomic S segment RNA was determined by quantitative real time RT-PCR. At this time point, mice infected with LCMV alone had ~2×107 genome copies per mg of spleen, and ~5×106 per mg of liver; mice that had been infected 2 days previously with wtCVB3 had a similar genome load in the spleen, but significantly elevated (~6-fold) load in the liver (Figure 10). These data suggest that CVB3 does not reduce the ability of LCMV to replicate within co-infected mice. Second, we examined whether splenic APCs from co-infected mice present LCMV antigen in a manner that can drive the proliferation of naïve LCMV-specific CD8+ transgenic T cells. We previously demonstrated that mice infected with rCVB3 expressing LCMV immunodominant epitopes mount very weak endogenous T cell responses in vivo, but the capacity of splenocytes from CVB3-infected animals to stimulate naïve T cells in vitro was not examined (Kemball et al., 2008; Slifka et al., 2001). Mice were infected with wtCVB3 and, 4 days later (the time point when DCs are strongly affected, Figure 2), the mice received LCMV. Three groups of control mice received CVB3 alone, LCMV alone, or no virus. Splenocytes were harvested from the infected mice at 2 days after LCMV infection and/or 6 days post-CVB3, and were incubated for 2–4 days in vitro with CFSE-labeled naïve P14 CD8+ transgenic T cells. Since APCs from the donor P14 spleen could potentially present viral antigens and drive CD8+ transgenic T cell proliferation independently of APCs from virus-infected spleens, P14 CD8+ T cells were purified by magnetic selection (see Materials & Methods), and these enriched CD8+ T cells (93% CD8+Thy1.1+) were incubated with splenocytes from virus-infected mice. CD11chi DCs were almost completely absent from this enriched CD8+ T cell fraction (0.03% of selected cells were CD11chiThy1.1−CD19−CD49b−, data not shown), and therefore a vanishingly small number of DCs in these cultures originated from the P14 donor mouse. P14 CD8+ T cell proliferation was analyzed by flow cytometry after 2, 3, or 4 days of in vitro culture. The vast majority of P14 cells did not divide (remained CFSEhi) following stimulation with splenocytes from uninfected mice, or from mice infected with wtCVB3 only (Figure 11A & B). A substantial proportion (~60%) of P14 cells divided (and diluted their CFSE) after 2 days of stimulation with splenocytes from mice infected with LCMV only; by day 3 almost all of the input P14 cells had divided, the majority several times, and they continued to proliferate through day 4 (Figure 11A & B). In contrast, relatively few P14 cells (~20%) had begun to proliferate on day 2 after stimulation with splenocytes from co-infected mice. In addition, prior infection with wtCVB3 altered the CFSE fluorescence profiles observed after 2 days of in vitro culture (Figure 11A top row; compare LCMV & co-infected panels): although, in both cases, the maximum number of observed divisions was 4, many more cells had undergone 1–4 divisions in response to splenocytes from mice infected with LCMV alone, compared to splenocytes from co-infected mice. Thus, wtCVB3 co-infection appears to diminish the division of naïve LCMV-specific CD8+ T cells in vitro, paralleling what we have observed in vivo. However, as we have also observed in vivo (Figure 8), the magnitude of the effect diminishes with time; after 3 days of stimulation by splenocytes from co-infected mice, the majority of naïve P14 cells in co-infected culture wells had proliferated, and almost all of the sensor cells had divided by day 4 (Figure 11A & B). To determine if P14 T cell proliferation increased when the level of viral antigen presentation by co-infected splenocytes was artificially increased, we setup an additional group of cultures in which P14 cells were incubated with co-infected splenocytes together with GP33–41 peptide. In these wells, a greater percentage of P14 cells (~70%) had divided after 2 days of culture, and this was similar to the proportion of P14 cells that divided following stimulation with splenocytes from mice infected with LCMV alone (Figure 11A & B). P14 cells stimulated with co-infected splenocytes + peptide continued to divide further by day 4 of culture. Thus, we conclude that the weaker proliferation of P14 cells in co-infected (no peptide) cultures on day 2 most likely resulted from a lower level of LCMV antigen presentation and/or fewer APCs presenting viral antigens to T cells; and that this deficiency could be overcome by provision of abundant peptide antigen. In support of this hypothesis, flow cytometric analysis of the stimulator splenocytes from the various groups showed that, when added to the wells at the beginning of the culture, the frequency of CD8α+CD11chi DCs was substantially lower in co-infected mice compared to mice infected with LCMV alone (Figure 11C). In summary, these data show that, in co-infected mice, endogenous LCMV antigen is presented sufficiently well to trigger naïve CD8+ T cell proliferation in vitro. However, T cell division is delayed compared to that observed with LCMV-only stimulator cells; this delay appears to result, in part, from a lower level of viral antigen presentation and/or fewer APCs; and it can be overcome by the addition of exogenous GP33–41 peptide to co-infected cultures.
Figure 10. CVB3 infection does not dampen LCMV replication in co-infected mice.
Mice were infected with wtCVB3 and then co-infected with LCMV 2 days later. Another group of mice received only LCMV. Five days after LCMV infection, RNA was isolated from the spleen and liver and the number of LCMV genomes was determined by quantitative real time RT-PCR. Data show the mean ± SEM of 3 or 6 mice per group, combined from 2 independent experiments (* p<0.05, determined by an unpaired t-test).
Figure 11. In vitro stimulation assay: wtCVB3 delays, but does not prevent, expansion of LCMV-specific CD8+ T cells.
Mice were infected with wtCVB3 and, 4 days later, were co-infected with LCMV. Control mice received CVB3 alone, LCMV alone, or no virus. Splenocytes were harvested from infected mice 2 days after LCMV infection and/or 6 days post CVB3, and were incubated in vitro with magnetically purified, CFSE-labeled, P14 CD8+ transgenic T cells. An additional set of cultures included P14 cells incubated with splenocytes from co-infected mice + 1 µM GP33–41 peptide. Duplicate wells were harvested on days 2, 3, or 4, and analyzed by flow cytometry. (A) Proliferation (loss of CFSE fluorescence) of P14 T cells. Histograms are gated on CD8+Thy1.1+ cells, and, for each histogram, the percentage of P14 cells that have divided is shown; the gate used to define P14 division is shown by dotted lines in the upper left panel. (B) Kinetics of P14 T cell division in vitro. Data show the percentage of total P14 cells in each well that have divided at least once, and are expressed as the mean ± SEM of 3–7 mice per group (n=1 for P14 cells incubated with uninfected control splenocytes). (C) The frequency of cDC subsets in the splenocyte stimulator populations (prior to in vitro culture) was determined by flow cytometry. Representative contour plots are shown, and are gated on CD11chi lineageneg cells. The numbers indicate the proportion of cells in each quadrant, as a percentage of all CD11chi lineageneg cells.
Purified dendritic cells from LCMV-infected or co-infected mice are similarly capable of stimulating T cell proliferation in vitro
The data presented in Figure 11 show that splenic APCs from co-infected mice can stimulate naïve CD8+ T cell proliferation in vitro, although the functionality of DCs was not assessed independently of other splenic APCs. It is unclear whether the delay in T cell expansion in vitro is due to decreased functionality of DCs or due to a reduction in their numbers alone. In order to isolate DC function from that of other APCs, groups of mice were infected with wtCVB3 and/or LCMV as in Figure 11, spleens were harvested (and pooled within mouse groups), and CD11c+ DCs were purified by magnetic selection. Flow cytometric analysis of these purified DCs populations showed a lower frequency of CD8α+CD11chi DCs in co-infected mice compared to mice infected with LCMV alone (Figure 12C), in agreement with the analysis of non-purified DCs (Figure 11C). An equal number of purified DCs from each group were incubated with purified, CFSE-labeled, P14 T cells for 2–4 days in vitro. The majority of P14 cells did not divide (remained CFSEhi) following stimulation with DCs from uninfected mice, or from mice infected with wtCVB3 only (Figure 12A & B). A substantial proportion (~30%) of P14 cells divided after 2 days of stimulation with DCs from mice infected with LCMV only; by day 3 most of the input P14 cells had divided, the majority several times, and they continued to proliferate through day 4 (Figure 12A & B). A similar response by P14 cells was observed using DCs from co-infected mice, and the kinetics of P14 cell division were comparable in wells containing DCs from co-infected mice or mice infected with LCMV alone (Figure 12A & B). To determine if P14 T cell proliferation increased when the level of viral antigen presentation by co-infected DCs was artificially increased, we setup an additional group of cultures in which P14 cells were incubated with GP33–41 peptide-pulsed, purified DCs from co-infected mice. In these wells, the vast majority of P14 cells (~95%) had divided after 2 days of culture, and continued to divide further on day 3 and 4 (Figure 12A & B). Taken together, these data show that when equal numbers of purified DCs (from LCMV-only and co-infected mice) are used as stimulators, both populations are similarly capable of stimulating in vitro T cell proliferation. Therefore, the delay in T cell division following stimulation with co-infected splenocytes (Figure 11A & B) is most likely due to the reduced number of DCs present in the culture wells (compared to splenocytes from mice infected with LCMV alone), rather than a decrease in DC functionality.
Discussion
In vitro studies have suggested that enteroviruses utilize strategies that impair the ability of DCs to trigger adaptive immunity, by inhibiting protein synthesis within DCs, and by limiting cellular maturation, antigen presentation, and proinflammatory cytokine secretion. However, it is not known whether picornaviruses exert similar effects on DCs in vivo, or if other effects of virus infection also modulate the stimulatory capacity of DCs. Although DC infection by coxsackievirus has been reported, the extent to which it occurs in vivo, and the level of protein expression in infected DCs, was unknown. In the present study, we evaluated the effects of wtCVB3 on DCs in vivo and in vitro, and we make several novel observations: first, viral protein synthesis is extremely limited in DCs and the relative frequency of infected DCs is very low, both in vitro and in vivo. Second, wtCVB3 nevertheless substantially affects DCs and reduces their numbers more profoundly than in other well-characterized viral systems. Third, the severe reduction in CD8α+ cDCs is associated with a reduced ability of the host to mount T cell responses to a subsequent viral infection. Finally, DCs obtained from wtCVB3-infected mice do present antigen in a manner that can trigger naïve CD8+ T cell responses, but the T cell responses are delayed, both in vivo and in vitro.
CVB3 does not establish a productive infection in murine DCs (Weinzierl et al., 2008b), and cannot replicate in human DCs upon entry via phagocytosis (Schulte et al., 2010). However, a recent study found that murine DCs can endocytose CVB3 virions, and reported that DCs isolated from infected mice contain positive and negative strand viral RNA (Weinzierl et al., 2008b). Thus, we considered the possibility that DCs may be non-productively infected by CVB3 in vivo; if viral proteins were generated in sufficient abundance within infected DCs, they might lead to cell death and/or inhibit the presentation of MHC class I-restricted CVB3 epitopes, thereby contributing to the weak primary CVB3-specific T cell response, and to the reduction in LCMV-specific T cell responses reported herein. However, very few virus-infected DCs expressing DsRed protein were detected in the spleen and pancreatic lymph nodes of DsRed-CVB3-infected mice (Figure 5). In addition, in vitro exposure of murine DCs to CVB3 at high multiplicity did not lead to viral protein expression (VP1 or GFP) at a level detectable by flow cytometry (Figure 6A), and the cells did not exhibit significant changes in their surface expression of MHC class I & II and CD86 (Figure 6B and data not shown). Furthermore, CVB3 did not cause detectable cytopathic effects and, as others have reported, productive infection did not take place. We cannot exclude the possibility that a small subset of DCs is infected by CVB3 in vivo and that, in these cells, direct presentation of MHC class I-restricted antigens is compromised by the virus; but these data suggest that CVB3 may elude naïve CD8+ T cells in large part by the simple expedient of avoiding DCs, rather than by inhibiting antigen presentation within these key APCs. However, this raises another issue: even if the virus does not infect DCs, one would expect that CVB-encoded epitopes would be presented to naïve CD8+ T cells by DCs that are specialized for cross presentation / cross-priming (CD8+CD205+ DCs). Our previous study suggested that this alternate mode of entry to the MHC class I pathway does not operate efficiently during CVB3 infection, but the molecular basis for the apparent failure of cross-priming in CVB3-infected mice has not been determined (Kemball et al., 2009). One possible explanation is that these DCs are indirectly affected by CVB3 infection. By this hypothesis, CVB3 not only evades DCs by failing to infect them, it also disrupts – probably by indirect means – the DCs that might otherwise present viral epitopes for cross-priming.
We found that, despite its inability to infect DCs in a widespread fashion, wtCVB3 causes a rapid and profound reduction in DC numbers (Figure 1), and alters the relative proportions of the various cDC subsets (Figure 2). From the data in Figure 5 and Figure 6, these changes are unlikely to be caused by direct infection of DCs; rather, the CVB3-related reduction in DC numbers is more likely to be mediated indirectly. Although changes in DC abundance and heterogeneity have been observed in other viral infection models, our findings suggest that the reduction in cDCs in CVB3-infected mice is more profound compared to other virus infections. For example, the decline in numbers of splenic CD8α+ and CD4+ cDCs is more modest following LCMV Armstrong infection (2–3 fold reduction by day 3 p.i.), and cDC numbers remain stable (or increase slightly) following murine cytomegalovirus and γ-herpesvirus 68 infections (Dalod et al., 2003; Montoya et al., 2005; Weslow-Schmidt et al., 2007). The reduction in cDCs that follows LCMV infection is mediated in part by type I IFN signaling (Montoya et al., 2005). Type I IFN levels peak in the serum of CVB3- and LCMV-infected mice at a similar time point (24–48 hrs p.i.) (Huhn et al., 2010; Lee et al., 2009; Wang et al., 2010; Zuniga et al., 2008), so we speculate that the reduction of cDCs in CVB3-infected mice may be mediated by type I IFNs; experiments to evaluate this hypothesis are particularly challenging in the CVB model because, in the absence of type I IFN signaling, CVB infection is invariably lethal within 2–4 days (ref Wessely et al., 2001, and Kemball, Flynn & Whitton, unpublished data).
pDC accumulation has been observed in lymphoid and non-lymphoid tissues during virus infections in mice and humans, and the recruitment of pDCs can either positively or negatively affect the antiviral immune response (Swiecki and Colonna, 2010a). Following CVB3 infection, we found that the total number and frequency of pDCs increases dramatically between day 4 and day 6 days p.i. (Figure 1 D & F). In addition, flow cytometric analysis of splenic pDCs in virus-infected mice (from day 4 onward) showed that these cells are considerably larger in size compared to pDCs in uninfected animals, and suggests that these cells have been activated, and are blasting and/or mature (data not shown). The dramatic increase in pDC numbers cannot be attributed to activated T cells that up-regulated the DC marker CD11c, because our pDC gating strategy excluded cells that expressed the T cell marker CD3 (Figure 1F). During CVB3 infection, pDC activation appears to require engulfment of virus-antibody complexes, and results in TLR-7-dependent production of type I IFN (Wang et al., 2007). Thus, the antiviral effects of pDCs in response to wtCVB3 may be delayed until antiviral antibodies have developed, and we speculate that the dramatic increase in pDC numbers at day 6 is temporally associated with the emergence of virus-specific antibodies that facilitate pDC activation. CVB3 antibodies are detectable by ~day 4 p.i. (Henke et al., 1995) which, as shown in Figure 1D, coincides with an increase in the number of pDCs in the spleen of infected mice. Unlike mature cDCs, activated pDCs do not down-regulate MHC class II ubiquitination and turnover, and as a result new MHC class II-peptide complexes are continually generated following pDC maturation (Villadangos and Young, 2008; Young et al., 2008). Therefore, we propose that, at ~day 4 and beyond, when cDCs are fewer in number, pDCs may play a role in CVB3 immunity by maintaining MHC class II presentation of CVB3 epitopes to CVB3-specific CD4+ T cells.
The loss of cDCs that occurs at 3–4 days after wtCVB3 infection is striking, and prompted us to question whether wtCVB3 infection diminishes the ability of an infected animal to mount T cell responses to a secondary infection. We have previously demonstrated that rCVB3 infection has no such effect (Kemball et al., 2009), but rCVB3 are generally attenuated in vivo, and we considered it vital to evaluate the effects of the wt virus. In stark contrast to rCVB3, we found that wtCVB3 infection dampened CD8+ and CD4+ T cell responses to subsequent LCMV infection; the magnitude of LCMV-specific T cell responses at day 7 post LCMV were considerably reduced (≥10-fold) in mice that had received wtCVB3 4 days prior to LCMV (Figure 7D & E). This suppressive effect of wtCVB3 infection was rapid, and could be observed even when LCMV was administered only 2 days after CVB3; the total number of LCMV-specific T cells was strongly reduced 5 days later. However, this rapid effect was not absolute because, by day 7 post LCMV, there was only a modest difference in the magnitude of the T responses in co-infected mice compared to those in mice infected with LCMV alone (Figure 8D–F). Thus, wtCVB3 infection delays LCMV-specific T cell expansion, but does not prevent it. Furthermore, dysfunctional cells were not identified in co-infected mice (Figure 8D, E, G, & H), indicating that, although slower to accumulate, the responding effector T cells in these mice are functionally competent and produce antiviral cytokines. Thus, wtCVB3 infection dampens, but does not ablate, host CD8+ T cell immunity.
The most obvious explanation for the lower LCMV-specific T cell responses in co-infected mice invokes the wtCVB3-related changes in DC quantity and quality. Our analysis of cDC numbers in co-infected mice, and mice infected with wtCVB3 alone, showed that these cells were significantly diminished and to a similar extent in both groups ((Figure 1, Figure 2, Figure 9). It is easy to imagine how the substantial reduction in DC numbers caused by wtCVB3 at 3 & 4 days p.i. (Figure 1, Figure 2) might diminish the host’s ability to mount primary T cell responses to subsequent LCMV infection, and our in vitro stimulation data (Figure 11) are consistent with this hypothesis. Compared with splenocytes from mice infected with LCMV alone, the frequency of CD8α+ DCs was substantially lower in co-infected stimulator splenocytes (day 2 post LCMV/day 6 post CVB3, data not shown), and these “CD8α+ DC-depleted” splenocytes induced fewer LCMV-specific T cells to proliferate after 2 days of in vitro culture (Figure 11C). In contrast, when equal numbers of purified DCs (from LCMV-only and co-infected mice) were used as stimulators, both populations were similarly capable of stimulating T cell proliferation (Figure 12A & B). Together, these data suggest that the delayed T cell responses to co-infected splenocytes in vitro are due to a reduction in DC numbers rather than a decrease in DC functionality. The impact of these quantitative differences may be amplified by the qualitative changes in the APCs that are rapidly induced by wtCVB3 infection in vivo. Qualitative changes (e.g., cell maturation) in CD8α+ DCs occur as early as day 2 p.i. (Figure 4), preceding their numerical collapse (Figure 2D & E), and we speculate that these changes may contribute both to the delayed T cell response when LCMV is administered at this time point (Figure 8) and to the delayed T cell response that is observed in vitro (Figure 11). However, qualitative changes alone may not have irredeemable effects because, in mice infected with LCMV at 2 days after wtCVB3 infection, the magnitude of the LCMV-specific CD8+ T cell response in these mice is only modestly reduced 7 days later (Figure 8); similarly, P14 T cell proliferation in vitro appears to progress normally by day 3–4 (Figure 11). Based on these findings we speculate that, in mice that receive LCMV shortly after CVB3 (2 days later, Figure 8), the kinetics of LCMV-specific T cell expansion are altered because wtCVB3 infection reduces DC quality, delaying (but not aborting) the expansion of LCMV-specific T cells. If the interval between these infections is increased (e.g. if LCMV is given 3 or 4 days after CVB3), both quality and quantity are markedly reduced, thereby amplifying the suppressive effect of wtCVB3 on the LCMV T cell response. In contrast, if LCMV is given 3 days after rCVB3 infection, LCMV-specific T cell responses are enhanced, which suggests that the less virulent recombinant virus does not exert these suppressive effects on DCs; rather, rCVB3 infection appears to establish a microenvironment that is capable of promoting strong T cell responses against a subsequent virus infection (Kemball et al., 2009). We considered the possibility that the weaker LCMV-specific T cell response in CVB3-infected mice might result from a reduced level of LCMV antigen, perhaps because LCMV replication was suppressed by ongoing host responses to the CVB3 infection. However, our quantitative assessment of LCMV genomes in the spleen and liver suggests that LCMV replication is not limited by CVB3 co-infection. On the contrary, the number of LCMV genomes in these organs was similar to, or slightly greater than, the number measured in mice infected with LCMV alone (Figure 10). Indeed, our finding that LCMV genome copy numbers were significantly higher in the livers of co-infected mice, which lack strong LCMV-specific T cell responses, is consistent with the fact that control of LCMV is T cell-dependent (Ahmed et al., 1988; Ahmed et al., 1984; Jamieson et al., 1987). It is unclear why LCMV replication is not inhibited by the innate immune response to CVB3. Levels of type I IFN peak in the serum and pancreas of CVB3-infected mice at day 2 p.i. (the time point at which mice were co-infected with LCMV, Figure 8), but this increase is not observed in the liver (Huhn et al., 2010; Wang et al., 2010). Therefore, it is conceivable that LCMV may escape the effects of CVB3-induced type I IFN in some sites including the liver and spleen. Furthermore, CVB3 can disrupt type I IFN signaling in vitro by cleaving the innate immune adaptor molecules MAVS and TRIF, and we speculate that antagonism of these signaling pathways by CVB3 in vivo could minimize the effects of type I IFN on concurrent LCMV replication (Mukherjee et al., 2011).
The effects of CVB3 on DCs, and in particular CD8+CD205+ DCs, may contribute to our previous findings that CD8+ T cell responses to CVB3 are barely detectable (Kemball et al., 2009; Kemball et al., 2008; Slifka et al., 2001). However, we feel that additional factors must play a role, for several reasons. First, activation of DCs is known to decrease their ability to cross-prime naïve virus-specific T cells (Segura and Villadangos, 2009; Wilson et al., 2006; Young et al., 2007); consequently, cross-priming is thought to occur very soon after the onset of infection, when naïve CD8+ T cells interact briefly with immature antigen-bearing DCs (Hickman et al., 2008; Kastenmuller et al., 2010; Norbury et al., 2002). One would expect that CVB3 proteins synthesized in the first hours p.i. would gain access to, and be cross-presented by, immature DCs, prior to their being activated by the infection. This appears not to occur. Second, we show here that a wtCVB3-infected host remains capable of mounting a strong (albeit slightly delayed) response as late as 2 days after wtCVB3 infection (Figure 8). At this time point, CD8α+ DCs are numerous, and wtCVB3 has replicated to high titers (Figure 5A), so viral proteins are abundant, yet mice do not develop a readily-detectable CVB3-specific CD8+ T cell response (Kemball et al., 2009); this suggests that the existing CD8α+ DCs do not effectively cross-prime in these wtCVB-infected mice. Third, even when DCs are largely depleted by wtCVB3 infection (day 4 p.i.) the host remains capable of mounting a detectable response to LCMV. We cannot exclude other, DC-independent, explanations that may contribute to the weak primary T cell response to CVB, and for the diminished LCMV-specific T cell response in wtCVB3-infected mice. For example, somewhat counter-intuitively, many viruses cause a rapid but transient lymphopenia (Bautista et al., 2003; Díaz-San Segundo et al., 2006; Dietz, Jr. et al., 1979; Geisbert et al., 2000; Grubman et al., 2008; Nfon et al., 2010; O'Donnell and Carrington, 2002; Okada et al., 2000; Peacock et al., 2003); the effect, which is mediated by interferons (Kamphuis et al., 2006; Schattner et al., 1983), has been shown to occur during enteroviral infections (Bautista et al., 2003; Nfon et al., 2010) and we have observed it in mice infected with wtCVB3 (Althof & Whitton, unpublished).
In conclusion, our evaluation of the effects of wtCVB3 on DCs in vitro and in vivo reveals that the infection dramatically alters the abundance, heterogeneity, and immunostimulatory capacity of conventional dendritic cells. These changes are unlikely to be the result of direct infection of DCs by CVB3, and instead the virus appears to induce DC maturation and decay in an indirect manner. The loss of CD8α+ DCs may deprive the host of the most potent inducers of virus-specific T cell responses, and other effects on cDCs, e.g., on activation/maturation, may augment the immunoevasive effects of CVB3 and related enteroviruses in vivo. These features also may weaken the host’s capacity to mount T cell responses to a secondary virus infection.
Materials & Methods
Mice, viruses, and infections
C57BL/6J mice were purchased from The Scripps Research Institute (TSRI) breeding facility. P14/Thy1.1 TCR transgenic mice specific for the H-2Db restricted LCMV epitope GP33–41 (Pircher et al., 1990) were bred and maintained by our laboratory as described (Whitmire et al., 2006; Whitmire et al., 2008). This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All experimental procedures with mice were reviewed and approved by TSRI Animal Care and Use Committee. The wtCVB3 used in these studies is a plaque-purified isolate (designated H3) of the myocarditic Woodruff variant of CVB3 (van Houten et al., 1991). Plasmid pH3, encoding a full length infectious clone of this virus (Knowlton et al., 1996), was provided by Kirk Knowlton (University of California, San Diego, CA). Recombinant CVB3 encoding eGFP or DsRed were generated as described previously (Feuer et al., 2002; Tabor-Godwin et al., 2010). wtCVB3, eGFP-CVB3, and DsRed-CVB3 were grown in HeLa cells, and virus stocks were generated as described previously (Harkins et al., 2005). Naïve adult male C57BL/6J mice were inoculated i.p. with 1×103 PFU of wtCVB3, 1×107 PFU of DsRed-CVB3, or 2×105 PFU of LCMV Armstrong. To confirm CVB3 infection, the virus titer in the feces was determined on day 2 post infection, as described previously (Kemball et al., 2009). Pancreas was isolated from wtCVB3-infected mice, weighed, and homogenized in 1 ml DMEM, and the titer of infectious virus in the lysate was determined. Plaque assays were performed on subconfluent HeLa cell monolayers, as described previously (Hunziker et al., 2007), and the virus titer (PFU/g) was calculated for each sample.
Lymphocyte isolation
Spleens and pancreatic lymph nodes were minced and digested with collagenase D (1 mg/ml, Roche) and DNase I (20 µg/ml, Roche) for 30 min at 37°C, and then filtered through a 70 µm cell strainer (BD Biosciences) to generate a single cell suspension. Red blood cells were lysed with 0.83% NH4Cl.
Isolation and culture of bone marrow-derived dendritic cells
Bone marrow was flushed from the femurs and tibiae of C57BL/6J mice using cold DPBS (Invitrogen). Marrow was passed repeatedly through an 18-gauge needle and then filtered through a 70 µm cell strainer to generate a single cell suspension. Viable cells were separated from dead cells and erythrocytes using Lympholyte-M (Cedarlane Labs), according to the manufacturer’s instructions. Cells were setup in 24-well plates (1–2×106 cells/well) in 1 ml of BMDC culture media (RPMI containing 10% FBS, L-glutamine, 2-mercaptoethanol, penicillin/streptomycin, sodium pyruvate, and HEPES) with recombinant murine GM-CSF (50 ng/ml, Peprotech), and cultured for 6–10 days before use. On day 2, 700 ul of media was removed from each well and replaced with fresh media + GM-CSF. On day 3, the entire volume of media was removed from the wells, then the cells were gently washed with 500 ul fresh media (to remove residual non-adherent cells), and 1 ml of culture media + GM-CSF was added to the wells. Thereafter, cells were fed daily with 1 ml culture media + GM-CSF.
In vitro antigen presentation assay
Transgenic T cells were used to detect the presentation of LCMV antigen by splenocytes isolated from virus-infected mice. To prepare these indicator cells, CD8+ T cells were purified from the spleens of P14/Thy1.1 TCR transgenic mice by MACS separation (negative selection) using a mouse CD8α+ T cell Isolation Kit II (Miltenyi Biotec), following the manufacturer’s instructions. The purity of enriched CD8+ T cells (93%) and the frequency of P14 transgenic T cells (CD8+Vα2+) were determined by flow cytometry. P14 cells were labeled with 3 µM CFSE. Single cell suspensions of spleens from virus-infected mice were prepared (see Lymphocyte isolation, above), and 106 stimulator spleen cells were incubated with 5×104 P14 transgenic T cells per well in 96-well flat bottom plates containing 0.2 ml culture media (RPMI containing 10% FBS, 50 µM 2-mercaptoethanol, and penicillin/streptomycin). One set of cultures included P14 cells, splenocytes from co-infected mice, and 1 µM LCMV GP33–41 peptide. Duplicate cultures were setup for each sample. Cells were fed on day 2 and 3 of culture; ~1/2 the volume of media from each well was removed, and replaced with 100 µl fresh culture media. Wells were harvested after 2, 3, and 4 days of culture, and transgenic T cell proliferation was determined by flow cytometry.
To analyze antigen presentation by DCs, these cells were enriched from the spleens of virus-infected mice by MACS separation (positive selection) using mouse CD11c microbeads (Miltenyi Biotec), following the manufacturer’s instructions. The spleens within each mouse group (3–6 mice/group) were pooled before MACS separation. 6×104 CD11chi cells were incubated with 1.2×105 CFSE-labeled P14 cells per well. In one set of cultures, DCs from co-infected mice were pulsed with 1 µM GP33–41 peptide for 30 min in 96-well culture wells (1.2×105 cells/well), then washed to remove unbound peptide, and incubated with P14 cells. Triplicate cultures were setup for each group. Wells were harvested after 2, 3, and 4 days of culture, and transgenic T cell proliferation was determined by flow cytometry.
Flow cytometry
Cells were incubated with Fc receptor antibody (FcBlock, BD Biosciences) in PBS containing 2% FBS and 0.1% sodium azide (FACS buffer) for 10 min on ice. Cells were then surface stained with fluorochrome-conjugated antibodies and/or MHC class I tetramers (NIH Tetramer Core Facility, Atlanta, GA) in FACS buffer for 30 min on ice, and washed 2X in FACS buffer. Following the surface staining, some samples were stained with eFluor780-conjugated fixable viability dye (eBioscience) following the manufacturer’s instructions, in order to identify dead cells during flow cytometry and exclude them from our analyses. Samples were fixed in PBS containing 1% paraformaldehyde and acquired on a FACSCalibur or LSR-II (BD Biosciences). Data were analyzed with FlowJo software (TreeStar).
For intracellular cytokine staining of splenocytes, 2×106 splenocytes were incubated for 5 hrs in 96-well plates in 0.2 ml/well RPMI containing 10% FBS, 50 µM 2-mercaptoethanol, penicillin/streptomycin, GolgiPlug (BD Biosciences), and synthetic peptides (1 µM LCMV GP33–41 or NP396–404, 10 µM GP61–80). After stimulation, cells were incubated with FcBlock, stained with CD8 or CD4 antibodies as described above, then stained for intracellular IFNγ with the Cytofix/Cytoperm kit (BD Biosciences). The total number of epitope-specific cytokine-producing T cells was determined by subtracting unstimulated cytokine+ T cells (no peptide) from peptide-stimulated cytokine+ T cells.
For intracellular staining of CVB3-infected primary BMDCs and cell lines [including DC2.4 cells (kindly provided by Ken Rock, University of Massachusetts) and HeLa cells], cells were detached from tissue culture wells by forceful pipetting or treatment with versene (Invitrogen), and then washed and resuspended in FACS buffer. Cells were permeabilized with the Cytofix/Cytoperm kit, and then stained with VP1 antibody in Permwash for 30 min on ice.
The following antibodies were used for the flow cytometric analyses described above. Fluorochrome-conjugated CD11c (clone N418), CD11b (clone M1/70), CD3ε (clone 145-2C11), CD19 (clone 6D5), CD49b (clone DX5), CD8α (clone 53-6.7), CD4 (clone RM4-5 and GK1.5), Thy1.1 (clone HIS51), Thy1.2 (clone 53-2.1), Vα2 (clone B20.1), MHC class I (clone 28-14-8), MHC class II (clone M5/114.15.2), CD40 (clone HM40-3), CD80 (clone 16-10A1), CD86 (clone GL1), IFNγ (clone XMG1.2), and isotype control antibodies were purchased from eBioscience or BioLegend. CD205 (clone NLDC-145) and mPDCA-1 (clone JF05-1C2.4.1) antibodies were purchased from Miltenyi Biotec. VP1 antibody (clone 5-D8/1) was purchased from Leica Microsystems as a lyophilized tissue culture supernatant. The antibody was reconstituted in water, column purified by TSRI Antibody Core Facility, and conjugated to Alexa Fluor 647 using a Molecular Probes monoclonal antibody labeling kit (Invitrogen).
Vibratome sectioning and confocal microscopy
Mice were perfused with DPBS, and the pancreas was harvested and fixed in buffered zinc formalin (Z-FIX, Anatech Ltd.) at RT for 5 days. The fixative was replaced with PBS and tissues were preserved at 4°C. Sections of pancreas (80 µm) were cut with a VT1000 S vibratome (Leica Microsystems, Inc.). Free floating sections were washed 3 times in PBS and permeabilized in PBS with 0.5% Triton X-100 for 30 min at RT. Next, the sections were washed twice in PBS with 0.1% Triton X-100 and then incubated with Alexa Fluor 488-conjugated phalloidin (Invitrogen) in PBS with 0.1% Triton X-100 overnight at 4°C (to label F-actin). After incubation, the sections were washed twice in PBS with 0.1% Triton X-100 and then 3 times in PBS. Lastly, sections were stained with Hoechst 33342 (Invitrogen) in PBS for 30 min at RT (to label nuclei) and then washed 3 times in PBS and mounted on glass slides with ProLong Gold mounting medium (Invitrogen). Three color images were captured using a LSM 710 laser scanning confocal microscope equipped with ZEN software (Carl Zeiss, Inc.). Representative regions within each vibratome section of pancreas were scanned at ×40 magnification as serial 8-bit optical sections with 0.5 µm interval step sizes (512 by 512 image size). Images were analyzed with Imaris software (BitPlane, Inc.) and maximum projected for presentation using the Easy 3D function.
Quantitative real time PCR
Spleen and liver samples were collected and immediately stored in RNAlater stabilization reagent (Qiagen). RNA was isolated from these organs with an RNeasy mini kit (Qiagen), following the manufacturer’s instructions, and genomic LCMV RNA (present in mature virions) was quantitated as follows. A reverse transcription (RT) primer (5’CAGGGTGCAAGTGGTGTGGTAAGA3’) complementary to residues 2865 to 2842 of the viral S segment genomic RNA was synthesized, and used in an RT reaction together with 1/10th of the total RNA sample, MultiScribe RT, 10× PCR Buffer II, MgCl2, RNAse Inhibitor, and GeneAmp dNTP Mix (all from Applied Biosystems). Additional control reactions were setup that omitted RT (“no RT”). The RT reaction was carried out in a thermocycler as follows: 25°C for 10 min, 48°C for 30 min, 95°C for 5 min. Next, Taqman quantitative real time PCR was performed using the following primers and probe: forward primer (Arm S 2275+), 5’CGCTGGCCTGGGTGAAT3’; reverse primer (Arm S 2338-), 5’ATGGGAAAACACAACAATTGATCTC3’, Taqman FAM/MGB-labeled probe, 5’TTGCTGCAGAGCTTA3’. PCR reactions contained primers & probe, TaqMan Universal PCR Master Mix (No AmpErase UNG, Applied Biosystems), and 1/10th of the total cDNA. Quantitative analysis of viral RNA was carried out using a BioRad iQ5 real time PCR system in 96-well optical reaction plates heated to 95°C for 10 min, followed by 40 cycles of: denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. All samples were evaluated in triplicate amplification reactions. Dilutions of the p-T7-S plasmid (encoding the S segment of the LCMV Armstrong genome, kindly provided by Juan Carlos de la Torre, TSRI) were analyzed (in quadruplicate) to generate a standard curve. The standard curve was based on threshold cycle (CT) values, and CT values from tissue samples were compared to the standard curve to determine viral RNA copy numbers. The limit of detection of this assay is 5 copies of viral RNA/reaction. Values are expressed as the average number of LCMV genomes per mg tissue; these were determined by calculating the total number of LCMV genomes per RNA sample, divided by the sample weight.
Statistical analyses
Statistical significance was determined by an unpaired two-tailed t-test, or by one-way ANOVA with Tukey’s post hoc test (GraphPad Prism software). A p value <0.05 was considered statistically significant.
Acknowledgements
We thank Annette Lord and Sheila Silverstein for excellent secretarial support. We also thank the NIH Tetramer Core Facility for providing MHC class I tetramers, and Dr. Ken Rock (University of Massachusetts) for providing the DC2.4 cell line. This work was supported by NIH grants AI042314 and HL093177 (to J.L.W.); T32 NS41219 and F32 AI078660 (to C.C.K.); T32 HL007195-34 (to M.P.H.), and F32 AI056827, P20 RR021905, and R21 AI088059 (to J.B.). This is manuscript number 21350 from the Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed R, Butler LD, Bhatti L. T4+ T helper cell function in vivo: differential requirement for induction of antiviral cytotoxic T-cell and antibody responses. J. Virol. 1988;62:2102–2106. doi: 10.1128/jvi.62.6.2102-2106.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MBA. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice: role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista EM, Ferman GS, Golde WT. Induction of lymphopenia and inhibition of T cell function during acute infection of swine with foot and mouth disease virus (FMDV) Vet. Immunol. Immunopathol. 2003;92:61–73. doi: 10.1016/s0165-2427(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Belz GT, Shortman K, Bevan MJ, Heath WR. CD8α+ dendritic cells selectively present MHC class I-restricted noncytolytic viral and intracellular bacterial antigens in vivo. J. Immunol. 2005;175:196–200. doi: 10.4049/jimmunol.175.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SS, Dodd DA, Kirkegaard K. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology. 2005;337:18–29. doi: 10.1016/j.virol.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Crocker SJ, Frausto RF, Whitmire JK, Benning N, Whitton JL. Amelioration of coxsackievirus B3-mediated myocarditis by inhibition of tissue inhibitors of matrix metalloproteinase-1. Am. J. Pathol. 2007;171:1762–1773. doi: 10.2353/ajpath.2007.070179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley AJ, Isaacs D, Dwyer DE, Gilbert GL. A cluster of cases of neonatal coxsackievirus B meningitis and myocarditis. J. Paediatr. Child Health. 1998;34:196–198. doi: 10.1046/j.1440-1754.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- Dalod M, Hamilton T, Salomon R, Salazar-Mather TP, Henry SC, Hamilton JD, Biron CA. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon alpha/beta. J. Exp. Med. 2003;197:885–898. doi: 10.1084/jem.20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitz SB, Dodd DA, Cooper S, Parham P, Kirkegaard K. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13790–13795. doi: 10.1073/pnas.250483097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-San Segundo F, Salguero FJ, de Avila A, de Marco MM, Sanchez-Martin MA, Sevilla N. Selective lymphocyte depletion during the early stage of the immune response to foot-and-mouth disease virus infection in swine. J. Virol. 2006;80:2369–2379. doi: 10.1128/JVI.80.5.2369-2379.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz WH, Jr, Peralta PH, Johnson KM. Ten clinical cases of human infection with venezuelan equine encephalomyelitis virus, subtype I-D. Am. J. Trop. Med. Hyg. 1979;28:329–334. doi: 10.4269/ajtmh.1979.28.329. [DOI] [PubMed] [Google Scholar]
- Dodd DA, Giddings TH, Jr, Kirkegaard K. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 2001;75:8158–8165. doi: 10.1128/JVI.75.17.8158-8165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez PM, Ardavin C. Differentiation and function of mouse monocyte-derived dendritic cells in steady state and inflammation. Immunol. Rev. 2010;234:90–104. doi: 10.1111/j.0105-2896.2009.00876.x. [DOI] [PubMed] [Google Scholar]
- Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, Steinman RM, Nussenzweig MC. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- Feuer R, Mena I, Pagarigan RR, Slifka MK, Whitton JL. Cell cycle status affects coxsackievirus replication, persistence, and reactivationin vitro . J. Virol. 2002;76:4430–4440. doi: 10.1128/JVI.76.9.4430-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuer R, Pagarigan RR, Harkins S, Liu F, Hunziker IP, Whitton JL. Coxsackievirus targets proliferating neuronal progenitor cells in the neonatal CNS. J. Neurosci. 2005;25:2434–2444. doi: 10.1523/JNEUROSCI.4517-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Gibb TR, Steele KE, Jaax NK, Jahrling PB. Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab Invest. 2000;80:171–186. doi: 10.1038/labinvest.3780021. [DOI] [PubMed] [Google Scholar]
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- Grubman MJ, Moraes MP, Diaz-San SF, Pena L, de los ST. Evading the host immune response: how foot-and-mouth disease virus has become an effective pathogen. FEMS Immunol. Med. Microbiol. 2008;53:8–17. doi: 10.1111/j.1574-695X.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- Harkins S, Cornell CT, Whitton JL. Analysis of translational initiation in Coxsackievirus B3 suggests an alternative explanation for the high frequency of R+4 in the eukaryotic consensus motif. J. Virol. 2005;79:987–996. doi: 10.1128/JVI.79.2.987-996.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath WR, Carbone FR. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 2009;10:1237–1244. doi: 10.1038/ni.1822. [DOI] [PubMed] [Google Scholar]
- Henke A, Huber SA, Stelzner A, Whitton JL. The role of CD8+ T lymphocytes in coxsackievirus B3-induced myocarditis. J. Virol. 1995;69:6720–6728. doi: 10.1128/jvi.69.11.6720-6728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman HD, Takeda K, Skon CN, Murray FR, Hensley SE, Loomis J, Barber GN, Bennink JR, Yewdell JW. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat. Immunol. 2008;9:155–165. doi: 10.1038/ni1557. [DOI] [PubMed] [Google Scholar]
- Huber S, Ramsingh AI. Coxsackievirus-induced pancreatitis. Viral Immunol. 2004;17:358–369. doi: 10.1089/vim.2004.17.358. [DOI] [PubMed] [Google Scholar]
- Huhn MH, McCartney SA, Lind K, Svedin E, Colonna M, Flodstrom-Tullberg M. Melanoma differentiation-associated protein-5 (MDA-5) limits early viral replication but is not essential for the induction of type 1 interferons after Coxsackievirus infection. Virology. 2010;401:42–48. doi: 10.1016/j.virol.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Hunziker IP, Cornell CT, Whitton JL. Deletions within the 5'UTR of coxsackievirus B3: consequences for virus translation and replication. Virology. 2007;360:120–128. doi: 10.1016/j.virol.2006.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Kuckelkorn U, Szalay G, Plotz M, Textoris-Taube K, Opitz E, Klingel K, Stevanovic S, Kandolf R, Kotsch K, Stangl K, Kloetzel PM, Voigt A. Differential interferon responses enhance viral epitope generation by myocardial immunoproteasomes in murine enterovirus myocarditis. Am. J. Pathol. 2009;175:510–518. doi: 10.2353/ajpath.2009.090033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson BD, Butler LD, Ahmed R. Effective clearance of a persistent viral infection requires cooperation between virus-specific Lyt2+ T cells and nonspecific bone marrow-derived cells. J. Virol. 1987;61:3930–3937. doi: 10.1128/jvi.61.12.3930-3937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood. 2006;108:3253–3261. doi: 10.1182/blood-2006-06-027599. [DOI] [PubMed] [Google Scholar]
- Kastenmuller W, Gerner MY, Germain RN. The in situ dynamics of dendritic cell interactions. Eur. J. Immunol. 2010;40:2103–2106. doi: 10.1002/eji.201040482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemball CC, Alirezaei M, Whitton JL. Type B coxsackieviruses and their interactions with the innate and adaptive immune systems. Future Microbiol. 2010a;5:1329–1347. doi: 10.2217/fmb.10.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemball CC, Fujinami RS, Whitton JL. Adaptive immune responses to picornaviruses. In: Ehrenfeld E, Domingo E, Roos RP, editors. The Picornaviruses. Washington, DC: ASM Press; 2010b. pp. 303–319. [Google Scholar]
- Kemball CC, Harkins S, Whitmire JK, Flynn CT, Feuer R, Whitton JL. Coxsackievirus B3 inhibits antigen presentation in vivo, exerting a profound and selective effect on the MHC class I pathway. PLoS Path. 2009;5:e1000618. doi: 10.1371/journal.ppat.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemball CC, Harkins S, Whitton JL. Enumeration and functional evaluation of virus-specific CD4+ and CD8+ T cells in lymphoid and peripheral sites of coxsackievirus B3 infection. J. Virol. 2008;82:4331–4342. doi: 10.1128/JVI.02639-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton KU, Jeon ES, Berkley N, Wessely R, Huber SA. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 1996;70:7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M, Schulte BM, Toonen LW, de Bruijni MA, Galama JM, Adema GJ, van Kuppeveld FJ. Echovirus infection causes rapid loss-of-function and cell death in human dendritic cells. Cell. Microbiol. 2007;9:1507–1518. doi: 10.1111/j.1462-5822.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Lee HK, Iwasaki A. Innate control of adaptive immunity: dendritic cells and beyond. Semin. Immunol. 2007;19:48–55. doi: 10.1016/j.smim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Lee LN, Burke S, Montoya M, Borrow P. Multiple mechanisms contribute to impairment of type 1 interferon production during chronic lymphocytic choriomeningitis virus infection of mice. J. Immunol. 2009;182:7178–7189. doi: 10.4049/jimmunol.0802526. [DOI] [PubMed] [Google Scholar]
- Leon B, Ardavin C. Monocyte-derived dendritic cells in innate and adaptive immunity. Immunol. Cell Biol. 2008;86:320–324. doi: 10.1038/icb.2008.14. [DOI] [PubMed] [Google Scholar]
- Lopez-Bravo M, Ardavin C. In vivo induction of immune responses to pathogens by conventional dendritic cells. Immunity. 2008;29:343–351. doi: 10.1016/j.immuni.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, Mann M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Mena I, Fischer C, Gebhard JR, Perry CM, Harkins S, Whitton JL. Coxsackievirus infection of the pancreas: evaluation of receptor expression, pathogenesis, and immunopathology. Virology. 2000;271:276–288. doi: 10.1006/viro.2000.0332. [DOI] [PubMed] [Google Scholar]
- Modlin JF, Rotbart HA. Group B coxsackie disease in children. Curr. Top. Microbiol. Immunol. 1997;223:53–80. doi: 10.1007/978-3-642-60687-8_4. [DOI] [PubMed] [Google Scholar]
- Montoya M, Edwards MJ, Reid DM, Borrow P. Rapid activation of spleen dendritic cell subsets following lymphocytic choriomeningitis virus infection of mice: analysis of the involvement of type 1 IFN. J. Immunol. 2005;174:1851–1861. doi: 10.4049/jimmunol.174.4.1851. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. The Coxsackievirus B 3C Protease Cleaves MAVS and TRIF to Attenuate Host Type I Interferon and Apoptotic Signaling. PLoS Pathog. 2011;7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neznanov N, Kondratova A, Chumakov KM, Angres B, Zhumabayeva B, Agol VI, Gudkov AV. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 2001;75:10409–10420. doi: 10.1128/JVI.75.21.10409-10420.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nfon CK, Toka FN, Kenney M, Pacheco JM, Golde WT. Loss of plasmacytoid dendritic cell function coincides with lymphopenia and viremia during foot-and-mouth disease virus infection. Viral Immunol. 2010;23:29–41. doi: 10.1089/vim.2009.0078. [DOI] [PubMed] [Google Scholar]
- Norbury CC, Malide D, Gibbs JS, Bennink JR, Yewdell JW. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat. Immunol. 2002;3:265–271. doi: 10.1038/ni762. [DOI] [PubMed] [Google Scholar]
- O'Connell JB. The role of myocarditis in end-stage dilated cardiomyopathy. Tex. Heart Inst. J. 1987;14:268–275. [PMC free article] [PubMed] [Google Scholar]
- O'Donnell DR, Carrington D. Peripheral blood lymphopenia and neutrophilia in children with severe respiratory syncytial virus disease. Pediatr. Pulmonol. 2002;34:128–130. doi: 10.1002/ppul.10140. [DOI] [PubMed] [Google Scholar]
- Okada H, Kobune F, Sato TA, Kohama T, Takeuchi Y, Abe T, Takayama N, Tsuchiya T, Tashiro M. Extensive lymphopenia due to apoptosis of uninfected lymphocytes in acute measles patients. Arch. Virol. 2000;145:905–920. doi: 10.1007/s007050050683. [DOI] [PubMed] [Google Scholar]
- Peacock CD, Kim SK, Welsh RM. Attrition of virus-specific memory CD8+ T cells during reconstitution of lymphopenic environments. J. Immunol. 2003;171:655–663. doi: 10.4049/jimmunol.171.2.655. [DOI] [PubMed] [Google Scholar]
- Pircher H, Moskophidis D, Rohrer U, Burki K, Hengartner H, Zinkernagel RM. Viral escape by selection of cytotoxic T cell-resistant virus variantsin vivo . Nature. 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- Probst HC, van den BM. Priming of CTLs by lymphocytic choriomeningitis virus depends on dendritic cells. J. Immunol. 2005;174:3920–3924. doi: 10.4049/jimmunol.174.7.3920. [DOI] [PubMed] [Google Scholar]
- Rahnefeld A, Ebstein F, Albrecht N, Opitz E, Kuckelkorn U, Stangl K, Rehm A, Kloetzel PM, Voigt A. Antigen-presentation capacity of dendritic cells is impaired in ongoing enterovirus myocarditis. Eur. J. Immunol. 2011 doi: 10.1002/eji.201041039. [DOI] [PubMed] [Google Scholar]
- Rhoades RE, Tabor-Godwin JM, Tsueng G, Feuer R. Enterovirus infections of the central nervous system. Virology. 2011;411:288–305. doi: 10.1016/j.virol.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero JR. Pediatric group B coxsackievirus infections. Curr. Top. Microbiol. Immunol. 2008;323:223–239. doi: 10.1007/978-3-540-75546-3_10. [DOI] [PubMed] [Google Scholar]
- Schattner A, Meshorer A, Wallach D. Involvement of interferon in virus-induced lymphopenia. Cell Immunol. 1983;79:11–25. doi: 10.1016/0008-8749(83)90046-1. [DOI] [PubMed] [Google Scholar]
- Schulte BM, Kramer M, Ansems M, Lanke KH, van Doremalen N, Piganelli JD, Bottino R, Trucco M, Galama JM, Adema GJ, van Kuppeveld FJ. Phagocytosis of enterovirus-infected pancreatic β-cells triggers innate immune responses in human dendritic cells. Diabetes. 2010;59:1182–1191. doi: 10.2337/db09-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E, Villadangos JA. Antigen presentation by dendritic cells in vivo. Curr. Opin. Immunol. 2009;21:105–110. doi: 10.1016/j.coi.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Shen Z, Reznikoff G, Dranoff G, Rock KL. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J. Immunol. 1997;158:2723–2730. [PubMed] [Google Scholar]
- Shortman K, Heath WR. The CD8+ dendritic cell subset. Immunol. Rev. 2010;234:18–31. doi: 10.1111/j.0105-2896.2009.00870.x. [DOI] [PubMed] [Google Scholar]
- Slifka MK, Pagarigan RR, Mena I, Feuer R, Whitton JL. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells during picornavirus infection. J. Virol. 2001;75:2377–2387. doi: 10.1128/JVI.75.5.2377-2387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka MK, Whitton JL. Functional avidity maturation of CD8+ T cells without selection of higher affinity TCR. Nat. Immunol. 2001;2:711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- Sole MJ, Liu P. Viral myocarditis: a paradigm for understanding the pathogenesis and treatment of dilated cardiomyopathy. J. Am. Coll. Cardiol. 1993;22:99A–105A. doi: 10.1016/0735-1097(93)90470-l. [DOI] [PubMed] [Google Scholar]
- Swiecki M, Colonna M. Accumulation of plasmacytoid DC: Roles in disease pathogenesis and targets for immunotherapy. Eur. J. Immunol. 2010a;40:2094–2098. doi: 10.1002/eji.201040602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M, Colonna M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol. Rev. 2010b;234:142–162. doi: 10.1111/j.0105-2896.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor-Godwin JM, Ruller CM, Bagalso N, An N, Pagarigan RR, Harkins S, Gilbert PE, Kiosses WB, Gude NA, Cornell CT, Doran KS, Sussman MA, Whitton JL, Feuer R. A novel population of myeloid cells responding to coxsackievirus infection assists in the dissemination of virus within the neonatal CNS. J. Neurosci. 2010;30:8676–8691. doi: 10.1523/JNEUROSCI.1860-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PE. Coxsackievirus myocarditis: Interplay between virus and host in the pathogenesis of heart disease. Viral Immunol. 2006;19:133–146. doi: 10.1089/vim.2006.19.133. [DOI] [PubMed] [Google Scholar]
- Tracy S, Chapman NM. Group B coxsackievirus diseases. In: Ehrenfeld E, Domingo E, Roos RP, editors. Picornaviruses: Molecular Biology, Evolution and Pathogenesis. ASM; 2010. pp. 353–368. [Google Scholar]
- van Houten N, Bouchard PE, Moraska A, Huber SA. Selection of an attenuated Coxsackievirus B3 variant, using a monoclonal antibody reactive to myocyte antigen. J. Virol. 1991;65:1286–1290. doi: 10.1128/jvi.65.3.1286-1290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Calvino R, Skowera A, Arif S, Peakman M. Identification of a naturally processed cytotoxic CD8 T-cell epitope of coxsackievirus B4, presented by HLA-A2.1 and located in the PEVKEK region of the P2C nonstructural protein. J. Virol. 2004;78:13399–13408. doi: 10.1128/JVI.78.24.13399-13408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos JA, Schnorrer P. Intrinsic and cooperative antigen-presenting functions of dendritic-cell subsets in vivo. Nat. Rev. Immunol. 2007;7:543–555. doi: 10.1038/nri2103. [DOI] [PubMed] [Google Scholar]
- Villadangos JA, Young L. Antigen-presentation properties of plasmacytoid dendritic cells. Immunity. 2008;29:352–361. doi: 10.1016/j.immuni.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Voigt A, Jakel S, Textoris-Taube K, Keller C, Drung I, Szalay G, Klingel K, Henklein P, Stangl K, Kloetzel PM, Kuckelkorn U. Generation of in silico predicted coxsackievirus B3-derived MHC class I epitopes by proteasomes. Amino Acids. 2010;39:243–255. doi: 10.1007/s00726-009-0434-5. [DOI] [PubMed] [Google Scholar]
- Wahid R, Cannon MJ, Chow M. Dendritic cells and macrophages are productively infected by poliovirus. J. Virol. 2005;79:401–409. doi: 10.1128/JVI.79.1.401-409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Asher DR, Chan M, Kurt-Jones EA, Finberg RW. Cutting Edge: Antibody-mediated TLR7-dependent recognition of viral RNA. J. Immunol. 2007;178:3363–3367. doi: 10.4049/jimmunol.178.6.3363. [DOI] [PubMed] [Google Scholar]
- Wang JP, Cerny A, Asher DR, Kurt-Jones EA, Bronson RT, Finberg RW. MDA5 and MAVS mediate type I IFN responses to Coxsackie B virus. J. Virol. 2010;84:254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinzierl AO, Rudolf D, Maurer D, Wernet D, Rammensee HG, Stevanovic S, Klingel K. Identification of HLA-A*01- and HLA-A*02-restricted CD8+ T-cell epitopes shared among group B enteroviruses. J. Gen. Virol. 2008a;89:2090–2097. doi: 10.1099/vir.0.2008/000711-0. [DOI] [PubMed] [Google Scholar]
- Weinzierl AO, Szalay G, Wolburg H, Sauter M, Rammensee HG, Kandolf R, Stevanovic S, Klingel K. Effective chemokine secretion by dendritic cells and expansion of cross-presenting CD4−/CD8+ dendritic cells define a protective phenotype in the mouse model of coxsackievirus myocarditis. J. Virol. 2008b;82:8149–8160. doi: 10.1128/JVI.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weslow-Schmidt JL, Jewell NA, Mertz SE, Simas JP, Durbin JE, Flano E. Type I interferon inhibition and dendritic cell activation during gammaherpesvirus respiratory infection. J. Virol. 2007;81:9778–9789. doi: 10.1128/JVI.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessely R, Klingel K, Knowlton KU, Kandolf R. Cardioselective infection with coxsackievirus B3 requires intact type I interferon signaling: implications for mortality and early viral replication. Circulation. 2001;103:756–761. doi: 10.1161/01.cir.103.5.756. [DOI] [PubMed] [Google Scholar]
- Whitmire JK, Benning N, Whitton JL. Precursor frequency, nonlinear proliferation, and functional maturation of virus-specific CD4+ T cells. J. Immunol. 2006;176:3028–3036. doi: 10.4049/jimmunol.176.5.3028. [DOI] [PubMed] [Google Scholar]
- Whitmire JK, Eam B, Whitton JL. Tentative T cells : memory cells are quick to respond, but slow to divide. PLoS Path. 2008;4:1–11. doi: 10.1371/journal.ppat.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton JL. Immunopathology during coxsackievirus infection. Springer Semin. Immunopathol. 2002;24:201–213. doi: 10.1007/s00281-002-0100-4. [DOI] [PubMed] [Google Scholar]
- Wilson NS, Behrens GM, Lundie RJ, Smith CM, Waithman J, Young L, Forehan SP, Mount A, Steptoe RJ, Shortman KD, de Koning-Ward TF, Belz GT, Carbone FR, Crabb BS, Heath WR, Villadangos JA. Systemic activation of dendritic cells by Toll-like receptor ligands or malaria infection impairs cross-presentation and antiviral immunity. Nat. Immunol. 2006;7:165–172. doi: 10.1038/ni1300. [DOI] [PubMed] [Google Scholar]
- Young LJ, Wilson NS, Schnorrer P, Mount A, Lundie RJ, La Gruta NL, Crabb BS, Belz GT, Heath WR, Villadangos JA. Dendritic cell preactivation impairs MHC class II presentation of vaccines and endogenous viral antigens. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17753–17758. doi: 10.1073/pnas.0708622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LJ, Wilson NS, Schnorrer P, Proietto A, ten BT, Matsuki Y, Mount AM, Belz GT, O'Keeffe M, Ohmura-Hoshino M, Ishido S, Stoorvogel W, Heath WR, Shortman K, Villadangos JA. Differential MHC class II synthesis and ubiquitination confers distinct antigen-presenting properties on conventional and plasmacytoid dendritic cells. Nat. Immunol. 2008;9:1244–1252. doi: 10.1038/ni.1665. [DOI] [PubMed] [Google Scholar]
- Zuniga EI, Liou LY, Mack L, Mendoza M, Oldstone MB. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe. 2008;4:374–386. doi: 10.1016/j.chom.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]