Abstract
Benzo[a]pyrene-7,8-dione (B[a]P-7,8-dione) is produced in human lung cells by the oxidation of (±)-B[a]P-7,8-trans-dihydrodiol, which is catalyzed by aldo-keto reductases (AKRs). However, information relevant to the cell-based metabolism of B[a]P-7,8-dione is lacking. We studied the metabolic fate of 2 µM 1,3-[3H2]-B[a]P-7,8-dione in human lung adenocarcinoma A549 cells, human bronchoalveolar H358 cells and immortalized human bronchial epithelial HBEC-KT cells. In these three cell lines, 1,3-[3H2]-B[a]P-7,8-dione was rapidly consumed and radioactivity was distributed between the organic and aqueous phase of ethyl acetate-extracted media, as well as in the cell lysate pellets. After acidification of the media, several metabolites of 1,3-[3H2]-B[a]P-7,8-dione were detected in the organic phase of the media by high performance liquid chromatography-ultraviolet-radioactivity monitoring (HPLC-UV-RAM). The structures of B[a]P-7,8-dione metabolites varied in the cell lines and were identified as: B[a]P-7,8-dione conjugates with glutathione (GSH) and N-acetyl-L-cysteine (NAC), 8-O-mono-methylated-catechol, catechol mono-sulfate and mono-glucuronide, and mono-hydroxylated-B[a]P-7,8-dione by liquid chromatography–tandem mass spectrometry (LC-MS/MS). We also obtained evidence for the first time for the formation of an adenine adduct of B[a]P-7,8-dione. Among these metabolites, the identity of the GSH-B[a]P-7,8-dione and the NAC-B[a]P-7,8-dione were further validated by comparison to authentic synthesized standards. The pathways of B[a]P-7,8-dione metabolism in the three human lung cell lines are formation of GSH and NAC conjugates, reduction to the catechol followed by phase II conjugation reactions leading to its detoxification, mono-hydroxylation as well as formation of the adenine adduct.
Keywords: Methylation, sulfation, glucuronidation, catechol-O-methyltransferase, sulfotransferases, uridine 5'-diphospho-glucuronosyltransferases
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants formed as a result of incomplete combustion of fossil fuels (e.g. coal and oil) and are present in car and diesel exhaust and smoked or charbroiled food.1–3 They are also found in cigarette smoke condensate and tobacco products and are suspect agents in the causation of human lung cancer.4, 5 To cause their carcinogenic effects, PAHs must be metabolically activated to reactive genotoxins.
Three metabolic activation pathways of benzo[a]pyrene (B[a]P), a representative PAH, have been proposed that lead to mutations. In the first pathway, B[a]P is activated by P450 peroxidase in the presence of peroxide to yield a radical cation,6 which forms depurinating DNA adducts that lead to abasic sites.7–10 In the second pathway, P4501A1/1B1 and epoxide hydrolase convert B[a]P to the proximate carcinogen (−)-7β,8α–dihydroxy-7,8-dihydrobenzo[a]pyrene ((−)-B[a]P-7,8-trans-dihydrodiol), which is further monooxygenated by P4501A1/1B1 to yield (+)-7β,8α-dihydroxy-7,8,9,10-tetrahydro-9α,10α-oxo-benzo[a]pyrene ((+)-anti-B[a]PDE).11–13 This diol-epoxide forms stable N2-2’-deoxyguanosine (dGuo) adducts in vitro and in vivo,14, 15 which leads to G to T transversions via trans-lesional by-pass DNA polymerases.16–18 In the third pathway, the intermediate B[a]P-7,8-trans-dihydrodiol is oxidized in a NAD(P)+-dependent manner by aldo-keto reductases (AKRs) to produce benzo[a]pyrene-7,8-dione (B[a]P-7,8-dione).19, 20 AKR1A1 and AKR1C1-AKR1C4 convert B[a]P-7,8-trans-dihydrodiol to a ketol which spontaneously rearranges to a catechol.19–21 The unstable catechol undergoes two subsequent one-electron oxidations to yield the B[a]P-7,8-dione and reactive oxygen species (ROS).22 B[a]P-7,8-dione is both electrophilic and redox-active. The electrophilic B[a]P-7,8-dione can undergo 1,4- or 1,6-Michael addition with GSH and NAC to form GSH and NAC conjugates in vitro.23 It can also react with guanine and adenine bases to form a stable N2-dGuo adduct, a stable N6-dAdo adduct, or a N7-guanine depurinating adduct in vitro.24–27 B[a]P-7,8-dione can also be reduced back to the catechol and can establish futile redox cycles that result in the amplification of ROS until cellular reducing equivalents (e.g. NADPH) are depleted.21, 28 The enzymes responsible for the two electron reduction of B[a]P-7,8-dione to the catechol appear to be NQO1 and AKR themselves, while carbonyl reductases (CBRs) play a minor role.29 We have shown that this pathway exists in A549 and H358 cells and that this results in the generation of the mutagenic lesion 8-oxo-dGuo.30, 31 During the redox-cycling the catechol could be intercepted by phase II conjugating enzymes (e.g., COMT, SULTs or UGTs). Using human recombinant COMT and SULT1A1 we have obtained evidence for the formation of the mono-O-methylated catechol32 and the formation of the mono-O-sulfated catechol.33
Human lung cells are major sites of inhalation exposure to B[a]P and generation of its metabolites. However, to our best knowledge, information relevant to the cell based metabolism of B[a]P-7,8-dione is lacking. The objective of this study was to elucidate the metabolic fate of B[a]P-7,8-dione in human lung cells. A549 (human lung adenocarcinoma cells), H358 (human bronchoalveolar cells) and HBEC-KT (immortalized human bronchial epithelial cells), were selected as three human lung cell models to investigate the comparative metabolism of B[a]P-7,8-dione.
We found that B[a]P-7,8-dione was rapidly metabolized in the three cell lines. The metabolic pathways of B[a]P-7,8-dione involved the formation of GSH and NAC conjugates, mono-hydroxylation, and reduction to the catechol followed by O-mono-methylation or O-mono-sulfation, as well as the formation of an adenine adduct. In particular, reduction to the catechol followed by O-glucuronidation was found to be an unique metabolic pathway of B[a]P-7,8-dione in A549 cells. We conclude that quinone reduction followed by phase II conjugation could be a potential detoxification pathway of B[a]P-7,8-dione.
Materials and Methods
Caution
All PAHs are potentially hazardous and should be handled in accordance with the NIH Guidelines for the Laboratory Use of Chemical Carcinogens.
Chemicals and Reagents
Cell culture medium and reagents were all obtained from Invitrogen Co. (Carlsbad, CA) except fetal bovine serum (FBS) which was purchased from Hyclone (Logan, UT). 1,3-[3H2]-B[a]P-7,8-dione (specific activity of 650 mCi/mmol, ≥ 95% pure by HPLC) was purchased from NCI Radiochemical Repository. B[a]P-7,8-dione was synthesized according to published methods.34 All other chemicals used were of the highest grade available and all solvents were HPLC grade.
Cell Culture
A549 (human lung adenocarcinoma cells) were obtained from American Type Culture Collection (ATCC #CCL-185) and maintained in F-12K nutrient mixture (Kaighn’s modification) with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin. H358 (human bronchoalveolar cells) were obtained from American Type Culture Collection (ATCC #CRL-5807) and maintained in RPMI 1640 nutrient mixture with 10% heat-inactivated FBS, 2 mM L-glutamine, 100 units/mL penicillin, and 100 µg/mL streptomycin. HBEC-KT (immortalized human bronchial epithelial cells) originated from a patient without lung cancer were a gift from Dr. John Minna at University of Texas Southwestern Medical Center and maintained in keratinocyte serum free medium with 0.1–0.2 ng/mL recombinant epidermal growth factor (rEGF), 20–30 µg/mL bovine pituitary extract (BPE) and 2 mM L-glutamine. Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2 and were passaged every 3 days at a 1:6 dilution. Cultured cells with a passage number of 10–20 were used in the experiments to reduce variability due to cell culture conditions.
Distribution of 1,3-[3H2]-B[a]P-7,8-dione in Human Lung Cells
The cells (~5 × 106) were cultured in 6-well plates. After the culture media was aspirated, the cells were washed twice and treated with 1,3-[3H2]-B[a]P-7,8-dione (2 µM, 1 × 105 cpm/nmol, 0.2 % DMSO) in HBSS buffer containing 1 mM sodium pyruvate as an energy source. The culture media and the cells were collected separately at 0, 3, 6, 12, 24 h. At each time point, media was removed and then extracted with 2 × 1.5-fold volume cold H2O-saturated ethyl acetate, with and without prior acidification using 0.1% formic acid. Aliquots from both aqueous and organic phases were counted by liquid scintillation counting. The cells left inside each well were scraped into H2O, homogenized with a glass cell homogenizer. Cell lysates were subsequently spun down to obtain the supernatant and the pellet. The supernatant from cell lysates was extracted twice and both aqueous and organic phases were counted as described above. The pellet from cell lysates was dissolved in Radio-Immunoprecipitation Assay buffer and then counted by liquid scintillation counting. All counts were obtained on a Packard Tri-Carb 2100TR liquid scintillation counter with a machine efficiency of 63% for tritium and radioactivity was reported as corrected cpm.
Analysis of 1,3-[3H2]-B[a]P-7,8-dione Metabolites in Human Lung Cells by HPLC-UV-RAM
The cells (~5 × 106) were treated with 1,3-[3H2]-B[a]P-7,8-dione (2 µM, 1 × 105 cpm/nmol, 0.2 % DMSO) in the same manner as described above. The culture media were collected at 0 h and 24 h respectively, and subsequently acidified with 0.1% formic acid before extraction with 2 × 1.5-fold volume of cold ethyl acetate saturated with H2O. The organic phases from the extracted culture media were combined and dried under vacuum. The residue was redissolved in 80 µL of methanol. A 60 µL aliquot was analyzed on a tandem Waters Alliance 2695 chromatographic system (Waters Corporation, Milford, MA) with a Waters 996 photodiode array (PDA) detector and a β-RAM inline radioactive detector (IN/US Systems Inc., Tampa, FL). Separations were accomplished on a reverse-phase (RP) column (Zorbax-ODS C18, 5 µm, 4.6 mm ×250 mm) (DuPont Co., Wilmington, DE) with a guard column at ambient temperature. The mobile phase consisted of 5 mM ammonium acetate and 0.1% trifluoroacetic acid (TFA) (v/v) in H2O (solvent A) and 5 mM ammonium acetate and 0.1% TFA in acetonitrile (solvent B) and was delivered at a flow rate of 0.5 mL/min. The linear gradient elution program was as follows: 50 % to 65 % B over 30 min, 65 % to 95 % B over 10 min, followed by an isocratic hold at 95 % B for another 10 min. At 50 min, B was returned to 50 % in 1 min and the column was equilibrated for 9 min before the next injection. The total run time for each analysis was 60 min. Eluants from the column were introduced separately into the PDA detector and the radiometric detector following mixture of the scintillant with the HPLC effluent (IN/US Systems Inc.) at a flow rate 1.5 mL/min. The IN/US detector was calibrated by injecting know amounts of B[a]P determined by scintillation counting directly into the detector to determine counting efficiency (59.5%). The counting efficiency of the detector was unaffected by the composition of the mobile phase. Radioactivity was reported as corrected cpm.
Synthesis of GSH-B[a]P-7,8-dione, NAC-B[a]P-7,8-dione and N7-Adenine-B[a]P-7,8-dione Depurinating Adduct
The GSH-B[a]P-7,8-dione conjugate was synthesized in a reaction system (1 mL) containing potassium phosphate buffer (10 mM, pH 7.0), 800 µM GSH, and 20 µM B[a]P-7,8-dione in 2% (v/v) DMSO and was sonicated for 20 min followed by incubation in a shaking water bath at 25 °C for 1 h. The NAC-B[a]P-7,8-dione conjugate was synthesized in a reaction system (1 mL) containing potassium phosphate buffer (10 mM, pH 7.0), 800 µM NAC, and 20 µM B[a]P-7,8-dione in 2% (v/v) DMSO and was incubated in a shaking water bath at 25 °C for 2 h. Reaction mixtures were acidified with 0.1% formic acid before extraction with 2 × 2 vols of cold ethyl acetate saturated with H2O. The organic phases were combined and dried under vacuum. The residues were redissolved in methanol and were subjected to analysis using HPLC-UV and LC-MS/MS. The identity of the GSH-B[a]P-7,8-dione and NAC-B[a]P-7,8-dione conjugates were validated using Q1 full scan and product ion scan modes. The N7-adenine-B[a]P-7,8-dione depurinating adduct was synthesized by heating B[a]P-7,8-dione with adenine in DMSO as previously described.36 The identity of the N7-adenine-B[a]P-7,8-dione depurinating adduct was validated using product ion scan and subsequent MS/MS/MS (MS3) modes.
Identification of B[a]P-7,8-dione Metabolites in Human Lung Cells by LC-MS/MS
The cells (~5 × 106) were treated with non-radiolabeled B[a]P-7,8-dione (2 µM, 0.2 % DMSO) in the same manner as described above. The culture media were collected at 0 h and 24 h respectively, and subsequently acidified with 0.1% formic acid before extraction with 2 × 1.5-fold volume of cold ethyl acetate saturated with H2O. The organic phases of culture media were combined and dried under vacuum. The residue was redissolved in 100 µL of methanol. A 20 µL aliquot was analyzed on a Waters Alliance 2690 HPLC system (Waters Corporation) coupled to a Finnigan TSQ Quantum Ultra spectrometer (Thermo Fisher Scientific, San Jose, CA). Separations were accomplished on a reversed-phase (RP) column (Zorbax-ODS C18, 5 µm, 4.6 mm ×250 mm) (DuPont Co., Wilmington, DE) with a guard cartridge at ambient temperature. The mobile phase, flow rate and the linear gradient elution program were the same as described in the analysis by HPLC-UV-RAM. During LC-MS/MS analysis, up to 5 min of the initial flow was diverted away from the mass spectrometer before evaluation of metabolites. The mass spectrometer was operated in the positive ion mode with a heated electrospray ionization (HESI) source. Eluants were monitored with the TSQ using selected reaction monitoring (SRM) and product ion scan modes. The mass spectrometry parameters, including spray voltage (4.5 kV), vaporizer temperature (250 °C), sheath gas pressure (35 arbitrary units), auxiliary gas pressure (10 arbitrary units), capillary temperature (350 °C), source CID (0 V), collision gas (1.0 mTorr), scan time (0.1 s), and scan width (m/z 0.002), were automatically optimized with a B[a]P-7,8-dione standard solution in methanol. A list of SRM transitions for the metabolites in Table 1 was used to cover the most common phase I and phase II biotransformation reactions of B[a]P-7,8-dione. The Q1, Q3, and collision energy (CE) values were based on the phase I and phase II predictions of B[a]P-7,8-dione biotransformation as well as the MS/MS spectra of synthetic GSH-B[a]P-7,8-dione and NAC-B[a]P-7,8-dione standards. The corresponding mass spectrum of each metabolite was acquired from a product ion scan and subsequently compared with that of B[a]P-7,8-dione to investigate the fragmentation patterns and obtain information on the metabolite structures. Identification of the metabolite peaks was also achieved by comparing chromatographic retention times and UV spectra of the metabolites with those obtained from HPLC-UV-RAM. The Xcalibur software version 2.0 (Thermo Fisher Scientific) was used to control the LC-MS/MS system and to process data.
Table 1.
SRM transition parameters for cellular metabolites of B[a]P-7,8-dione.
| Potential metabolites | Mass gain (amu) |
Parent ion Q1 (amu) |
Daughter ions Q3 (amu) |
CE (eV) |
|---|---|---|---|---|
| O-sulfate-catechol | 80 | 365 | 285 | 20 |
| O-glucuronide-catechol | 176 | 461 | 285 | 20 |
| Hydroxylated-B[a]P-7,8-dione | 16 | 299 | 202, 230, 283 | 30 |
| GSH-B[a]P-7,8-dione | 305 | 588 | 145, 283, 459 | 20 |
| NAC-B[a]P-7,8-dione | 161 | 444 | 130, 283, 315 | 15 |
| Cys-Gly-B[a]P-7,8-dione | 176 | 459 | 283, 385, 402 | 20 |
| Cys-B[a]P-7,8-dione | 119 | 402 | 315, 357, 283 | 20 |
| GSH-catechol | 305 | 590 | 145, 285 | 20 |
| NAC-catechol | 161 | 446 | 130, 285 | 20 |
| Cys-catechol | 119 | 404 | 285, 317 | 20 |
| Gly-catechol | 57 | 342 | 285 | 20 |
| O-methylated-catechol | 14 | 299 | 239, 267, 285 | 15 |
| Acetylated-catechol | 42 | 327 | 267, 285 | 20 |
| GSH-methylated-catechol | 319 | 604 | 145, 285, 475, 572 | 20 |
| NAC-methylated-catechol | 175 | 460 | 130, 285, 331, 428 | 20 |
| Cys-methylated-catechol | 133 | 418 | 285, 331, 386 | 20 |
| GSH-acetylated-catechol | 347 | 632 | 145, 267, 285, 503 | 20 |
| NAC-acetylated-catechol | 203 | 488 | 130, 285, 359, 428 | 20 |
| Cys-acetylated-catechol | 161 | 446 | 285, 359, 386 | 20 |
| GSH-glucuronide-catechol | 481 | 766 | 145, 285, 590, 637 | 20 |
| GSH-sulfate-catechol | 385 | 670 | 145, 285, 541, 590 | 20 |
| NAC-glucuronide-catechol | 337 | 622 | 130, 285, 446, 493 | 20 |
| NAC-sulfate-catechol | 241 | 526 | 130, 285, 397, 446 | 20 |
| Cys-glucuronide-catechol | 295 | 580 | 285, 404, 493 | 20 |
| Cys-sulfate-catechol | 199 | 484 | 285, 397, 404 | 20 |
| GSH-glycine-catechol | 362 | 647 | 145, 285, 590 | 20 |
| NAC-glycine-catechol | 218 | 503 | 130, 285, 446 | 20 |
| Cys-Gly-catechol | 176 | 461 | 285, 374, 404 | 20 |
The samples extracted from culture media were also analyzed on a Waters Alliance 2690 HPLC system (Waters Corporation) coupled to a Finnigan LTQ spectrometer (Thermo Fisher Scientific, San Jose, CA). The column, mobile phase, flow rate and the linear gradient elution program were the same as described above. The mass spectrometer was operated in the positive ion mode with an electrospray ionization (ESI) source. Eluants were monitored with LTQ using product ion scan and subsequent MS/MS/MS (MS3) modes. The mass spectrometry parameters, including spray voltage (4 kV), sheath gas flow rate (35 arbitrary units), auxiliary gas flow rate (18 arbitrary units), capillary temperature (220 °C), capillary voltage (9 V), and tube lens (80 V), were automatically optimized with a B[a]P-7,8-dione standard solution in methanol. The Xcalibur software version 2.0 (Thermo Fisher Scientific) was used to control the LC-MS/MS system and to process data.
Results
Strategy
A549, H358 and HBEC-KT cells were treated with radiolabeled and non-radiolabeled B[a]P-7,8-dione. Radiotracer metabolism studies were performed to determine mass-balance and distribution by liquid scintillation counting, subsequently the radioactivity of interest was analyzed by HPLC-UV-RAM to obtain the retention times and UV spectra of potential radioactive metabolites. The UV spectra can be used to determine whether metabolites have the characteristic chromophore of either a catechol or o-quinone. Metabolism studies were then replicated with non-radiolabeled B[a]P-7,8-dione to identify the structures of potential metabolites by LC-MS/MS, and to validate the structures by comparison to authentic synthesized standards. As GSH is the most abundant small molecule cellular nucleophile with concentrations in the mM range, the GSH-B[a]P-7,8-dione conjugate was predicted to be a major cellular metabolite which could be further metabolized into the NAC-B[a]P-7,8-dione conjugate. Thus, both conjugates were synthesized as authentic standards for structure validation. Also, since B[a]P-7,8-dione has the potential to form depurinating DNA adducts, the N7-adenine depurinating adduct was also synthesized as a standard.
Distribution of 1,3-[3H2]-B[a]P-7,8-dione in Human Lung Cells
After treatment of A549, H358 and HBEC-KT cells with 1,3-[3H2]-B[a]P-7,8-dione, it was found that the distribution profiles of radioactivity were similar (Figure S-1). Radioactivity was mainly found in the organic and aqueous phase of the ethyl acetate-extracted media, as well as in the cell lysate pellets. Only a small amount of radioactivity was distributed in the organic and aqueous phase of the ethyl acetate-extracted cell lysate supernatants. Radioactivity in the organic phase of ethyl acetate-extracted media was markedly decreased within the first 3 h, suggesting that 1,3-[3H2]-B[a]P-7,8-dione was consumed rapidly. Radioactivity in the aqueous phase of the media increased concomitantly within the first 6 h, suggesting that 1,3-[3H2]-B[a]P-7,8-dione was converted to water soluble metabolites. Radioactivity retained in the cell lysate pellets indicated the formation of macromolecule adducts. Acidification of the cell culture media before extraction released most of the radioactivity from the aqueous phase into the organic phase, suggesting that the water soluble metabolites were charged at neutral pH but extractable with ethyl acetate at low pH.
Detection of 1,3-[3H2]-B[a]P-7,8-dione Metabolites in Human Lung Cells by HPLC-UV-RAM
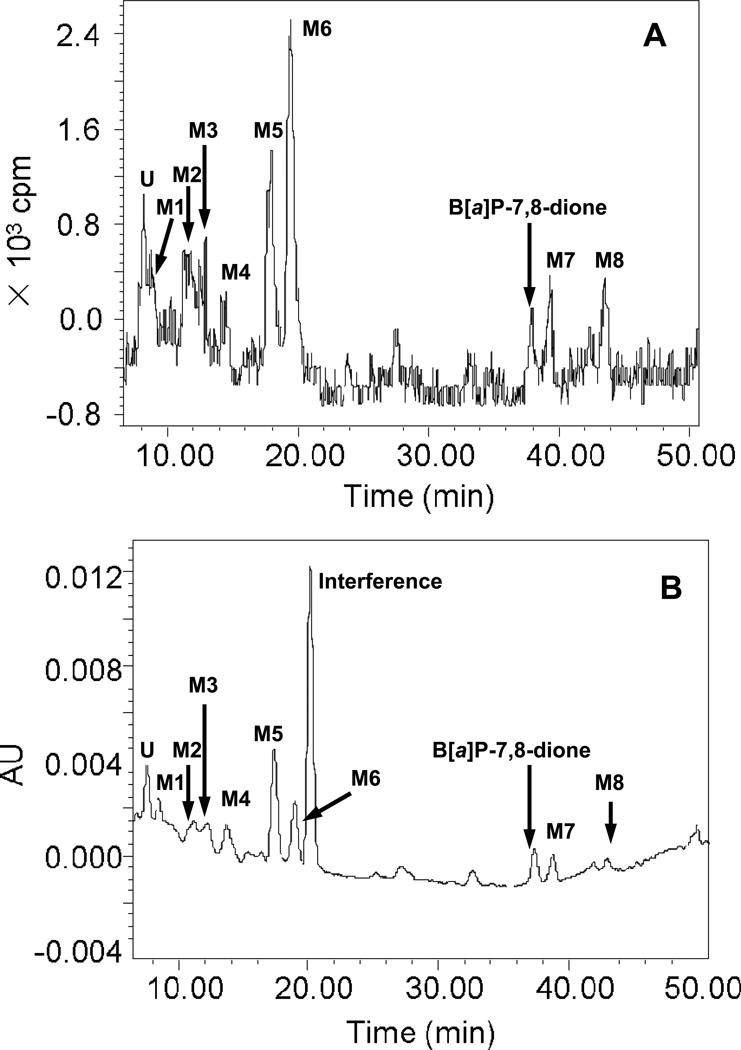
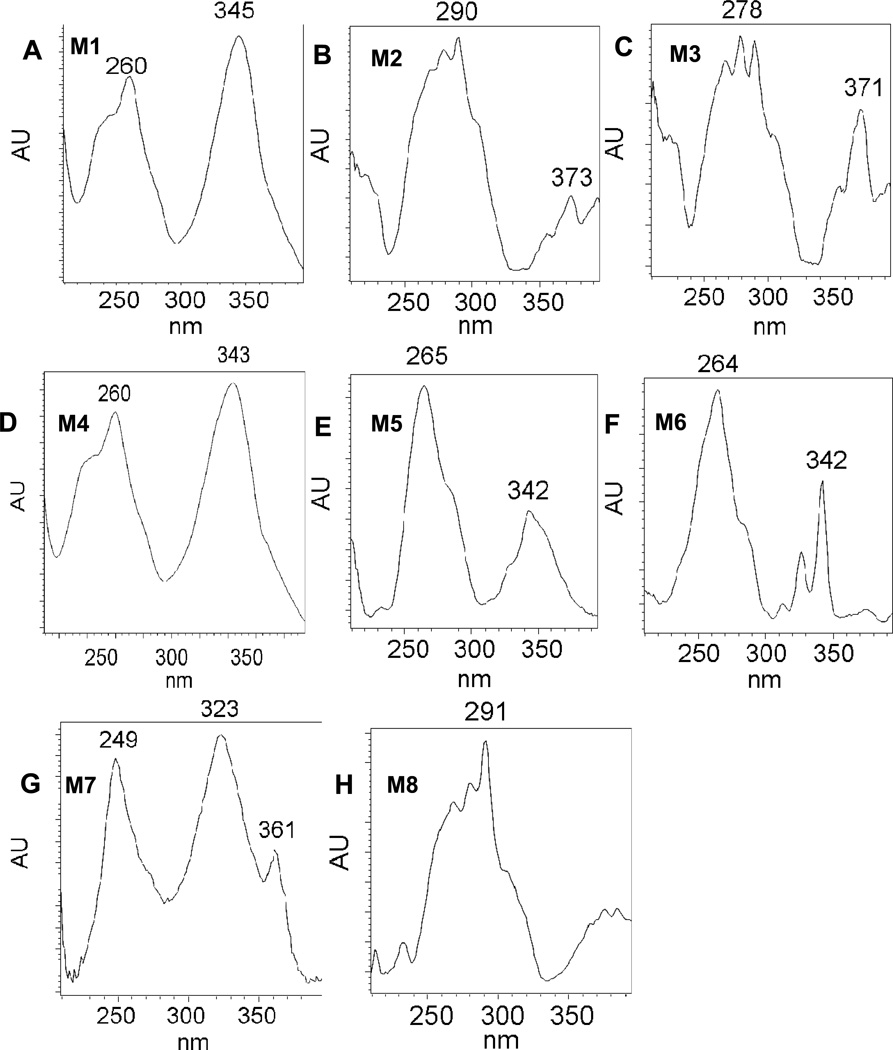
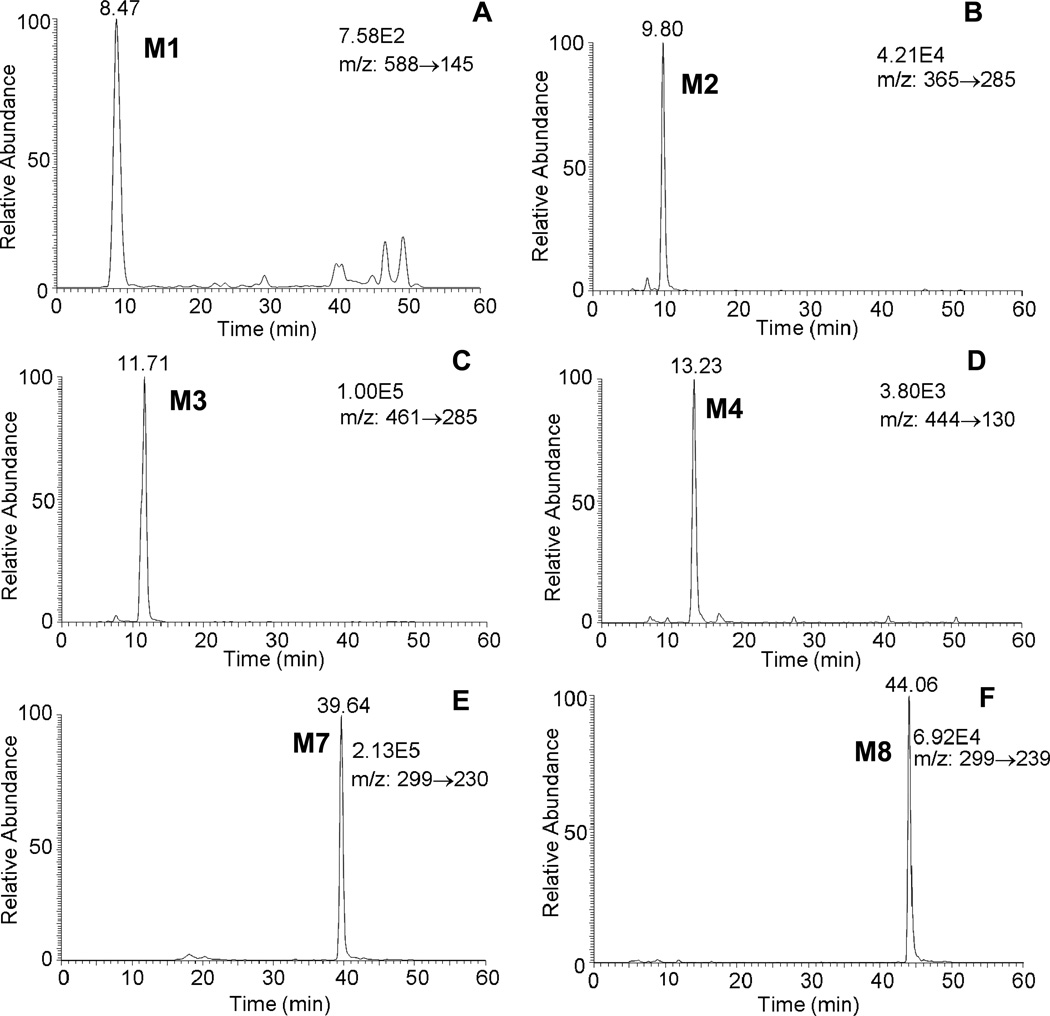
Comparison of chromatographic profiles of the 0 h and 24 h showed that several metabolites of 1,3-[3H2]-B[a]P-7,8-dione were detected in the organic phase of the ethyl acetate-extracted media by HPLC-UV-RAM after acidification of the media from three human lung cells. The metabolite profile from organic phase of the extracted media in A549 cells at 24 h was the most comprehensive among the three cells studied and the corresponding HPLC radio-chromatogram and UV chromatogramsare shown in Figure 1. The peak with retention time of 37.97 min was attributed to 1,3-[3H2]-B[a]P-7,8-dione, but was barely detectable at 24 h and had much lower intensity than the peak at 0 h, suggesting that [1,3-3H]-B[a]P-7,8-dione was rapidly metabolized by A549 cells over this time course. The HPLC radiochromatogram (Figure 1A) showed that there were eight radioactive peaks with retention times of 8.50 min (M1), 11.25 min (M2), 12.91 min (M3), 14.13 min (M4), 17.98 min (M5), 19.47 min (M6), 39.31 min (M7) and 43.47 min (M8) respectively, indicating that these peaks were derived from 1,3-[3H2]-B[a]P-7,8-dione. The peak areas showed that M5 and M6 were two major metabolites of 1,3-[3H2]-B[a]P-7,8-dione. The corresponding UV spectra of these eight radioactive metabolites were extracted from the UV chromatogram (Figure 1B) and are shown in Figure 2. These spectra indicated that M2, M3 and M8 were catechol metabolites and M1, M4, M5, M6 and M7 were quinone metabolites based upon their chromophores. Another radioactive peak with retention times of 8.10 min (U) eluted first was a minor polar metabolite. One non-radioactive peak with a retention time of 20.50 min appeared in the UV chromatogram and was found to be an interference peak from the cell culture media. Similar HPLC-UV-RAM results from organic phase of media were obtained from H358 and HBCE-KT cells and are provided in the supporting information. M1, M2, M4, M5, M6, M7 and M8 were detected in H358 cells whereas only M2, M4, M5, M6, M7 and M8 were detected in HBCE-KT cells. The distribution of 1,3-[3H2]-B[a]P-7,8-dione in three human lung cells at 24 h is summarized in Table 2.
Figure 1.
HPLC radio-chromatogram (A) and UV chromatogram (348 nm) (B) of extracts from organic phase of media in A549 cells at 24 h. A549 cells (~5 × 106) were treated with 1,3-[3H2]-B[a]P-7,8-dione (2 µM, 1 × 105 cpm/nmol, 0.2 % (v/v) DMSO) in HBSS buffer containing 1 mM sodium pyruvate. The culture media were collected at 24 h, and subsequently acidified with 0.1% formic acid before extraction with ethyl acetate. The extracts were analyzed on a HPLC-UV-RAM.
Figure 2.
UV spectra of metabolites from organic phase of media in A549 cells at 24h. (A): M1, GSH-B[a]P-7,8-dione; (B): M2, O-mono-sulfate-catechol; (C): M3, O-mono-glucuronide-catechol; (D): M4, NAC-B[a]P-7,8-dione; (E): M5, unknown metabolite with m/z 444; (F): M6, adenine-B[a]P-7,8-dione adduct; (G): M7, Mono-hydroxylated-B[a]P-7,8-dione; (H): M8, O8-mono-methylated-catechol.
Table 2.
Distribution of 1,3-[3H2]-B[a]P-7,8-dione in three human lung cells at 24h.
| A549 | H358 | HBEC-KT | |
|---|---|---|---|
| Aqueous phase of medium | 14.7 % | 29.6 % | 15.0 % |
| Organic phase of medium | 15.6 % | 19.5 % | 13.9 % |
| Aqueous phase of cell lysate supernatant | 0.8 % | 1.0 % | 1.7 % |
| Organic phase of cell lysate supernatant | 1.9 % | 5.1 % | 2.8 % |
| Cell lysate pellet | 11.7 % | 21.7 % | 25.9 % |
| Total recovery | 44.7 % | 76.9 % | 59.3 % |
100% is the amount that could be recovered from the combined isolates at zero time for each cell line.
Synthesis of GSH-B[a]P-7,8-dione
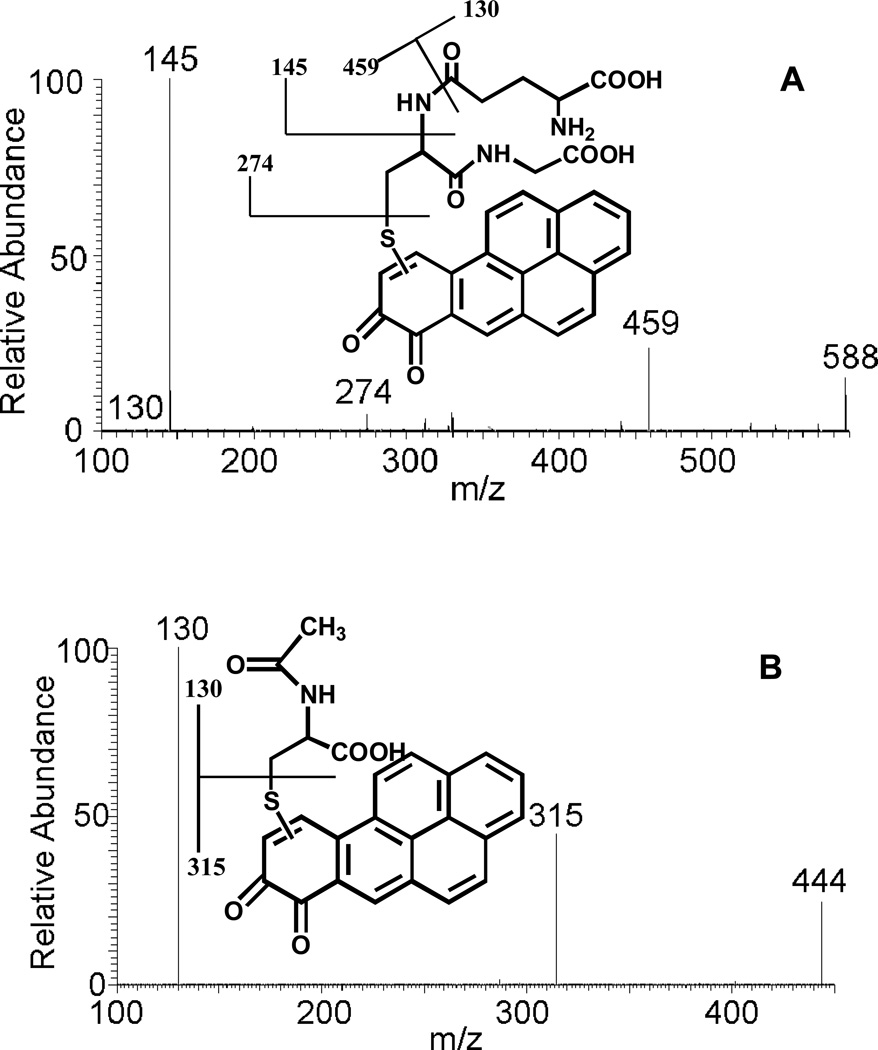
The conjugation of purple B[a]P-7,8-dione with GSH generated a yellow solid after extraction and solvent evaporation. Subsequent HPLC-UV analysis showed that B[a]P-7,8-dione was essentially completely consumed and a significant peak was observed which eluted earlier than B[a]P-7,8-dione, with the same retention time of M1 in Figure 1. This metabolite showed a protonated molecule (MH+) at m/z 588 under Q1 full scan positive mode, corresponding to the molecular weight of a protonated GSH-B[a]P-7,8-dione conjugate. The fragmentation pattern of this metabolite obtained from the product ion scan confirmed the structure of the GSH-B[a]P-7,8-dione conjugate. The proposed fragmentation scheme and product ions of the protonated GSH-B[a]P-7,8-dione conjugate (m/z 588) are shown in Figure 3 (A). The specific position of GSH conjugation could not be ascertained based on MS data only. According to the two-dimensional proton nuclear magnetic resonance (NMR) of the synthetic GSH-B[a]P-7,8-dione conjugate reported previously,23 the position of GSH conjugation is C10.
Figure 3.
MS/MS spectra of the protonated GSH-B[a]P-7,8-dione conjugate (m/z 588) (A) and the protonated NAC-B[a]P-7,8-dione conjugate (m/z 444) (B) prepared synthetically as well as the proposed origin of key product ions.
Synthesis of NAC-B[a]P-7,8-dione
The conjugation of purple B[a]P-7,8-dione with NAC generated a light yellow solid after extraction and solvent evaporation. Subsequent HPLC-UV analysis showed that B[a]P-7,8-dione disappeared and a significant peak was observed which eluted earlier than B[a]P-7,8-dione, with the same retention time of M4 in Figure 1. This metabolite showed a MH+ at m/z 444 under Q1 full scan positive mode, corresponding to the molecular weight of a protonated NAC-B[a]P-7,8-dione conjugate. The fragmentation pattern of this metabolite obtained from the product ion scan confirmed the structure of the NAC-B[a]P-7,8-dione conjugate. The proposed fragmentation scheme and product ions of the protonated NAC-B[a]P-7,8-dione conjugate (m/z 444) are shown in Figure 3 (B). The specific position of NAC conjugation could not be ascertained based on mass spectrometry data only. According to the two-dimensional proton nuclear magnetic resonance (NMR) of the synthetic NAC-B[a]P-7,8-dione reported previously, 23 the position of NAC conjugation is C10.
Synthesis of the N7-Adenine-B[a]P-7,8-dione Adduct
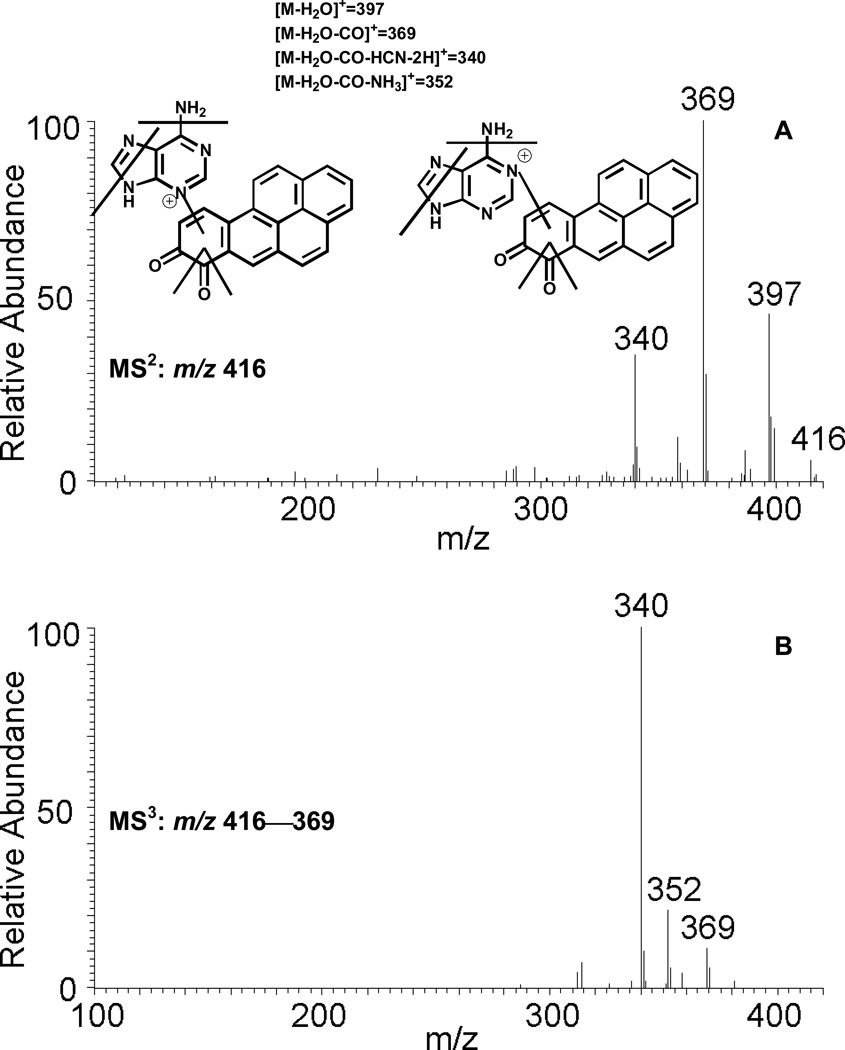
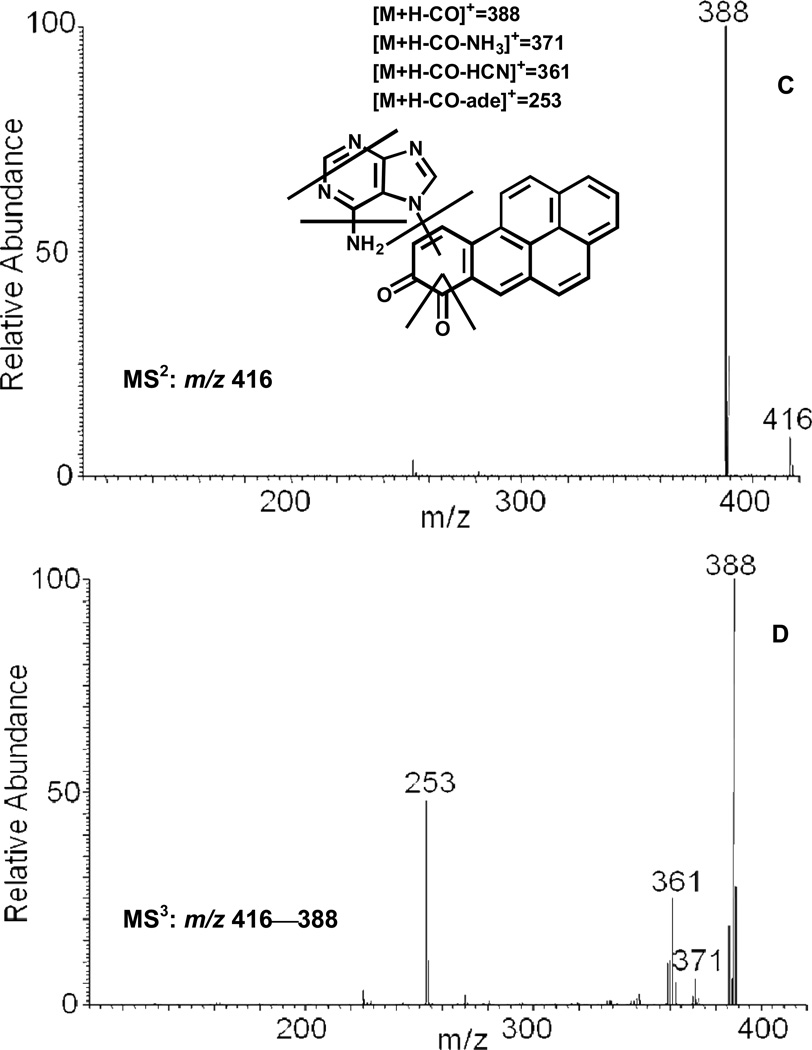
The reaction of purple B[a]P-7,8-dione with adenine generated a light yellow solid after extraction and solvent evaporation. Subsequent HPLC-UV analysis showed that B[a]P-7,8-dione was essentially completely consumed and a significant peak was observed which eluted earlier than B[a]P-7,8-dione. This peak showed a protonated molecule (MH+) at m/z 416 corresponding to the molecular weight of a protonated adenine-B[a]P-7,8-dione adduct. The fragmentation pattern obtained from the MS2 and MS3 spectra confirmed the structure of the adenine-B[a]P-7,8-dione adduct. The proposed fragmentation scheme and product ions of the protonated adenine-B[a]P-7,8-dione adduct (m/z 416) are shown in Figure 6 (C) and (D). The specific position of adenine addition could not be ascertained based on MS data only. According to the NMR of the synthetic adenine-B[a]P-7,8-dione adduct reported previously, 36 the position of adenine addition is either N7 or N9.
Figure 6.
MS2 (A) and MS3 (B) spectra of the protonated adenine-B[a]P-7,8-dione adduct (m/z 416) and MS2 (C) and MS3 (D) spectra of the protonated N7-adenine-B[a]P-7,8-dione depurinating adduct (m/z 416) as well as origin of key product ions.
Identification of B[a]P-7,8-dione Metabolites in Human Lung Cells
Attempts were made to identify the cellular metabolites by Q1 full scan in combination with product ion scans on the TSQ but failed due to the limited sensitivity to detect the potential metabolites. The most sensitive scan type, selected reaction monitoring (SRM) based on Table 1, was used to search for B[a]P-7,8-dione metabolites in human lung cells. The extracted ion chromatograms of SRM transitions for metabolites from the acidified organic phase of media in A549 cells at 24h are shown in Figure 4. The results showed that significant metabolites in A549 cells were detected with six SRM transitions i.e. 588→145 for GSH-B[a]P-7,8-dione, 365→285 for O-mono-sulfate-catechol, 461→285 for O-mono-glucuronide-catechol, 444→130 for NAC-B[a]P-7,8-dione, 299→230 for hydroxylated-B[a]P-7,8-dione and 299→239 for O-methylated-catechol, respectively. These metabolites were absent at 0 h. Comparison of the retention times of these metabolites with the corresponding metabolites on HPLC-UV-RAM showed an excellent concordance.
Figure 4.
Extracted ion chromatograms of SRM transitions for metabolites from the organic phase of media in A549 cells at 24h. (A): GSH-B[a]P-7,8-dione; (B): O-mono-sulfate-catechol; (C): O-mono-glucuronide-catechol; (D): NAC-B[a]P-7,8-dione; (E): mono-hydroxylated-B[a]P-7,8-dione; (F): O8-mono-methylated-catechol.
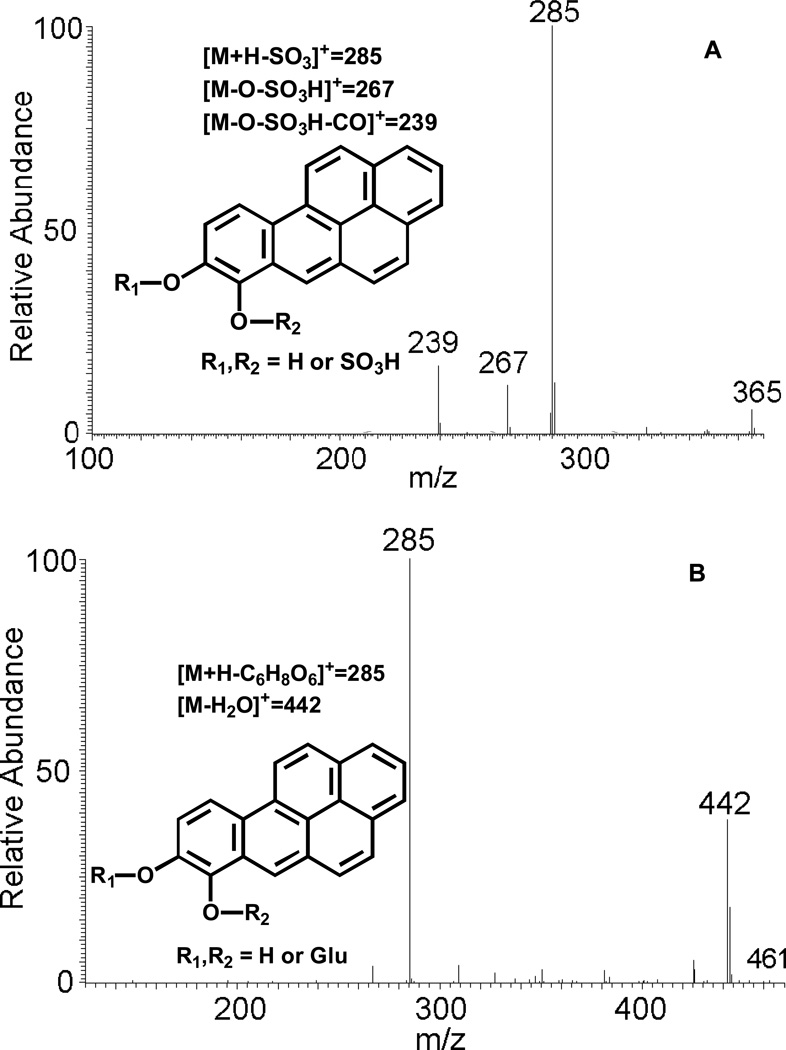
SRM transitions 365→285 and 461→285 showed an increase of 80 amu and 176 amu over that observed for the catechol (m/z 285), strongly indicating the occurrence of O-sulfation and O-glucuronidation of the catechol. In addition, MS/MS spectra of the O-mono-sulfate-catechol and O-mono-glucuronide-catechol were obtained successfully. The proposed fragmentation scheme and product ions of protonated O-sulfate-catechol (m/z 365) and O-glucuronide-catechol (m/z 461) are shown in Figure 5 (A) and (B), respectively. However, the specific position of the sulfate group or the glucuronide group could not be assigned based on mass spectrometry data only.
Figure 5.
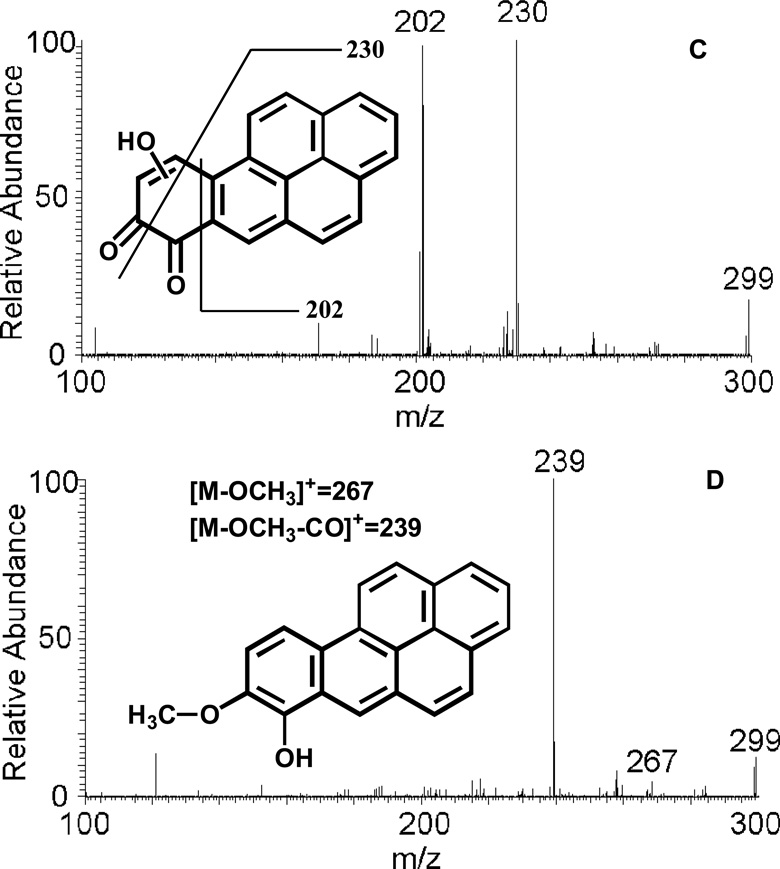
MS/MS spectra of protonated O-mono-sulfate-catechol (m/z 365) (A), protonated O-mono-glucuronide-catechol (m/z 461) (B), protonated mono-hydroxylated-B[a]P-7,8-dione (m/z 299) (C) and protonated O8-mono-methylated-catechol (m/z 299) (D) as well as origin of key product ions.
MS/MS spectra of hydroxylated-B[a]P-7,8-dione and O-methylated-catechol were successfully obtained as well. The proposed fragmentation scheme and product ions of protonated hydroxylated-B[a]P-7,8-dione (m/z 299) and the O-methylated-catechol (m/z 299) were shown in Figure 5 (C) and (D), respectively. The specific position of hydroxyl group or methyl group could not be assigned based on mass spectrometry data only.
The results of product ion scans on LTQ at 0 h and 24 h in A549 cells showed that two metabolites with m/z 444 and 416 respectively were potential metabolites of B[a]P-7,8-dione. Comparison of the retention times of these two metabolites with M5 and M6 on HPLC-UV-RAM in Figure 1 showed an excellent concordance as well. Subsequent MS3 spectra of these two metabolites were successfully obtained. The metabolite with m/z 444 showed the same molecular weight as NAC-B[a]P-7,8-dione. However, MS2 and MS3 spectra of this metabolite did not show any characteristic fragmentation from NAC (data not shown) and thus its structure remains unassigned.
Identification of the Adenine-B[a]P-7,8-dione Adduct
An unknown metabolite with m/z 416 gave a protonated MH+ predicted for the adenine-B[a]P-7,8-dione adduct. MS2 and MS3 spectra of this metabolite (Figure 6) showed reasonable fragmentation patterns from both adenine and B[a]P-7,8-dione. In particular, the loss of NH3 (17 amu) and HCN (27 amu) are characteristic of the fragmentation of the adenine ring. 35 The loss of NH3 (17 amu) shown in the MS3 spectrum strongly indicates that the covalent binding did not occur at the N6 position of adenine. 35 The covalent binding at N7 position of adenine could also be ruled out since the spectra of the synthetic N7-adenine adduct differed from the cellular adenine adduct. 36 Thus addition via N1 or N3-adenine adduct most likely accounts for the formation of this adduct. Although the specific position of N1 or N3 attack to B[a]P-7,8-dione could not be ascertained based on mass spectrometry data only, previous studies on the N7 guanine adducts of phenanthrene-1,2-dione and B[a]P-7,8-dione suggests addition at C10. 25
Quantitation of B[a]P-7,8-dione Metabolites in Human Lung Cells
By correlating the recovery of 1,3-[3H2]-B[a]P-7,8-dione under the HPLC peaks for each metabolite with those identified by LC-MS/MS, it was possible to quantify the percentage of B[a]P-7,8-dione that was converted to each of the metabolites (Table 3). It was found that the major metabolites formed varied by cell-line. In A549 cells, the major metabolite was a O-mono-sulfated catechol followed by mono-hydroxylated B[a]P-7,8-dione. In H358 cells, the major metabolite was again the O-mono-sulfated catechol, but the mono-hydroxylated B[a]P-7,8-dione was formed in small amounts. In HBEC-KT cells, the major metabolites were the NAC-B[a]P-7,8-dione conjugate followed by mono-hydroxylated B[a]P-7,8-dione. The O-mono-glucuronide-catechol was only detected in A549 cells. Also, the GSH-B[a]P-7,8-dione conjugate was one of the least abundant metabolites detected and was not present in HBEC-KT cells. In all cell lines, no metabolite comprised more than 15% of the total accounted for. The compound of most abundance in the cell culture media was the adenine adduct.
Table 3.
Relative amounts of 1,3-[3H2]-B[a]P-7,8-dione metabolites and adduct in three human lung cells at 24h.
| Metabolites and adduct | Metabolite | A549 | H358 | HBEC-KT |
|---|---|---|---|---|
| GSH-B[a]P-7,8-dione | M1 | 0.7 % | 0.8 % | — |
| O-mono-sulfate-catechol | M2 | 12.2 % | 14.9 % | 3.5 % |
| O-mono-glucuronide-catechol | M3 | 4.3 % | — | — |
| NAC-B[a]P-7,8-dione | M4 | 1.4 % | 2.4 % | 11.1 % |
| Unknown metabolite with m/z 444 | M5 | 16.2 % | 25.3 % | 6.4 % |
| adenine-B[a]P-7,8-dione | M6 | 30.1 % | 25.7 % | 48.4 % |
| B[a]P-7,8-dione | — | 5.7 % | 4.2 % | 2.1 % |
| Mono-hydroxylated-B[a]P-7,8-dione | M7 | 7.8 % | 1.7 % | 10.1 % |
| O-methylated-catechol | M8 | 6.6 % | 1.2 % | 5.8 % |
| Other unknown metabolites | — | 15.0 % | 23.8 % | 12.6 % |
Discussion
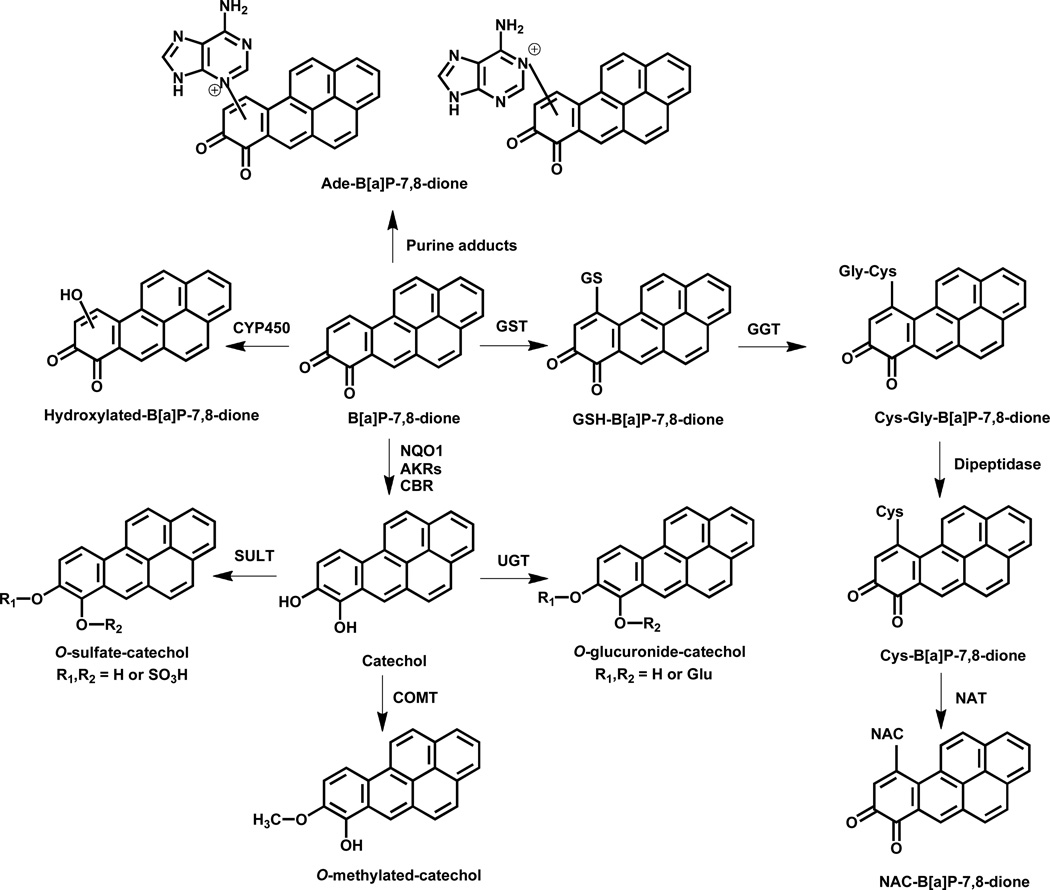
This study provides a comprehensive account of the metabolism of radiolabeled and unlabeled B[a]P-7,8-dione in three different human lung cell lines A549, H358 and HBEC-KT. B[a]P-7,8-dione is the signature metabolite of the AKR pathway of PAH activation but little is known about its downstream cellular metabolism. B[a]P-7,8-dione metabolites were detected from the culture media by HPLC-UV-RAM and by LC-MS/MS. The metabolite profile in A549 cells was the most comprehensive among the three human lung cells studied (Scheme 1). It is proposed that GSH-B[a]P-7,8-dione and NAC-B[a]P-7,8-dione conjugates are formed and that B[a]P-7,8-dione undergoes mono-hydroxylation. In addition, reduction of B[a]P-7,8-dione back to the catechol followed by O-methylation or O-mono-sulfation occurs. Reduction of B[a]P-7,8-dione to the catechol to form the O-mono-glucuronide was found to be an unique metabolic pathway for B[a]P-7,8-dione in A549 cells. We also obtained evidence for the formation of the adenine-B[a]P-7,8-dione adduct.
Scheme 1.
Proposed metabolic pathway of B[a]P-7,8-dione in three human lung cells. (Abbreviations: Ade, adenine; AKR, aldo-keto reductase; CBR, carbonyl reductase; COMT, catechol-O-methyltransferase; CYP450, cytochrome P450; Cys, cysteine; Cys-Gly, cysteinylglycine; GGT, γ–glutamyltranspeptidase; GST, glutathione S-transferase; GSH, glutathione; NAC, N-acetyl-L-cysteine; NAT, N-acetyl transferase; NQO1, NAD(P)H: quinone oxidoreductase 1; SULTs, sulfotransferases; UGTs, uridine 5'-diphospho-glucuronosyltransferases.)
Structural elucidation of the metabolites was conducted where possible by the use of reference standards. In vitro synthesis of the GSH-B[a]P-7,8-dione and NAC-B[a]P-7,8-dione conjugates was previously reported by our group.23 The structures of these two conjugates were identified by two-dimensional proton nuclear magnetic resonance (NMR) and the positions of GSH conjugation and NAC conjugation were found to be both at C10. The specific methylation position of O-methylated-catechol was also identified to be at O8 by comparison to an authentic metabolite produced by enzymatic synthesis and characterized by 1H-NMR.32 However, the specific positions at which phase I mono-hydroxylation of B[a]P-7,8-dione and phase II conjugation of the catechol occur (O-mono-sulfation and O-mono-glucuronidation) were not assignable by LC-MS/MS. NMR spectroscopy, preferably nuclear overhauser effect (NOE) experiments, would be needed to characterize the definite structures of these metabolites, but the approach is not feasible with the limited material isolated from cells and will have to wait for their enzymatic synthesis using recombinant SULTs and UGTs which is the subject of a separate study.
Reduction of B[a]P-7,8-dione to the catechol followed by formation of phase II conjugates such as O-mono-methylated-catechol, O-mono-glucuronide-catechol and O-mono-sulfate-catechol could represent detoxification pathways of B[a]P-7,8-dione due to loss of its electrophilicity and redox activity. In contrast, other metabolites such as GSH-B[a]P-7,8-dione, NAC-B[a]P-7,8-dione and mono-hydroxylated-B[a]P-7,8-dione have lost their electrophilicity but are still capable of redox-cycling to produce oxidative stress and oxidative DNA damage. Knowledge of the major metabolic pathways of B[a]P-7,8-dione in human lung cells can be used to identify biomarkers of the AKR pathway of PAH activation by measuring their presence in human urine and plasma.
The enzyme isoforms responsible for B[a]P-7,8-dione metabolism in A549, H358 and HBEC-KT cells remain to be completely identified. However, the presence of catechol conjugates suggests that enzymatic two electron reduction of the quinone to the catechol is occurring. In a recent study we provided evidence that of the two electron quinone reductases responsible for the conversion of B[a]P-7,8-dione to the catechol (NQO1, AKRs, and CBR1 and CBR3). AKRs likely play a dominant role in this reaction in A549 cells. Thus AKRs are not only responsible for the formation of B[a]P-7,8-dione but they are also responsible for the redox-cycling of this quinone.29 Formation of an O-mono-methylated-catechol indicates that catechol-O-methyltransferase (COMT) is present and is in agreement with our studies which show that human recombinant COMT can form the O-8-mono-methylated catechol of B[a]P-7,8-dione.30 The detection of this metabolite is supported by our earlier findings that oxidative stress and the formation of 8-oxo-dGuo is exacerbated in A549 cells treated with B[a]P-7,8-trans-dihydrodiol when a COMT inhibitor is present.30 Detection of mono-hydroxylated-B[a]P-7,8-dione indicates mono-oxygenation by cytochrome P450 in the three cell lines. H358 cells contain glutathione S-transferase (GSTs) and sulfotransferases (SULTs) but lack uridine 5'-diphospho-glucuronosyltransferases (UGTs), 37 which would explain why GSH-B[a]P-7,8-dione, NAC-B[a]P-7,8-dione and O-mono-sulfate-catechol are detected and while an O-mono-glucuronide-catechol was not present. The structure of the O-mono-sulfate-catechol was recently reported when human recombinant SULT1A3 was used to generate the conjugate and was found to be 8-hydroxy-B[a]P-7-mono-sulfate. 41
GSH conjugates can be further metabolized into cysteinylglycine (Cys-Gly) conjugates by γ–glutamyltranspeptidase (GGT), and then converted to cysteine (Cys) conjugates by dipeptidase, and eventually the NAC conjugates are formed by the action of N-acetyl transferase (NAT).38 Since NAC-B[a]P-7,8-dione was detected in all three cells, Cys-Gly-B[a]P-7,8-dione and Cys-B[a]P-7,8-dione were expected to be formed as well. These two intermediates were not detectable probably due to their trace amounts in the cells. In addition, the GSH-B[a]P-7,8-dione conjugate was only detected in A549 and H358 cells and not HBEC-KT cells, which could be accounted for by more extensive metabolism of GSH-B[a]P-7,8-dione in HBEC-KT cells.
Distribution studies of 1,3-[3H2]-B[a]P-7,8-dione in three cells (Figure S-1) also showed that a significant amount of radioactivity was distributed in the cell lysate pellets, indicating the formation of macromolecule adducts especially protein and DNA adducts. A subcellular protein fractionation kit was used to subsequently separate the radioactivity retained in the cell lysate pellets. The results showed that most of radioactivity was located in membrane protein and cytoskeletal protein (data not shown). A proteomics approach would be required to further investigate the protein adducts of B[a]P-7,8-dione in these cells but would require the use of “click”-chemistry by modification of B[a]P-7,8-dione with either an azide or alkynyl group. 39, 40
A unique feature of the present work was the detection of an adenine B[a]P-7,8-dione adduct in the cell culture media. It is not possible to determine the source of this adduct unequivocally. It could arise from two sources. First, nucleotide excision repair of a bulky stable B[a]P-7,8-dione would yield a B[a]P-7,8-dione-deoxyribose adduct. Acidification of the aqueous media followed by extraction would likely cleave the N-glycoside bond to yield an adenine adduct. Second, most cellular cofactors such as NAD, FAD, ATP, ADP, etc. contain a nucleophilic adenine which could react with electrophilic B[a]P-7,8-dione. The acidification during the sample preparation would again facilitate glycosidic bond cleavage and could lead to the formation of the cellular adenine adduct. Previously, stable bulky DNA adducts with B[a]P-7,8-dione have been observed in vitro,24, 26, 27 but there was a failure to detect these adducts in the A/J mouse lung model of B[a]P carcinogenesis, when animals were treated with B[a]P, B[a]P-7,8-trans-dihydrodiol or B[a]P-7,8-dione.42 By contrast, B[a]P and B[a]P-7,8-trans-dihydrodiol gave anti-B[a]PDE-DNA adducts. One explanation for this discrepancy is that the adenine-adduct observed in this study is not derived from DNA.
The conventional approach to detect metabolites is to employ full MS scan in combination with a product ion scan (MS/MS) but this was not successful due to the limited sensitivity to detect potential metabolites, which could result from the relatively low substrate concentration i.e. 2 µM and the high background signal from the cell media matrices. A triple quadrupole set to perform selected reaction monitoring (SRM) offers significantly higher sensitivity and selectivity and enables a wide range of potential transitions to be targeted as a result of the rapid cycle times, SRM screening could be an alternative approach for metabolite detection, utilizing metabolite prediction and knowledge of the MS/MS fragmentation of the potential metabolites. In our study, SRM led to considerable success in detecting metabolites from the cell culture media. However, this approach was unable to detect and identify unexpected and unusual metabolites as compared to the conventional full MS scan in combination with a product ion scan (MS/MS). This disadvantage was surmounted by addition of possible SRM transitions based on the knowledge of the predicted metabolites and their likely MS/MS fragmentation patterns.
In summary, we have conducted both radiolabeled and non-radiolabeled metabolism studies of B[a]P-7,8-dione in three human lung cells. The metabolic pathways of B[a]P-7,8-dione in human lung cells included the formation of GSH and NAC conjugates, reduction to the catechol followed by phase II conjugation, mono-hydroxylation as well as the formation of an adenine adduct. Among these metabolic pathways, reduction to the catechol followed by phase II conjugation could be a potential detoxification pathway of B[a]P-7,8-dione. We also report a novel adenine-B[a]P-7,8-dione adduct.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by NIH grant by 1R01-CA39504 and P30-ES013508 awarded to T.M.P and 1R01-CA130038 awarded to IAB. Mary E. Kushman was supported in part by a Cancer Pharmacology Training Grant R25-CA101871.
Abbreviations
- Ade
adenine
- AKR
aldo-keto reductase
- B[a]P
benzo[a]pyrene
- (+)-anti-B[a]PDE
(+)-7β,8α-dihydroxy-7,8,9,10-tetrahydro-9α,10α-oxo-benzo[a]pyrene
- dGuo
deoxyguanosine
- dAdo
deoxyadenosine
- (±)-B[a]P-7,8-trans-dihydrodiol
(−)-7β,8α-dihydroxy-7,8-dihydrobenzo[a]pyrene
- B[a]P-7,8-dione
benzo[a]pyrene-7,8-dione
- CBR
carbonyl reductase
- COMT
catechol-O-methyltransferase
- P450
cytochrome P450
- Cys
cysteine
- Cys-Gly
cysteinylglycine
- ESI
electrospray ionization
- FBS
fetal bovine serum
- GGT
γ–glutamyltranspeptidase
- GST
glutathione S-transferase
- GSH
glutathione
- HBSS
Hank’s balanced salt solution
- HPLC-UV-RAM
high performance liquid chromatography-ultraviolet-radioactivity monitoring
- HESI
heated electrospray ionization
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- MH+
protonated molecular ion
- NAC
N-acetyl-L-cysteine
- NAT
N-acetyl transferase
- NQO1
NAD(P)H: quinone oxidoreductase 1
- PAH
polycyclic aromatic hydrocarbon
- QR
quinone reductase
- ROS
reactive oxygen species
- SULTs
sulfotransferases
- UGTs
uridine 5'-diphospho-glucuronosyltransferases
Footnotes
Supporting Information Available
Distribution of 1,3-[3H2]-B[a]P-7,8-dione radioactivity in three human lung cell lines (Figure S-1).
HPLC radio-chromatograms and UV chromatograms (348 nm) of extracts from organic phase of ethyl acetate-extracted media in H358 cells (Figure S-2) and HBEC-KT cells (Figure S-3).
References
- 1.Grimmer G, Bohnke H. Polycyclic aromatic hydrocarbon profile analysis of high-protein foods, oils, and fats by gas chromatography. J Assoc Off Anal Chem. 1975;58:725–733. [PubMed] [Google Scholar]
- 2.Dipple A. Formation, metabolism, and mechanism of action of polycyclic aromatic hydrocarbons. Cancer Res. 1983;43:2422s–2425s. [PubMed] [Google Scholar]
- 3.Rothman N, Poirier MC, Baser ME, Hansen JA, Gentile C, Bowman ED, Strickland PT. Formation of polycyclic aromatic hydrocarbon-DNA adducts in peripheral white blood cells during consumption of charcoal-broiled beef. Carcinogenesis. 1990;11:1241–1243. doi: 10.1093/carcin/11.7.1241. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 6.Cavalieri EL, Rogan EG. Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25:677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- 7.Devanesan PD, RamaKrishna NV, Todorovic R, Rogan EG, Cavalieri EL, Jeong H, Jankowiak R, Small GJ. Identification and quantitation of benzo[a]pyrene-DNA adducts formed by rat liver microsomes in vitro. Chem Res Toxicol. 1992;5:302–309. doi: 10.1021/tx00026a024. [DOI] [PubMed] [Google Scholar]
- 8.Devanesan PD, Higginbotham S, Ariese F, Jankowiak R, Suh M, Small GJ, Cavalieri EL, Rogan EG. Depurinating and stable benzo[a]pyrene-DNA adducts formed in isolated rat liver nuclei. Chem Res Toxicol. 1996;9:1113–1116. doi: 10.1021/tx9600513. [DOI] [PubMed] [Google Scholar]
- 9.Rogan EG, Devanesan PD, RamaKrishna NV, Higginbotham S, Padmavathi NS, Chapman K, Cavalieri EL, Jeong H, Jankowiak R, Small GJ. Identification and quantitation of benzo[a]pyrene-DNA adducts formed in mouse skin. Chem Res Toxicol. 1993;6:356–363. doi: 10.1021/tx00033a017. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarti D, Pelling JC, Cavalieri EL, Rogan EG. Relating aromatic hydrocarbon-induced DNA adducts and c-H-ras mutations in mouse skin papillomas: the role of apurinic sites. Proc Natl Acad Sci U S A. 1995;92:10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 12.Shimada T, Gillam EM, Oda Y, Tsumura F, Sutter TR, Guengerich FP, Inoue K. Metabolism of benzo[a]pyrene to trans-7,8-dihydroxy-7-8-dihydrobenzo[a]pyrene by recombinant human cytochrome P450 1B1 and purified liver epoxide hydrolase. Chem Res Toxicol. 1999;12:623–629. doi: 10.1021/tx990028s. [DOI] [PubMed] [Google Scholar]
- 13.Shimada T, Oda Y, Gillam EM, Guengerich FP, Inoue K. Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab Dispos. 2001;29:1176–1182. [PubMed] [Google Scholar]
- 14.Jennette KW, Jeffrey AM, Blobstein SH, Beland FA, Harvey RG, Weinstein IB. Nucleoside adducts from the in vitro reaction of benzo[a]pyrene-7,8-dihydrodiol 9,10-oxide or benzo[a]pyrene 4,5-oxide with nucleic acids. Biochemistry. 1977;16:932–938. doi: 10.1021/bi00624a019. [DOI] [PubMed] [Google Scholar]
- 15.Koreeda M, Moore PD, Wislocki PG, Levin W, Yagi H, Jerina DM. Binding of benzo[a]pyrene-7,8-diol-9,10-epoxides to DNA, RNA, and protein of mouse skin occurs with high stereoselectivity. Science. 1978;199:778–781. doi: 10.1126/science.622566. [DOI] [PubMed] [Google Scholar]
- 16.Shibutani S, Margulis LA, Geacintov NE, Grollman AP. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)-or dG-(−)-anti-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Braithwaite E, Guo D, Zhao B, Geacintov NE, Wang Z. Mutagenesis of benzo[a]pyrene diol epoxide in yeast: requirement for DNA polymerase zeta and involvement of DNA polymerase eta. Biochemistry. 2003;42:11253–11262. doi: 10.1021/bi0346704. [DOI] [PubMed] [Google Scholar]
- 19.Palackal NT, Burczynski ME, Harvey RG, Penning TM. The ubiquitous aldehyde reductase (AKR1A1) oxidizes proximate carcinogen trans-dihydrodiols to o-quinones: potential role in polycyclic aromatic hydrocarbon activation. Biochemistry. 2001;40:10901–10910. doi: 10.1021/bi010872t. [DOI] [PubMed] [Google Scholar]
- 20.Palackal NT, Lee SH, Harvey RG, Blair IA, Penning TM. Activation of polycyclic aromatic hydrocarbon trans-dihydrodiol proximate carcinogens by human aldo-keto reductase (AKR1C) enzymes and their functional overexpression in human lung carcinoma (A549) cells. J Biol Chem. 2002;277:24799–24808. doi: 10.1074/jbc.M112424200. [DOI] [PubMed] [Google Scholar]
- 21.Penning TM, Burczynski ME, Hung CF, McCoull KD, Palackal NT, Tsuruda LS. Dihydrodiol dehydrogenases and polycyclic aromatic hydrocarbon activation: generation of reactive and redox active o-quinones. Chem Res Toxicol. 1999;12:1–18. doi: 10.1021/tx980143n. [DOI] [PubMed] [Google Scholar]
- 22.Penning TM, Ohnishi ST, Ohnishi T, Harvey RG. Generation of reactive oxygen species during the enzymatic oxidation of polycyclic aromatic hydrocarbon trans-dihydrodiols catalyzed by dihydrodiol dehydrogenase. Chem Res Toxicol. 1996;9:84–92. doi: 10.1021/tx950055s. [DOI] [PubMed] [Google Scholar]
- 23.Murty VS, Penning TM. Characterization of mercapturic acid and glutathionyl conjugates of benzo[a]pyrene-7,8-dione by two-dimensional NMR. Bioconjug Chem. 1992;3:218–224. doi: 10.1021/bc00015a003. [DOI] [PubMed] [Google Scholar]
- 24.Shou M, Harvey RG, Penning TM. Reactivity of benzo[a]pyrene-7,8-dione with DNA. Evidence for the formation of deoxyguanosine adducts. Carcinogenesis. 1993;14:475–482. doi: 10.1093/carcin/14.3.475. [DOI] [PubMed] [Google Scholar]
- 25.McCoull KD, Rindgen D, Blair IA, Penning TM. Synthesis and characterization of polycyclic aromatic hydrocarbon o-quinone depurinating N7-guanine adducts. Chem Res Toxicol. 1999;12:237–246. doi: 10.1021/tx980182z. [DOI] [PubMed] [Google Scholar]
- 26.Balu N, Padgett WT, Lambert GR, Swank AE, Richard AM, Nesnow S. Identification and characterization of novel stable deoxyguanosine and deoxyadenosine adducts of benzo[a]pyrene-7,8-quinone from reactions at physiological pH. Chem Res Toxicol. 2004;17:827–838. doi: 10.1021/tx034207s. [DOI] [PubMed] [Google Scholar]
- 27.Balu N, Padgett WT, Nelson GB, Lambert GR, Ross JA, Nesnow S. Benzo[a]pyrene-7,8-quinone-3'-mononucleotide adduct standards for 32P postlabeling analyses: detection of benzo[a]pyrene-7,8-quinone-calf thymus DNA adducts. Anal Biochem. 2006;355:213–223. doi: 10.1016/j.ab.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Flowers-Geary L, Bleczinki W, Harvey RG, Penning TM. Cytotoxicity and mutagenicity of polycyclic aromatic hydrocarbon ortho-quinones produced by dihydrodiol dehydrogenase. Chem Biol Interact. 1996;99:55–72. doi: 10.1016/0009-2797(95)03660-1. [DOI] [PubMed] [Google Scholar]
- 29.Shultz CA, Quinn AM, Park JH, Harvey RG, Bolton JL, Maser E, Penning TM. Specificity of human aldo-keto reductases, NAD(P)H:quinone oxidoreductase and carbonyl reductases to redox cycle polycyclic aromatic hydrocarbon diones and 4-hydroxyequilenin-o-quinone. Chem Res Toxicol. 2011;24:2153–2166. doi: 10.1021/tx200294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park JH, Mangal D, Tacka KA, Quinn AM, Harvey RG, Blair IA, Penning TM. Evidence for the aldo-keto reductase pathway of polycyclic aromatic trans-dihydrodiol activation in human lung A549 cells. Proc Natl Acad Sci U S A. 2008;105:6846–6851. doi: 10.1073/pnas.0802776105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangal D, Vudathala D, Park JH, Lee SH, Penning TM, Blair IA. Analysis of 7,8-dihydro-8-oxo-2'-deoxyguanosine in cellular DNA during oxidative stress. Chem Res Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Jin Y, Chen M, Huang M, Harvey RG, Blair IA, Penning TM. Detoxication of structurally diverse polycyclic aromatic hydrocarbon (PAH) o-quinones by human recombinant catechol-O-methyltransferase (COMT) via O-methylation of PAH catechols. J Biol Chem. 2011;286:25644–25654. doi: 10.1074/jbc.M111.240739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Blair IA, Penning TM. Detoxification of benzo[a]pyrene-7,8-dione by human recombinant SULTs via sulfation of B[a]P-7,8-catechol. Division of Chemical Toxicology; 242th American Chemical Society (ACS) National Meeting; 2011. http://abstracts.acs.org/chem/242nm/program/view.php. [Google Scholar]
- 34.Harvey RG, Dai Q, Ran C, Penning TM. Synthesis of the o-quinones and other oxidized metabolites of polycyclic aromatic hydrocarbons implicated in carcinogenesis. J Org Chem. 2004;69:2024–2032. doi: 10.1021/jo030348n. [DOI] [PubMed] [Google Scholar]
- 35.Rice JM, Dudek GO. Mass spectra of nucleic acid derivatives. II. Guanine, adenine, and related compounds. J Am Chem Soc. 1967;89:2719–2725. doi: 10.1021/ja00987a039. [DOI] [PubMed] [Google Scholar]
- 36.Harvey RG, Dai Q, Ran CZ, Lim K, Blair I, Penning TM. Syntheses of adducts of active metabolites of carcinogenic polycyclic aromatic hydrocarbons with 2'-deoxyribonucleosides. Polycyclic Aromatic Compounds. 2005;25:371–391. [Google Scholar]
- 37.Wiebel FJ, Kiefer F, Krupski G, Schuller HM. Expression of glutathione S-transferase and phenol sulfotransferase, but not of UDP-glucuronosyltransferase, in the human lung tumor cell lines NCI-H322 and NCI-H358. Biochem Pharmacol. 1986;35:1337–1343. doi: 10.1016/0006-2952(86)90279-0. [DOI] [PubMed] [Google Scholar]
- 38.Blair IA. Analysis of endogenous glutathione-adducts and their metabolites. Biomed Chromatogr. 24:29–38. doi: 10.1002/bmc.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 40.Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem Res Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Huang M, Blair IA, Penning TM. Formation and structural characterization of a catechol-O- sulfate metabolite of benzo[a]pyrene-7,8-dione in three human lung cells. 51st Society of Toxicology Annual Meeting; 2012. Abstract 1552. [Google Scholar]
- 42.Nesnow S, Nelson G, Padgett WT, George MH, Moore T, King LC, Adams LD, Ross JA. Lack of contribution of covalent benzo[a]pyrene-7,8-quinone-DNA adducts in benzo[a]pyrene-induced mouse lung tumorigenesis. Chem Biol Interact. 2010;186:157–165. doi: 10.1016/j.cbi.2010.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.