Abstract
Gap junctions are proteinaceous channels that reside at the plasma membrane and permit the exchange of ions, metabolites, and second messengers between neighboring cells. Connexin proteins are the subunits of gap junction channels. Mutations in zebrafish cx43 cause the short fin (sof b123) phenotype which is characterized by short fins due to defects in length of the bony fin rays. Previous findings from our lab demonstrate that Cx43 is required for both cell proliferation and joint formation during fin regeneration. Here we demonstrate that semaphorin3d (sema3d) functions downstream of Cx43. Semas are secreted signaling molecules that have been implicated in diverse cellular functions such as axon guidance, cell migration, cell proliferation, and gene expression. We suggest that Sema3d mediates the Cx43-dependent functions on cell proliferation and joint formation. Using both in situ hybridization and quantitative RT-PCR, we validated that sema3d expression depends on Cx43 activity. Next, we found that knockdown of Sema3d recapitulates all of the sof b123 and cx43-knockdown phenotypes, providing functional evidence that Sema3d acts downstream of Cx43. To identify the potential Sema3d receptor(s), we evaluated gene expression of neuropilins and plexins. Of these, nrp2a, plxna1, and plxna3 are expressed in the regenerating fin. Morpholino-mediated knockdown of plxna1 did not cause cx43-specific defects, suggesting that PlexinA1 does not function in this pathway. In contrast, morpholino-mediated knockdown of nrp2a caused fin overgrowth and increased cell proliferation, but did not influence joint formation. Moreover, morpholino-mediated knockdown of plxna3 caused short segments, influencing joint formation, but did not alter cell proliferation. Together, our findings reveal that Sema3d functions in a common molecular pathway with Cx43. Furthermore, functional evaluation of putative Sema3d receptors suggests that Cx43-dependent cell proliferation and joint formation utilize independent membrane-bound receptors to mediate downstream cellular phenotypes.
Keywords: Cx43, semaphorin, bone, fin regeneration, zebrafish
Introduction
Connexins are the subunits of gap junction channels that direct cell-cell communication of ions and metabolites (≤1200 Da). Each connexin is a four-pass transmembrane spanning domain protein. Six connexins oligomerize to form a hemichannel, or connexon. Two connexons dock at the plasma membrane of adjacent cells to form a complete gap junction channel. Gap junction intercellular communication (GJIC) contributes to numerous developmental processes, including skeletogenesis. For example, mutations in human CX43 result in oculodentodigital dysplasia (ODDD, Paznekas et al., 2003). ODDD is an autosomal dominant disease causing craniofacial bone deformities and limb abnormalities (Paznekas et al., 2003; Flenniken et al., 2005). Skeletal defects in the CX43−/− knockout mouse include delayed ossification of the axial and craniofacial skeletons (Lecanda et al., 2000). However, the underlying mechanisms by which Cx43-based GJIC leads to skeletal disease phenotypes are largely unknown.
Importantly, the function of Cx43 in skeletal morphogenesis is conserved. Indeed, our lab has found that mutations in zebrafish cx43 cause the short fin (sof b123) phenotype (Iovine et al., 2005). The sof b123 mutant is characterized by defects in the length of the bony fin ray segments, leading to short fins. The sof b123 allele exhibits reduced cx43 mRNA levels without a lesion in the coding sequence (Iovine et al., 2005). However, three additional alleles generated by non-complementation express missense mutations that cause reduced GJIC (Hoptak-Solga et al., 2007). During fin regeneration, the cx43 mRNA is up-regulated in the population of dividing cells. Indeed, all four sof alleles exhibit reduced levels of cell proliferation in addition to short segments (Hoptak-Solga et al., 2008). Furthermore, morpholino-mediated cx43 knockdown completely recapitulates the reduced fin length, reduced segment length, and reduced cell proliferation phenotypes observed in the sof alleles (Hoptak-Solga et al., 2008). Together, these data reveal that reduced mRNA expression (sof b123), reduced protein expression (sof b123 and morpholino-mediated knockdown), or reduced Cx43-based GJIC (three missense alleles) cause the same set of phenotypes. Thus, we refer to any loss of Cx43 function as a loss of Cx43 activity.
Given the observation that any loss of Cx43 activity leads to both reduced cell proliferation and short segments, it may be natural to speculate that reduced cell proliferation causes short segments. However, reduced signaling via the Shh or Fgfr1 signaling pathways also causes reduced cell proliferation and reduced fin length, but does not influence segment length (Quint et al., 2002; Lee et al., 2005). Thus, reducing the level of cell proliferation is not sufficient to impact segment length. We suggest instead that Cx43 plays an additional role in the regulation of segment length, perhaps by regulating joint formation. Our analyses of the another long fin (alf dty86) mutant supports this hypothesis. In contrast to sof, the alf dty86 mutant exhibits fin overgrowth and stochastic joint failure/overlong segments (van Eeden et al., 1996), phenotypes opposite to sof. Interestingly, our analyses revealed that alf dty86 mutants over-express cx43 mRNA (Sims et al., 2009). Furthermore, cx43-knockdown in alf dty86 fins rescues overgrowth and segment length, suggesting that cx43 over-expression is responsible for the alf dty86 phenotypes (Sims et al., 2009). Based on these loss-of-function and gain-of-function phenotypes, we suggest that Cx43 activity both promotes cell proliferation and suppresses joint formation, thereby coordinating bone growth and skeletal patterning.
A long-standing question with regard to connexin mutations is, how do gap junctions influence tangible cellular outcomes such as cell proliferation and cell differentiation? One hypothesis is that Cx43-based GJIC influences gene expression (Stains et al., 2003). To identify global changes in gene expression occurring downstream of cx43, we utilized a novel microarray strategy. We focused on the subset of genes both down-regulated in sof b123 and up-regulated in alf dty86 to enable the identification of cx43-dependent genes. Here we provide molecular and functional validation of one gene identified by this microarray, semaphorin3d (sema3d). Semas comprise a large family of evolutionarily conserved signaling molecules initially found to provide axonal guidance cues during patterning of the central nervous system (Kolodkin et al., 1992). More recent studies have revealed that semaphorins are expressed in most cell types and, in addition to patterning the nervous system, also contribute to vasculature, heart, lung, kidney, bone and tooth development (reviewed in Roth et al., 2009). Class 3 Semas, such as Sema3d, are secreted ligands that interact with several possible cell surface receptors in order to mediate downstream cellular outcomes including cell adhesion, cell migration, cell proliferation, cell viability, and gene expression (reviewed in Yazdani and Terman, 2006). Thus, the finding that a Semaphorin acts functionally downstream of Cx43 provides tangible insights into how skeletal morphogenesis may be influenced by Cx43 activity.
Materials and Methods
Fish maintenance
Zebrafish were raised at constant temperature of 25 °C with 14 light: 10 dark photoperiod (Westerfield, 1993). Wild-type (C32), sof b123 (Iovine and Johnson, 2000) and alf dyt86 (van Eeden et al., 1996) were used in this study.
RNA isolation, fluorescent cRNA synthesis and microarray hybridization
Total RNA of wild-type, sof b123 and alf dty86 5 dpa regenerating fins were extracted using Trizol (Invitrogen, San Diego). RNA quantity and quality were determined by nanodrop and Bioanalyzer 2100 (Agilent) analyses. Only samples in yield higher than 50 ng/uL RNA, having sharp 60S and 40S rRNA peaks shown in the Bioanalyzer electropheretogram, and 260/280 ratios > 1.7 were used. Fluorescent cRNAs were generated using the Agilent Low RNA Input Linear Amplification Kit and Qiagen RNeasy mini columns to purify the fluorescent target. Experimental Cy5 labeled samples (alf dty86, sof b123) were competitively hybridized against equal amounts of Cy3 labeled wild-type cRNAs on an Agilent 4×44K zebrafish 60-mer oligo microarray (G25190F-015064). After careful washing the microarray was scanned in an Agilent microarray scanner and red (Cy5) and green (Cy3) signal intensities were evaluated and processed with Agilent Feature Extraction software (v 7.5). The relative expression value of a gene for two different samples was represented by base 2 log ratios of the two signal intensities. Further data normalization, transformation and filtering for differential gene expression were performed using Agilent Genespring GX (v7.5).
In situ hybridization
Probes were prepared from linear DNA generated from PCR products where the reverse primer contained the binding site for the T7 RNA polymerase (see Table I for sequences). Five days post amputation (dpa) regenerating fins from wild-type, sof b123, or alf dty86 were fixed overnight with 4% paraformaldehyde in PBS and dehydrated in 100% methanol at −20 °C. Gradual aqueous washes were completed in methanol/PBST. Fins were then treated with 5 μg/ml proteinase K for 45 minutes and re-fixed in 4% paraformaldehyde in PBS for 20 minutes. Extensive washes in PBST were followed by prehybridization process in HYB+ solution (HYB+ solution is consisted of 50% formamide, 5 X SSC, 10 mM citric acid, 0.1% Tween20, heparin and tRNA) at 65 °C for 30 minutes–1 hour. Hybridization in the presence of digoxigenin-labeled antisense probes was completed overnight at 65 °C. The next day, the fins were washed gradually in HYB- to 2X SSC to 0.2X SSC and finally to PBST. Anti-digoxigenin Fab fragments (pre-absorbed against zebrafish tissue) were used at 1:5,000 overnight. On day 3, extensive washes in PBST were performed before three short washes in staining buffer (100 mM Tris, 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1 % Tween20, pH 9.0). Fins were next transferred to staining solution (staining buffer plus NBT and BCIP) and development proceeded until a purple color was observed. For final result, fins were then washed with PBST, pre-fixed in 4% paraformaldehyde in PBS and mounted onto microscope slides. Labeled fins were examined on a Nikon Eclipse 80i microscope. Images were collected using a digital Nikon camera.
Table I.
Primer and morpholino sequences
| Gene | Primers for ISH | Morpholinos |
|---|---|---|
| sema3d | F-CGAAGTGTAGTACCATTTACG RT7-TAATACGACTCACTATAGGG-TATGAGGATCATATGTCC |
MO-TGTCCGGCTCCCCTGCAGTCTTCAT 5MM-TGTGCCGCTGCCCTCCACTCTTCAT |
| nrp2a | F-CCAGTCCAGTAACCAGCG RT7-TAATACGACTCACTATAGGG-TCAAGCCTCGGAGCAGCAGC |
MO-CCAGAAATCCATCTTTCCGAAATGT 5MM-CCACAAAACGATCTTTGCCAAATGT |
| plxna1 | F-AAGTGTTCTCTGCGGCAG RT7-TAATACGACTCACTATAGGG-TTGCCCACCTCCGAAAAACC |
MO-GCCACATATCTGCACTGGTCCTTGA 5MM-GCCAGATATGTGGACTGCTCCTTCA |
| plxna3 | F-AGTGTCTCCTAAAGCAAC RT7-TAATACGACTCACTATAGGG-CCGCTTTCTGGAGCCTC |
MO-ATACCAGCAGCCACAAGGACCTCAT 5MM-ATACGACCACCCAGAAGCACCTCAT |
__ The RNA polymerase T7 binding site is underlined in the reverse primers.
MO = targeting morpholino; 5MM = control morpholino with 5 mismatch pairs to target sequence
Following whole mount in situ hybridization, fins were embedded in 1.5% agarose/5% sucrose, and equilibrated overnight in 30% sucrose. Fins were mounted in OCT and cryosectioned (18–20 μm sections) using a Reichert-Jung 2800 Frigocut cryostat. Sections were collected on Superfrost Plus slides (Fisher) and mounted in 100% glycerol.
Morpholino Knockdown and Electroporation
Injection and Electroporation experiments were performed as described (Thummel et al., 2006; Hoptak-Solga et al., 2008; Sims et al., 2009). Targeting morpholinos were targeted against the start codon and modified with fluorescein (Gene Tools, LLC) to provide a charge and for detection. Sequences for the targeting and control morpholinos can be found in Table I.
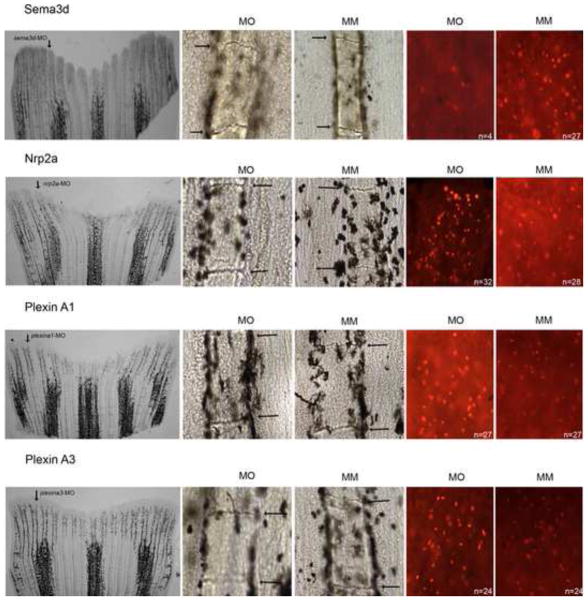
Adult fish were first anesthetized using Tricane-S. Fin amputation was performed under a dissecting microscope. At 3 dpa, morpholinos were injected using a Narishige IM 300 Microinjector. Approximately 50 nl of morpholino (i.e. targeting or control 5MM morpholinos) was injected per ray (5–6 fin rays per fin, the other rays were uninjected control). Immediately following injection, both dorsal and ventral halves were electroporated using a CUY21 Square Wave Electroporator (Protech International, Inc.). The following parameters were used: ten 50-ms pulses of 15 V with a 1 s pause between pulses. At 24 hpe (hours post electroporation), success was evaluated by monitoring fluorescein uptake under fluorescence microscope. Fins were harvested either at 1 dpe for H3P detection and for qRT-PCR or at 4 dpe for ZNS5 detection. Three to five fins were injected per morpholino (i.e. targeting or mismatch); the un-injected side served as an independent control. Each morpholino was tested in at least three independent experiments to ensure reproducibility. The graphs in Figure 2 are based combined data from two comparable experiments (n = 7). Statistical significance was determined using the student’s t-test (p<0.05).
Figure 2.
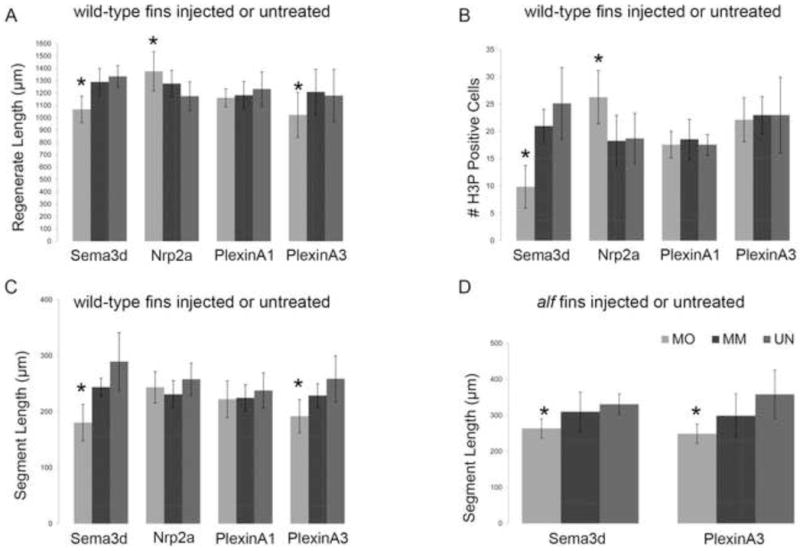
Morpholino-mediated gene knockdown of sema3d and its putative receptors. In all graphs, MO represents the particular gene-targeting morpholino; MM represents the particular 5 mis-match/control morpholino; UN represents uninjected/untreated fins. (A) Total regenerate length was measured. sema3d-knockdown and plxna3-knockdown cause reduced fin length (*); nrp2a-knockdown causes increased fin length (*). (B) The total number of H3P positive cells were counted. sema3d-knockdown causes reduced levels of cell proliferation (*); nrp2a-knockdown causes increased levels of cell proliferation (*). (C) Segment length was measured in treated wild-type fins. sema3d-knockdown and plxna3-knockdown cause reduced segment length (*). (D) Segment length was measured in treated alf dty86 fins. sema3d-knockdown and plxna3-knockdown cause reduced segment length and rescue joint formation in alf dty86 (*). Statistically different data sets (*) were determined by the student’s t-test where p < 0.05. By the student’s t-test, there is no statistical difference between MM and UN for any mismatch morpholino. Error bars represent the standard deviation.
Immunofluorescence
Fins were harvested after morpholino knockdown experiments (for ZNS5 detection staining, fins were harvested 4 dpe; for H3P detection staining, fins were harvested 1 dpe). Fins were then fixed in 4% paraformaldehyde in PBS for 2 hours at RT and then dehydrated in methanol. During processing, fins were washed gradually in methanol/PBS followed by 3 washes in block solution (50 ml PBS, 1 g BSA, 250 μl triton). Either the mouse ZNS5 (Zebrafish International Resource Center: http://zebrafish.org/zirc/home/guide.php, 1:200) or the rabbit antibody against histone-3-phosphate (anti-H3P, Millipore, 1:100) were incubated with fins overnight at 4 °C. Next day, antibodies were removed and fins were washed in block solution 3×10 minutes. Secondary antibodies (i.e. anti-mouse Alexa 488 for ZNS5 detection or anti-rabbit Alexa 546 for H3P detection) were diluted in 1:200 blocking solution and incubated overnight at 4 °C. Following 3×10 minutes treatment in blocking solution, fins were washed in PBS and then mounted onto slides in glycerol. Labeled fins were examined on a Nikon Eclipse 80i microscope. Images were collected using a digital Nikon camera.
Measurements
Fins were imaged on a Nikon SMZ1500 dissecting microscope at 4X (regenerate length) or 10X (segment length or the number of dividing cells). Photographs were taken using a Nikon DXM1200 digital camera. For regenerate length, segment length, and the number of dividing cells, all measurements were taken from only the longest fin rays (i.e. the third ray from either the dorsal or ventral end) since that was previously established as a standard (Iovine and Johnson, 2000). Student’s t-tests were performed in Excel to determine if experimental conditions were significantly different from control conditions.
Regenerate and segment length was measured using ImagePro software. Fin ray length was measured from the amputation plane (clearly visible in bright field) to the end of the fin. Segment length is measured as the distance between two joints where joints are identified (i.e. and clearly distinguished from breaks) following ZNS5 staining (Sims et al., 2009).
The mitotic cells were first detected by H3P staining as described above (i.e. Histone-3 is phosphorylated on Ser10 only during mitosis, Wei et al., 1999). H3P positive cells were counted from within the distal-most 250 μm of each ray (Hoptak-Solga et al., 2008).
qRT-PCR Analysis
qRT-PCR analysis was performed as described (Sims et al., 2009). In brief, Trizol reagent was used to isolate mRNA from 5 dpa wild-type, sof b123, or alf dty86 regenerating fins and 1 dpe (i.e. cx43-knockdown fins) (5 fins per pool). First strand cDNA was synthesized using oligodT (12–15) and reverse transcriptase. Dilution of template cDNA (1:10) was prepared. Oligos flanking introns were designed for sema3d (F-5′ TGGATGAGGAGAGAAGCCGAT 3′; R-5′ GCAGGCCAGCTCAACTTTTT 3′) using Primer Express software (primers for cx43 and keratin4 can be found in Sims et al., 2009). The sema3d, cx43, and keratin4 amplicons were amplified independently using the Power SYBR green PCR master mix (Applied Biosystems). Samples were run in triplicate on the ABI7300 Real Time PCR system and the average cycle number (CT) was determined for each amplicon. Delta CT (ΔCT) values represent normalized sema3d levels with respect to keratin4, the internal control. The relative level of gene expression was determined using the delta delta CT (ΔΔCT) method (i.e. 2−ΔΔCT). A minimum of three trials were run to ensure the reproducibility of the results.
Results
sema3d functions downstream of cx43
We completed a novel microarray strategy designed to identify genes acting downstream of cx43. We took advantage of our findings that cx43 expression is reduced in sof b123 and increased in alf dty86 in order to identify a group of genes that are both down-regulated when cx43 is down-regulated (i.e. in sof b123) and up-regulated when cx43 is up-regulated (i.e. in alf dty86). Importantly, the cx43 gene is found among the top 50 genes identified using this strategy, strongly suggesting that this approach identified relevant genes of interest (supplemental data, Table S1). Another gene found in the top half of the microarray was the secreted semaphorin gene, sema3d. Given the importance of semaphorins in a diversity of signaling pathways, we were intrigued at the possibility that Sema3d signaling mediates Cx43 activities.
In order to validate sema3d as a downstream target of cx43, we first examined sema3d expression in wild-type, sof b123 and alf dty86 regenerating fins by whole mount in situ hybridization. As anticipated, sema3d mRNA expression appeared down-regulated in sof b123 and up-regulated in alf dty86 (Figure 1). Next, we determined the tissue-specific expression of sema3d as revealed by cryosectioning. The outer cell layers of the fin are epidermis; the basal layer of the epidermis is identified as a row of cuboidal-shaped cells closest to the mesenchymal compartment. Within the mesenchyme, the skeletal precursor cells (i.e. pre-osteoblasts and pre-joint-forming cells) are located laterally. The regeneration blastema, the specialized population of dividing cells contributing to new fin outgrowth, is medially adjacent to the skeletal precursors. This population of cells up-regulates cx43 expression during fin regeneration (Hoptak-Solga et al., 2008). In contrast, cryosectioning of stained sema3d-positive fins revealed that sema3d is expressed in both the lateral skeletal precursor cells and in the lateral basal layer of the epidermis (Figure 1). Since sema3d expression is not up-regulated in the cx43-positive cells, sema3d appears not to be a direct target of Cx43 activity.
Figure 1.
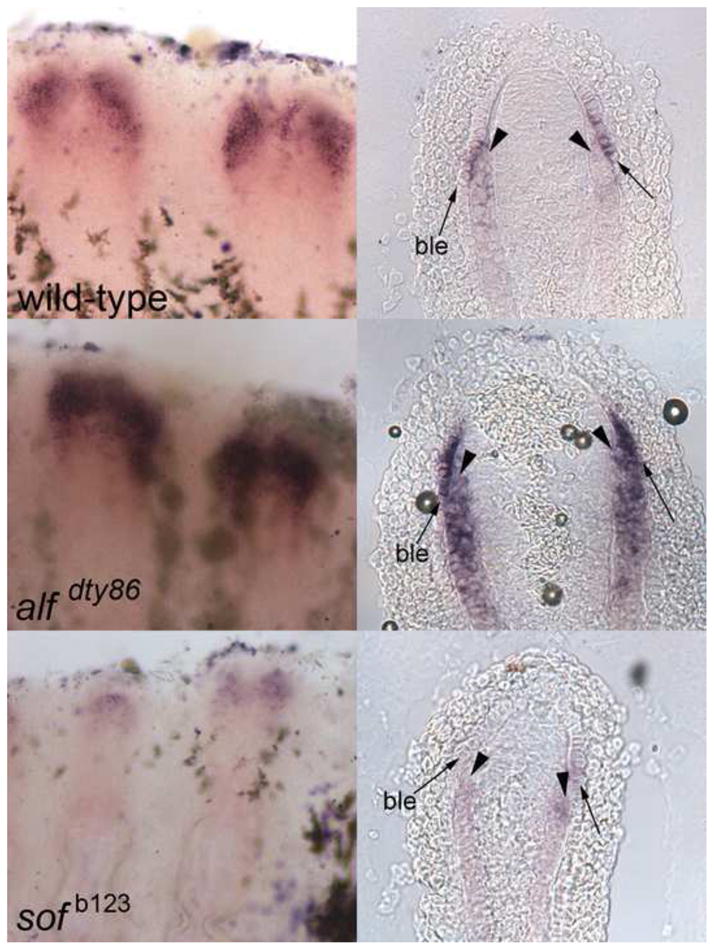
sema3d is differentially expressed in wild-type (top), alf dty86 (middle) and sof b123 (bottom). Left: whole mount in situ hybridization shows increased expression in alf dty86 and decreased expression in sof b123 compared to wild-type. Right: Cryosections reveal the tissue-specific localization of sema3d-expressing cells. Arrowheads point to skeletal precursor cells; arrows point to the basal layer of the epidermis (ble).
To confirm the observed qualitative differences in sema3d expression we performed quantitative RT-PCR (qRT-PCR). We find that sema3d is reduced in sof b123 and increased in alf dty86 (Table II). Moreover, we find that sema3d expression is reduced in wild-type fins treated for cx43-knockdown, providing independent evidence that sema3d expression is influenced by Cx43 activity. Together, these data support the hypothesis that sema3d expression is regulated by the level of Cx43.
Table II.
Expression of sema3d via qRT-PCR
| strain | trial 1 | trial 2 | trial 3 |
|---|---|---|---|
| wild-type | 1 | 1 | 1 |
| sof b123 | 0.6 | 0.65 | 0.71 |
| alf dty86 | 1.95 | 1.42 | 1.90 |
| cx43-KD fins | 0.7 | 0.4 | 0.55 |
The fold-difference with respect to wild-type is shown for each of three trials.
Sema3d mediates Cx43-dependent cell proliferation and joint formation
To determine if sema3d mediates Cx43-dependent phenotypes, we completed morpholino-mediated gene knockdown of sema3d (as described for cx43 knockdown in Hoptak-Solga et al., 2008 and in Sims et al., 2009). Briefly, fins were injected with either a gene-specific targeting morpholino (MO) or with an altered morpholino that includes five mismatches (MM) to the target sequence. Following injection into the distal region of the regenerate, fins were electroporated to permit cellular uptake. Morpholinos were modified with fluorescein, which both provides a requisite charge for electroporation and provides a method to confirm cellular uptake. Interestingly, we find that sema3d-knockdown exhibits all of the same loss-of-function phenotypes as sof b123 mutants (Iovine et al., 2005; Hoptak-Solga et al., 2008) and as cx43-knockdown (Hoptak-Solga et al., 2008; Sims et al., 2009). Thus, sema3d knockdown fins exhibit reduced fin length, reduced segment length, and reduced cell proliferation (Figure 2A–C and Figure 3 for representative images). The level of cell proliferation was evaluated by counting the number of cells in mitosis, detected using an antibody against histone-3-phosphate (i.e. H3P). These data demonstrate that sema3d mediates cx43-dependent fin phenotypes influencing growth and joint formation. To provide additional evidence that sema3d functions in a common pathway with cx43, we next attempted to rescue the joint formation phenotype of alf dty86. Indeed, sema3d knockdown rescued the joint failure phenotype of alf dty86, causing reduced segment length (Figure 2D). Until now, only reduced cx43 function has been associated with segment length phenotypes and with rescue of joint formation in alf dty86. Therefore, the finding that sema3d knockdown caused short segments in wild-type and rescued segment length in alf dty86is striking. Together these data indicate that cx43 and sema3d function in a common pathway to regulate cell proliferation and joint formation. Thus, Sema3d signaling mediates Cx43-specific effects.
Figure 3.
Representative images of morpholino-induced phenotypes. From left to right: representative whole fins following injection in the dorsal-most 5–6 fin rays (arrow); segment length in targeting morpholino-injected (MO) and in 5 mis-match morphlino-injected (MM); H3P-positive cells in targeting morpholino-injected (MO) and in 5 mis-match morphlino-injected (MM).
Identification of putative Sema3d receptors
Neuropilins (Nrps) and Plexins (Plxns) are likely receptors for Semaphorin signaling (reviewed in Zhou et al., 2008). Indeed, Nrps and Plxns may hetero-oligomerize to transduce Sema signals. Nrps are believed to bind Semas directly (although Plxns also contain a sema domain), but have a very short intracellular domain that may not be sufficient to transduce intracellular signals. Plxns, on the other hand, have an extensive intracellular domain (reviewed in Zhou et al., 2009). Since both Nrps and Plxns are the best known receptors for Semas, we initiated a candidate gene search of these gene families. The zebrafish genome contains 4 neuropilin (nrp) genes (nrp1a, nrp1b, nrp2a, and nrp2b, Yu et al., 2004). In addition, Plexins in the A and D families are candidate receptors for secreted Semas (Zhou et al., 2008). The zebrafish genome contains plexina1 (plxna1), plxna3, plxna4, and plxnd1. Of these 8 candidate genes, only nrp2a, plxna1, and plxna3 appear to be expressed in regenerating fins by in situ hybridization (Figure 4). The expression of nrp2a appears mainly in the blastema, perhaps more heavily localized distally. The distal-most blastema has been proposed to regulate fin outgrowth during regeneration (Nechiporuk and Keating, 2002). There is also apparent staining in the skeletal precursor cells, and sporadic but strong staining in individual cells of the outer layers of the epithelium. The identity of these cells is not known. The plxna1 gene is expressed primarily in the distal blastema and also in the distal basal layer of the epidermis. In contrast, plxna3 appears to be expressed primarily in the skeletal precursor cells and throughout the medial compartment of the regenerate.
Figure 4.
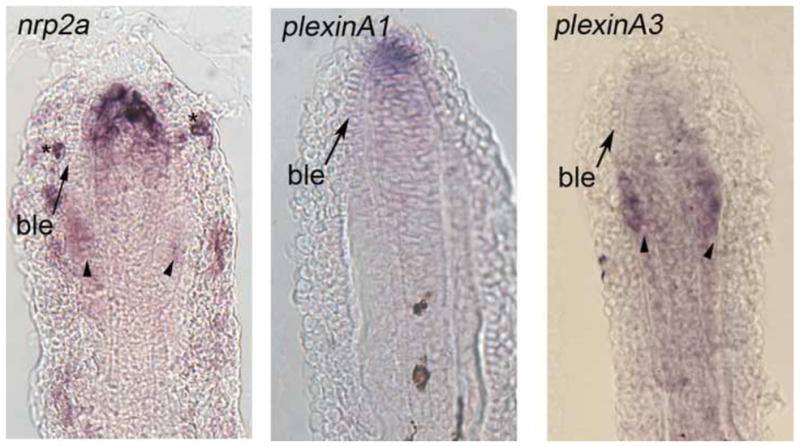
Gene expression of candidate receptors for Sema3d. Left: expression of nrp2a is primarily located in the distal blastema and in skeletal precursor cells. Staining of individual cells of the outer epithelial cells is also observed (*). Middle: expression of plxna1 is primarily in the distal blastema and in the distal cells of the basal epidermis. Right: expression of plxna3 is located in both the skeletal precursor cells and in the blastema. Arrows identify the basal layer of the epidermis (ble), arrowheads identify skeletal precursor cells.
PlxnA3 and Nrp2a mediate independent Sema3d functions
Next we completed functional analyses to determine which, if any, of these receptors contribute to the Cx43-Sema3d pathway. Receptors that mediate Sema3d function are expected to exhibit similar knockdown phenotypes as cx43 and sema3d. However, knockdown of plxna1 did not appear to influence either cell proliferation or joint formation (Figures 2 and 3), suggesting that PlxnA1 does not participate in Cx43-Sema3d-dependent skeletal morphogenesis. In contrast, knockdown of plxna3 caused short segments (Figure 2C and Figure 3) but had no effect on cell proliferation (Figure 2B). There is some influence of plxna3 knockdown on fin length, as the length of the regenerate was statistically shorter than the controls (Figure 2A). Since there was no effect on cell proliferation, we conclude that the small plxna3-dependent effect on fin length is due to its influence on segment length, and not on fin growth. To provide further support for the functional relationship between PlxnA3 and cx43-dependent joint formation, we evaluated the effect of plxna3-knockdown in alf dty86 regenerating fins. As anticipated, plxna3-knockdown rescued the joint formation phenotype, recapitulating the cx43-and sema3d-knockdown effects (Figure 2D). These data suggest that PlnxA3 contributes to Sema3d-mediated joint formation. Therefore, we have now identified a third gene (i.e. plxna3), predicted to function downstream of Cx43-Sema3d, whose function is required for appropriate joint formation.
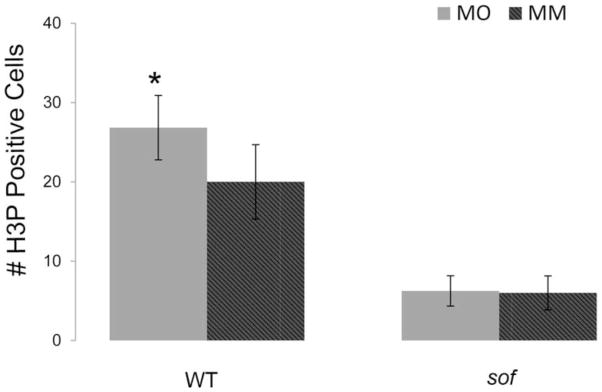
Knockdown of nrp2a caused increased fin growth and increased cell proliferation (Figure 2A,B and Figure 3), suggesting that signaling via Nrp2a negatively influences cell division. There was no effect on segment length following nrp2a gene knockdown (Figure 2C), indicating that Nrp2a signaling does not mediate Sema3d effects on joint formation. Since knockdown of cx43 and sema3d both cause reduced growth and reduced cell proliferation, it was anticipated that Nrpa2a knockdown would similarly cause reduced growth and cell proliferation. Since this was not the case, we suggest instead that Sema3d binding to the Nrp2a receptor inactivates its activity, thereby positively regulating cell division by inhibiting a negative signal. We attempted to provide evidence for this hypothesis by evaluating Nrpa2a knockdown in sof b123 fins, which express less sema3d (Figure 1 and Table II). For example, if Sema3d is required to block the effects of Nrp2a signaling, then the increase in cell proliferation associated with Nrp2a knockdown should be attenuated when Sema3d is reduced, as in sof b123. This is what we find. Nrp2a knockdown in wild-type regenerating fins causes a 30 % increase in dividing cells, while Nrp2a knockdown in sof b123 regenerating fins has no effect on the number of dividing cells (Figure 5). Further studies will be required to demonstrate unequivocally that Sema3d acts as a ligand for Nrp2a. However, our current findings provide support for the conclusion that Sema3d can mediate negative regulation of Nrp2a and thereby promote cell proliferation. Note that the observed Nrp2a effects may be mediated in conduction with an as yet unidentified Plxn co-receptor since Nrps appear not to encode intracellular signaling domains.
Figure 5.
Nrp2a-knockdown effects are abrogated in sof b123. Nrp2a-mediated gene knockdown causes an increase in cell proliferation when Sema3d is present at typical levels. In sof b123, where sema3d expression is reduced, Nrp2a is unable to enhance the level of cell proliferation. MO, gene-targeting morpholino. MM, 5 mis-match/control morpholino.
Together, our analyses of the plxna1, plxna3, and nrp2a genes suggest that Nrp2a and PlxnA3 mediate Sema3d-dependent events, while PlxnA1 does not appear to function in Sema3d-mediated events. Moreover, we suggest that Nrp2a and PlxnA3 mediate distinct Cx43- and Sema3d-dependent phenotypes, where Nrp2a mediates the Cx43-dependent effects on cell proliferation and PlexinA3 mediates the Cx43-dependent effects on joint formation.
Discussion
Much of what is known about sema3d has been determined during development of the central nervous system in zebrafish. In the embryonic nervous system sema3d has been found to exert multiple diverse functions. For example, sema3d may act as an axonal repellent or as an axonal attractant (Wolman et al., 2004). Alternatively, sema3d function can modify cell adhesion via influencing the expression of the adhesion protein L1 (Wolman et al., 2007). Further, sema3d has been found to influence the population of neural crest cells by promoting proliferation (Berndt and Halloran, 2006), and by regulating their migration (Yu and Moens, 2005). It has been suggested that the different functions of sema3d may depend on the receptors expressed on the responding cells. Indeed, depending on the cell-type, sema3d has been found to interact with nrp1a (Wolman et al., 2004), with nrp1a/nrp2b (Wolman et al., 2004), or via nrp-independent mechanisms (Wolman et al., 2007). Thus, Sema3d appears to interact with a variety of receptors in order to mediate a diversity of downstream cellular events. It is therefore not possible to predict a specific receptor complement/pathway for Sema3d function. However, it is also not difficult to envision how Sema3d signaling could be responsible for mediating multiple independent signaling events during fin regeneration.
The finding that Sema3d functions downstream of Cx43 is supported by multiple independent lines of evidence. First, the sema3d gene exhibits differential expression in sof b123 and alf dty86 regenerating fins by in situ hybridization and by qRT-PCR. Second, cx43-knockdown in wild-type fins is sufficient to reduce sema3d gene expression. Third, we provide functional evidence that sema3d acts downstream of cx43 since sema3d-knockdown recapitulates all of the cx43-dependent phenotypes, including rescue of joint formation in alf dty86. Thus, sema3d is both molecularly and functionally downstream of Cx43. Moreover, we identify two putative Sema3d receptors, Nrp2a and PlxnA3. Remarkably, these receptors appear to independently mediate Cx43-Sema3d-dependent cell proliferation and joint formation. The described functional analyses for Sema3d and its putative receptors utilized translation-blocking morpholinos. Since antibodies are not currently available, we are unable to demonstrate that protein translation of the targets is inhibited following morpholino-mediated gene knockdown. However, the specificity of sema3d- and plxna3- knockdown to Cx43-dependent phenotypes is provided by our findings that sema3d- and plxna3-knockdown both cause short segments and also rescue joint failure in alf dty86 (i.e. prior to this report, these findings were specific for cx43 mutations or knockdown). Similarly, the finding that sof b123 abrogates the effects of nrp2a-knockdown provides specificity for the role of Nrp2a in Cx43-Sema3d-dependent cell proliferation. We did not observe Cx43-dependent phenotypes following plxna1-knockdown. However, until we can demonstrate that the PlxnA1 protein has been successfully reduced, we cannot formally rule out the possibility that PlxnA1 also contributes to Sema3d signaling events.
Based on our current and published findings (summarized in Table III), we suggest the following model for Cx43 activity during fin regeneration (Figure 6). Prior studies from our lab have shown that Cx43 both promotes cell proliferation and suppresses joint formation (Hoptak-Solga et al., 2008; Sims et al., 2009). Here we find that Sema3d signaling contributes to these Cx43-dependent activities in a pathway that bifurcates after Sema3d (Figure 6A). Indeed, functional analyses of Cx43 and Sema3d provide evidence that cell proliferation and joint formation are coupled, while functional analyses of the putative Sema3d receptors demonstrate effects on either cell proliferation (i.e. Nrp2a) or joint formation (i.e. PlxnA3). Thus, Cx43 coordinates skeletal growth and patterning via Sema3d signaling, which in turn regulates cell proliferation and joint formation in distinct downstream signaling pathways.
Table III.
Phenotypes associated with altered expression of Cx43 and genes proposed to function downstream of Cx43.
| mutant/morphant proliferation | cx43 | sema3d | fin length | segment length | cell |
|---|---|---|---|---|---|
| sof b123 | low | low | short | short | reduced |
| alf dty86 | high | high | long | long | increased |
| cx43-KD | low | low | short | short | reduced |
| sema3d-KD | n/c | low | short | short | reduced |
| plxnA3-KD | n/d | n/d | short | short | n/c |
| nrp2a-KD | n/d | n/d | long | n/c | increased |
Changes in cx43 and sema3d gene expression were evaluated by qRT-PCR. All knockdowns (KD) listed were completed in wild-type regenerating fins. No change (n/c); not done (n/d).
Figure 6.
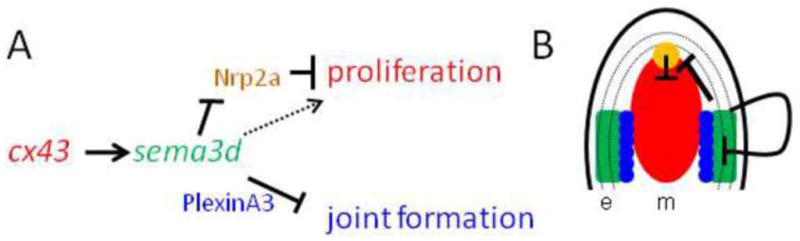
Model of how Cx43-Sema3d influences skeletal morphology. (A) Proposed pathway of Cx43-Sema3d and downstream receptors (text colors are coordinated with the cartoon in B). Cx43 activity in the dividing cells influences sema3d gene expression in the lateral skeletal precursors and basal layer of the epidermis. Secreted Sema3d promotes cell proliferation (dotted arrow) in the cx43-positive compartment by inhibiting a negative signal from Nrp2a in the distal blastema. Sema3d suppresses joint formation in the skeletal precursor cells by its interaction with PlxnA3. (B) Cartoon illustrating the compartments of gene expression in the Cx43-Sema3d pathway (e, epithelium; m, mesenchyme; basal layer of the epidermis is dotted). The cx43 mRNA is up-regulated in the blastema (red), adjacent to the sema3d-positive cells in the skeletal precursor cells and in the lateral basal layer of the epidermis (green). Cx43-dependent up-regulation of sema3d in the lateral compartment allows secreted Sema3d to signal back to the blastema via Nrp2a (yellow), relieving the Nrpa2a inhibition of cell proliferation. Sema3d signaling via PlxnA3 in the skeletal precursor cells (blue circles) inhibits joint formation in the skeletal precursor cells, perhaps by influencing osteoblast/joint-forming cell differentiation.
It is possible to visualize the steps of this molecular pathway by considering the location of gene expression of the molecular players (Figure 6B). For example, Cx43 is expressed in the medially located dividing cells during fin regeneration (Iovine et al., 2005). These cells are directly adjacent to the skeletal precursor cells that will differentiate as either osteoblasts or joint forming cells (Brown et al., 2009; Borday et al., 2001). We suggest that Cx43 activity in the dividing cells influences gene expression of sema3d in the adjacent lateral compartments, which in turn mediates independent signaling pathways that regulate cell division and joint formation. It remains unknown how gap junctions contribute to tangible changes in gene expression. One possibility is that Cx43-dependent GJIC influences the concentration of a second messenger that directly regulates the activity of a relevant transcription factor in the cx43-positive cells. These changes in gene expression in the cx43-positive compartment lead to changes in gene expression in the adjacent sema3d-positive compartment. Once the expression of sema3d is up-regulated in the lateral skeletal precursor cells, Sema3d will be secreted where it may interact with its receptors. Conveniently, Nrp2a and PlxnA3, which mediate independent events, are expressed in distinct populations of cells. For example, the Nrp2a receptor is expressed in the distal blastema where it may influence cell proliferation in the cx43-positive cells. We suggest that Sema3d binding to Nrp2a prevents the inhibition of cell proliferation, thereby promoting growth. Similarly, the expression of plxna3 in the skeletal precursor cells suggests that secreted Sema3d binds to the PlxnA3 receptor and initiates an autocrine response to influence the expression of genes that will determine joint formation (i.e. promoting osteoblast differentiation, suppressing joint formation, or both). However, recall that nrp2a and plxna3 are expressed in more than one cellular compartment during fin regeneration. Thus, it remains possible that Sema3d signaling events are more complicated than this model suggests.
The model we propose suggests that Sema3d initiates a typical signal transduction pathway that directly influences cell proliferation or joint formation in the cells expressing the putative receptors. This model is consistent with our examination of gene expression patterns and on functional analyses. Alternate models are also possible. For example, it has been suggested that Sema3A influences innervation and/or vasularization of endochondral bones in mammals, which in turn impacts bone growth (Gomez et al., 2005). The fin rays contain both nerve axons and blood vessels, although it is not known if Sema3d and/or its receptors are expressed in either of those cell populations. Future immunohistochemical analyses may provide new insights into the possibility that Cx43-Sema3d drives growth and/or patterning via the vasculature or nervous system. Moreover, others have found evidence that Sema3F may influence the localization of Cx43 to the plasma membrane in rat liver epithelial cell lines, perhaps regulating Cx43-based GJIC (Kawasaki et al., 2007). Our findings do not support this type of role for Sema3d during fin regeneration since cx43 and sema3d are not co-expressed in the same population of cells. However, it remains possible that additional Semas may contribute to the expression and/or localization of Cx43.
Conclusions
The identification of Sema3d acting downstream of Cx43 provides tangible insights into how cellular outcomes are coupled in order to coordinate bone growth with skeletal patterning. We find that the Cx43-Sema3d pathway diverges via distinct receptors to influence two cellular outcomes: cell proliferation and joint formation. Continued validation of additional genes identified by our microarray will fill the gaps of molecular events occurring both between Cx43 activity and Sema3d signaling as well as events occurring downstream of the putative Sema3d receptors that mediate changes in cell division and joint formation.
Supplementary Material
Highlights.
Sema3d acts downstream of Cx43 during fin regeneration.
Sema3d mediates Cx43-dependent cell proliferation and joint formation.
Nrp2a is a putative Sema3d receptor that regulates cell proliferation.
PlxnA3 is a putative Sema3d receptor that regulates joint formation.
Acknowledgments
The authors would like to thank Rebecca Jefferis for maintenance and care of the fish colony, and members of the Iovine lab for critical discussions regarding this manuscript. Isha Jain completed the microarray analysis in collaboration with Dr. Jutta Marzillier. This research was funded in part from the NIH (HD047737) and by Lehigh’s Department of Biological Sciences. The ZNS5 antibody was purchased from the Zebrafish International Resource Center, which is supported by P40 RR12546.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Borday V, Thaeron C, Avaron F, Brulfert A, Casane D, Laurenti P, Geraudie J. evx1 transcription in bony fin rays segment boundaries leads to a reiterated pattern during zebrafish fin development and regeneration. Dev Dyn. 2001;220:91–8. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1091>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Brown AM, Fisher S, Iovine MK. Osteoblast maturation occurs in overlapping proximal-distal compartments during fin regeneration in zebrafish. Dev Dyn. 2009;238:2922–8. doi: 10.1002/dvdy.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flenniken AM, Osborne LR, Anderson N, Ciliberti N, Fleming C, Gittens JE, Gong XQ, Kelsey LB, Lounsbury C, Moreno L, Nieman BJ, Peterson K, Qu D, Roscoe W, Shao Q, Tong D, Veitch GI, Voronina I, Vukobradovic I, Wood GA, Zhu Y, Zirngibl RA, Aubin JE, Bai D, Bruneau BG, Grynpas M, Henderson JE, Henkelman RM, McKerlie C, Sled JG, Stanford WL, Laird DW, Kidder GM, Adamson SL, Rossant J. A Gja1 missense mutation in a mouse model of oculodentodigital dysplasia. Development. 2005;132:4375–86. doi: 10.1242/dev.02011. [DOI] [PubMed] [Google Scholar]
- Gomez C, Burt-Pichat B, Mallein-Gerin F, Merle B, Delmas PD, Skerry TM, Vico L, Malaval L, Chenu C. Expression of Semaphorin-3A and its receptors in endochondral ossification: potential role in skeletal development and innervation. Dev Dyn. 2005;234:393–403. doi: 10.1002/dvdy.20512. [DOI] [PubMed] [Google Scholar]
- Hoptak-Solga AD, Klein KA, Derosa AM, White TW, Iovine MK. Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett. 2007;581:3297–302. doi: 10.1016/j.febslet.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR, Iovine MK. Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol. 2008;317:541–8. doi: 10.1016/j.ydbio.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278:208–19. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics. 2000;155:1321–9. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Kubomoto A, Yamasaki H. Control of intracellular localization and function of Cx43 by SEMA3F. J Membrane Biology. 2007;217:53–61. doi: 10.1007/s00232-007-9051-y. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Matthes DJ, O’Connor TP, Patel NH, Admon A, Bentley D, Goodman CS. Fasciclin IV: sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron. 1992;9:831–45. doi: 10.1016/0896-6273(92)90237-8. [DOI] [PubMed] [Google Scholar]
- Lecanda F, Warlow PM, Sheikh S, Furlan F, Steinberg TH, Civitelli R. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J Cell Biol. 2000;151:931–44. doi: 10.1083/jcb.151.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–83. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, Jabs EW. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–18. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, Laforest L, Miles J, Gaffield W, Akimenko MA. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 2002;99:8713–8. doi: 10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth L, Koncina E, Satkauskas S, Cremel G, Aunis D, Bagnard D. The many faces of semaphorins: from development to pathology. Cell Mol Life Sci. 2009;66:649–66. doi: 10.1007/s00018-008-8518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims K, Jr, Eble DM, Iovine MK. Connexin43 regulates joint location in zebrafish fins. Dev Biol. 2009;327:410–8. doi: 10.1016/j.ydbio.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stains JP, Lecanda F, Screen J, Towler DA, Civitelli R. Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J Biol Chem. 2003;278:24377–87. doi: 10.1074/jbc.M212554200. [DOI] [PubMed] [Google Scholar]
- Thummel R, Bai S, Sarras MP, Jr, Song P, McDermott J, Brewer J, Perry M, Zhang X, Hyde DR, Godwin AR. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–46. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Nusslein-Volhard C. Genetic analysis of fin formation in the zebrafish, Danio rerio. Development. 1996;123:255–62. doi: 10.1242/dev.123.1.255. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish (Brachydanio rerio) University of Oregon Press; Eugene, OR: 1993. [Google Scholar]
- Wolman MA, Liu Y, Tawarayama H, Shoji W, Halloran MC. Repulsion and attraction of axons by semaphorin3D are mediated by different neuropilins in vivo. J Neurosci. 2004;24:8428–35. doi: 10.1523/JNEUROSCI.2349-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolman MA, Regnery AM, Becker T, Becker CG, Halloran MC. Semaphorin3D regulates axon axon interactions by modulating levels of L1 cell adhesion molecule. J Neurosci. 2007;27:9653–63. doi: 10.1523/JNEUROSCI.1741-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani U, Terman JR. The semaphorins. Genome Biol. 2006;7:211. doi: 10.1186/gb-2006-7-3-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Houart C, Moens CB. Cloning and embryonic expression of zebrafish neuropilin genes. Gene Expr Patterns. 2004;4:371–8. doi: 10.1016/j.modgep.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Yu HH, Moens CB. Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev Biol. 2005;280:373–85. doi: 10.1016/j.ydbio.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Gunput RA, Pasterkamp RJ. Semaphorin signaling: progress made and promises ahead. Trends Biochem Sci. 2008;33:161–70. doi: 10.1016/j.tibs.2008.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.