Abstract
Neuronal circuit development and function require proper synapse formation and maintenance. Genetic screens are one powerful method to identify the mechanisms shaping synaptic development and stability. However, genes with essential roles in non-neural tissues may be missed in traditional loss-of-function screens. In an effort to circumvent this limitation, we used neuron-specific RNAi knock down in Drosophila and assayed the formation, growth, and maintenance of the neuromuscular junction (NMJ). We examined 1970 Drosophila genes, each of which has a conserved ortholog in mammalian genomes. Knock down of 158 genes in post-mitotic neurons led to abnormalities in the neuromuscular system, including misapposition of active zone components opposite postsynaptic glutamate receptors, synaptic terminal overgrowth and undergrowth, abnormal accumulation of synaptic material within the axon, and retraction of synaptic terminals from their postsynaptic targets. Bioinformatics analysis demonstrates that genes with overlapping annotated function are enriched within the hits for each phenotype, suggesting that the shared biological function is important for that aspect of synaptic development. For example, genes for proteasome subunits and mitotic spindle organizers are enriched among the genes whose knock down leads to defects in synaptic apposition and NMJ stability. Such genes play essential roles in all cells, however the use of tissue- and temporally-restricted RNAi indicates that the proteasome and mitotic spindle organizers participate in discrete aspects of synaptic development. In addition to identifying functional classes of genes shaping synaptic development, this screen also identifies candidate genes whose role at the synapse can be validated by traditional loss-of-function analysis. We present one such example, the dynein-interacting protein NudE, and demonstrate that it is required for proper axonal transport and synaptic maintenance. Thus, this screen has identified both functional classes of genes as well as individual candidate genes that are critical for synaptic development and will be a useful resource for subsequent mechanistic analysis of synapse formation and maintenance.
Keywords: Drosophila, RNA interference screen, synaptic development, synaptic maintenance, Gene Ontology analysis
Introduction
The Drosophila neuromuscular junction (NMJ) is a powerful system for identifying genes that regulate neural development (Collins and DiAntonio 2007). Genes have been identified that control many aspects of synapse formation, developmental plasticity, and synaptic maintenance. Both traditional loss-of-function screens and reverse genetic analysis of genes of interest have identified many of these key molecules (for example, Banovic et al. 2010; Liebl et al. 2006; Dickman and Davis 2009; Reeve et al. 2008; Graf et al. 2009). While extremely useful, these approaches are limited in their ability to identify the synaptic role of genes that play essential roles early in development or in multiple tissues. In the eye or mushroom body, this problem can be overcome by mosaic analysis (Lee and Luo 1999), but this is not efficient for analysis of early born motoneurons. An alternative is to use tissue-specific gene expression to target the motoneuron. For example, a large-scale overexpression screen in the nervous system identified many genes that can regulate NMJ development (Kraut et al 2001). With the recent development of large collections of fly lines carrying transgenic, inducible RNA interference (RNAi) constructs (Dietzl et al. 2007), it is now feasible to perform loss-of-function screens in a tissue- and temporally-selective manner (Cronin et al. 2009; Neely, et al. 2010a; Neely et al. 2010b).
Simple anatomical screens at the Drosophila NMJ can identify mutations affecting many aspects of neural development. With the development of antibodies that recognize presynaptic active zone components and postsynaptic neurotransmitter receptor subunits (Wagh et al. 2006; Marrus et al. 2004), it is now straightforward to screen for mutations affecting the formation and maintenance of active zones, their apposed receptors, and the apposition between the two (for example, Wairkar et al. 2009; Viquez et al. 2009; Graf et al. 2009; Owald et al. 2010; Fouquet et al. 2009; Johnson et al 2009; Nieratschker et al 2009; Cheng et al 2011). Furthermore, staining for these proteins also highlights the morphology of the entire synaptic terminal encompassing hundreds of individual active zones, indirectly assays the efficiency of axonal transport of active zone proteins, and reveals defects in synaptic stability leading to retraction of the presynaptic terminal from the postsynaptic specialization. We have combined such an anatomical analysis with RNAi-mediated knock down of target genes in postmitotic neurons.
Using transgenic RNAi, we screened a collection of almost two thousand genes, each of which has a clear mammalian ortholog. We found that approximately 8% of the genes screened disrupt normal NMJ development. We categorized the hits based on their phenotypes into those leading to 1) misapposition of active zones and glutamate receptors, 2) NMJ undergrowth, 3) NMJ overgrowth, 4) synaptic retractions, and 5) axonal transport defects. We then performed bioinformatics analysis to identify whether particular functional gene classes were enriched among the hits for any particular phenotype. Of note, proteasome subunits and mitotic spindle organizers were strongly enriched among the hits leading to both misapposition of active zones and synaptic retraction. The identification of these essential genes demonstrates the utility of the targeted RNAi approach. In addition to identifying functional classes of genes participating in synaptic development, this screen also identifies candidate genes whose role can be validated by a traditional loss-of-function analysis. We have performed such validation for one hit, the dynein-interacting protein NudE, and find that both targeted RNAi knock down as well as traditional loss-of-function mutants lead to defects in axonal transport and to synaptic retraction. Hence, this RNAi screen is useful for identifying functional classes of genes as well as single genes that regulate synaptic development and maintenance.
Materials and Methods
Fly strains and genetic screen
Flies were maintained at 25°C on standard fly food. Control flies were Canton S (CS) outcrossed to UAS-Dicer2; elav-Gal4 (Yao and White 1994). All the RNAi lines were obtained from the Vienna VDRC (Dietzl et al. 2007). The NudE39A mutant was a gift from Michael Goldberg (Wainman et al. 2009). Transgenic RNAi males were crossed to UAS-Dicer2; Elav-Gal4 virgin females. The lines were incubated at 25°C for 6 days and the offspring were tested for NMJ phenotypes. Immunostained 3rd instar larval NMJs from each RNAi and Dicer; Elav-Gal4 cross were analyzed by eye for morphology defects. The NMJs were scored for apposition defects, NMJ size, axonal transport and retractions. The lines that were different from wild-type animals were repeated.
Immunohistochemistry and Imaging
Third-instar larvae were dissected in PBS and fixed in either Bouin’s fixative for 5 min or 4% paraformaldehyde for 30 min. Larvae were washed with PBS containing 0.1% Triton X-100 (PBT) and blocked in 5% NGS in PBT for 30 min, followed by overnight incubation in primary antibodies in 5% NGS in PBT, three washes in PBT, incubation in secondary antibodies in 5% NGS in PBT for 45 min, three final washes in PBT, and equilibration in 70% glycerol in PBS. Samples were mounted in VectaShield (Vector, Burlingame, CA). The following primary antibodies were used: mouse α-Brp, 1:250 (Developmental Studies Hybridoma Bank), rabbit α-DGluRIII, 1:2000 (Marrus et al. 2004) and DyLight649-conjugated α-Horseradish Peroxidase (HRP) 1:1000 (Jackson ImmunoResearch). Goat Cy3-, and FITC-conjugated secondary antibodies against mouse and rabbit IgG were used at 1:1000 and were obtained from Jackson ImmunoResearch. Antibodies obtained from the Developmental Studies Hybridoma Bank were developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences of the University of Iowa, Iowa City, IA. Samples were imaged using a Nikon (Tokyo, Japan) C1 confocal microscope. All genotypes for an individual experiment were imaged at the same gain and set such that signals from the brightest genotype for a given experiment were not saturating.
Quantification of the phenotypes
Synaptic apposition was quantified as previously described (Viquez et al. 2009). Briefly, unapposed DGluRIII puncta were defined as occurring in the absence of adjacent BRP positive active zones. The total number of DGluRIII puncta per muscle 4 type Ib NMJ was counted as well as the total number of unapposed DGluRIII puncta. To calculate the percentage of unapposed DGluRIII clusters, we divided the number of unapposed DGluRIII puncta by the total number of DGluRIII clusters for each NMJ counted. The size of the NMJ was measured by counting the number of boutons on the muscle 4 type Ib NMJ of the segments A2–A3 as previously described in Schuster et al. 1996. Synaptic retractions were identified when a part or all of the presynaptic terminal was missing while postsynaptic glutamate receptors remained. We quantified the percentage of NMJs with retractions as described in Graf et al., 2011. Briefly, NMJs on the surface muscles of the segments A2–A5 were imaged at 40x magnification. The total number of assessed NMJs was counted as well as the total number of NMJs with retractions in each animal to generate the percentage of NMJs with retractions. Axonal transport defects were quantified as described in Viquez et al. 2009. Briefly, images of axons at segment A3 were taken with the same gain for all the genotypes in each experiment. The region of the nerve was defined by anti-HRP staining and then BRP signal/area was calculated using Image J. The values were normalized to BRP intensity in control larvae. Statistical analysis was performed using a paired t-test. All histograms and measurements are shown as mean ± SEM.
Gene ontology (GO) enrichment analysis
GO analysis was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7) bioinformatics resources (http://david.abcc.ncifcrf.gov/home.jsp). The list of screened genes was used as a background. Genes in each phenotypic class were analyzed separately. For each phenotypic class, a list of overrepresented GO terms was generated. For each GO term, the binomial P value was generated along with Bonferroni P value to correct for multiple testing. The list of overrepresented GO terms was manually sorted and the summary is presented in Table 1.
Table 1.
DAVID bioinformatics resources sorting of genes for each of the phenotypic classes
| GO process term | Enriched genes | p1 value | Bonferroni p value2 | fold change |
|---|---|---|---|---|
| Synaptic Apposition (61 genes) | ||||
| 1 proteasome | 17 out of 61 (27.9%) | 0.00000 | 0.00000 | 22 |
| 2 mitotic spindle organization | 14 out of 62 (23%) | 0.00000 | 0.00000 | 8 |
| 3 chaperone | 5 out of 61 (8.2%) | 0.00410 | 0.25000 | 7.2 |
| NMJ undergrowth (49 genes) | ||||
| 1 spindle organization | 10 out of 49 (20.4%) | 0.00002 | 0.01000 | 6 |
| NMJ overgrowth (16 genes) | ||||
| 1 cell differentiation | 8 out of 16 (50%) | 0.00007 | 0.02000 | 5.3 |
| Synaptic retractions (60 genes) | ||||
| 1 proteasome | 6 out of 60 (10%) | 0.00012 | 0.04500 | 8.5 |
| 2 mitotic spindle organization | 8 out of 60 (13.3%) | 0.00094 | 0.46000 | 4.7 |
| 3 Ribosome | 4 out of 60 (6.7%) | 0.00970 | 0.15000 | 7.9 |
| Axonal transport (30 genes) | ||||
| 1 cytoskeletal part | 6 out of 30 (20.0%) | 0.00410 | 0.45000 | 5 |
The p value represents the confidence with which we can conclude that the GO term is overrepresented in each of the phenotypic class compared to the collection of the screened genes.
The p value is derived from a binomial distribution and the Bonferroni p value results from multiplying this p value by the number of tests performed.
Results and discussion
A Drosophila Assay to Identify Genes Involved in Proper Formation of the NMJ
To identify genes that may participate in synapse formation and maintenance, we used transgenic RNAi to selectively knock down genes of interest in postmitotic neurons and assayed larval NMJ anatomy. Third instar larvae were dissected and stained with antibodies against the active zone component Bruchpilot (Brp) (Wagh et al. 2006) and the essential glutamate receptor subunit DGluRIII (Marrus et al. 2004). Many aspects of synaptic development, growth, and maintenance can be analyzed by visualizing active zones and glutamate receptors. In wild type larvae, each active zone is apposed to a cluster of glutamate receptors (Marrus and DiAntonio 2004), and so mutants can be identified in which there is a defect in active zones, glutamate receptors, or their apposition. Visualizing these synaptic components also highlights the morphology of the entire neuromuscular junction, making identification of both overgrowth and undergrowth mutants possible. Bruchpilot is transported to the synapse from the cell body, and accumulation of Bruchpilot within the motoneuron is consistent with defects in axonal transport (Pack-Chung et al. 2007). Finally, the presynaptic terminal is maintained opposite the postsynapse throughout larval life. The presence of the postsynaptic specialization without a presynaptic terminal is indicative of presynaptic retraction and defects in synaptic terminal stability (Eaton et al. 2002). For each line tested we scored synaptic apposition, synaptic terminal overgrowth, synaptic terminal undergrowth, Brp accumulation as a proxy for axonal transport, and synaptic terminal retractions.
Transgenic RNAi lines were obtained from the Vienna Drosophila RNAi Center (Dietzl et al. 2007). All genes tested in the screen have mammalian orthologs as defined by NCBI’s HomoloGene. We chose this subset in order to enrich for genes with conserved function, while recognizing that important but non-conserved genes will be missed (Rohrbough et al. 2007). The expression of each RNAi construct is controlled by the upstream activating sequence (UAS), which is activated in the presence of Gal4 (Duffy 2002). We drove expression with the postmitotic pan-neuronal Elav-Gal4 driver and co-expressed Dicer2 to increase the efficiency of RNAi knockdown (Dietzl et al. 2007). We screened a total of 2092 transgenic RNAi lines corresponding to 1970 unique genes. The gene list (Figure S7) is a non-random representation of the genome because particular functional groups are more conserved between flies and mammals and because RNAi lines are not available for all genes. In order to adjust for gene enrichment in the initial gene set, subsequent analyses used the tested genes as a comparison group rather than the entire Drosophila genome. For each RNAi line that gave an apparent phenotype, the analysis of the NMJ was repeated. The initial screen yielded 304 lines that were re-tested. Among these, 146 had weak phenotypes and/or did not repeat. The remaining 158 lines have a consistent phenotype and were included in subsequent analysis.
Among the genes tested were several with known roles in the development and/or maintenance of the NMJ. These genes include ATG1, Smad, Alpha spectrin, Kinesin, ether a go-go (Eag), dLiprin-alpha and Brp. These lines allowed us to test whether the screen could identify previously described phenotypes following knockdown of important synaptic proteins. Following knock down we observed synaptic apposition defects (ATG1 and dLiprin-alpha), NMJ undergrowth (Medea and ATG1), NMJ overgrowth (Eag), and synaptic retractions and axonal transport defects (Alpha spectrin and Kinesin). These phenotypes are consistent with those described in traditional loss-of-function mutants for these genes (Wairkar et al. 2009; Mccabe 2003; Budnik, Zhong, and Wu 1990; Pielage et al. 2005; Hurd and Saxton 1996; Owald et al. 2010). Knock down of Bruchpilot leads to a dramatic decrease in the levels of Brp, directly demonstrating the efficacy of the RNAi in this case. These findings indicate that this RNAi screen is sensitive enough to pick up defects in synaptic development and maintenance (Table S1). While RNAi presents many advantages for this screen, there are also limitations. A phenotype may be due to an off target effect, and so the demonstration of a role for any particular gene requires subsequent analysis. In addition, knockdown of any particular line may be inefficient, and so a negative result cannot be interpreted. For example, our screen included lines targeting the PP2A subunit Well-rounded, the protein tyrosine phosphatase LAR, and the membrane trafficking protein Ema, each of which has a known morphological phenotype at the NMJ (Viquez et al. 2009, Kaufmann et al. 2002, Kim et al. 2010), however none were identified in the screen.
Phenotypic Categorization of the Hits
The screen generated 158 hits that had an NMJ phenotype consistently different from UAS-Dcr2; elav-Gal4 control (Table S1). To separate the genes according to their potential role in synapse development, we divided them into six phenotypic classes: synaptic apposition defects, NMJ undergrowth, NMJ overgrowth, synaptic retractions, axonal transport defects and other. For each RNAi line, the phenotype can fall into one or more phenotypic class.
Synaptic apposition defects: In these lines, a fraction of the glutamate receptor clusters are not apposed to a Brp-positive active zone (Figure 1). This could reflect either the absence of an active zone or the presence of an aberrant active zone missing the important scaffolding protein Brp. Often the apposition defect is more severe in Type Is than in Type Ib synapses. This could reflect a true difference in the biology between these two classes of motoneurons or a difference in the levels of Gal4 expression.
NMJ undergrowth: This phenotype is defined by fewer synaptic boutons and actives zones per NMJ (Figure 2A-B). Some of these lines have an increase in the size of individual synaptic boutons.
NMJ overgrowth: The NMJs in this class are expanded and the bouton size is typically decreased (Figure 2C). This phenotype is characterized by an increased number of synaptic boutons per NMJ.
Synaptic retractions: Here, part or all of the presynaptic terminal is missing while postsynaptic glutamate receptors remain (Figure 4). These differ from the synaptic apposition mutants because presynaptic boutons or branches are lost in a distal to proximal manner within the NMJ. With synaptic apposition mutants, unapposted glutamate receptors are present in a salt-and-pepper pattern throughout the NMJ. With synaptic retractions, the frequency and severity of retractions is often higher in the posterior segments of the larvae. The glutamate receptor staining is denser in the retracted branches, consistent with previously published description of the phenotype (Pielage et al. 2011). Phenotypes can range from the retraction of single boutons to the loss of the entire presynaptic terminal.
Axonal transport defects: Lines are placed in this group when Bruchpilot protein accumulates inappropriately in the axons (Figure 5). Accumulation of synaptic proteins in axons is a hallmark of axonal transport mutants (Hurd and Saxton 1996), but it can occur for other reasons such as premature T-bar assembly in motor axons (Johnson et al 2009; Nieratschker et al. 2009). Since axonal transport defects are the most common cause of this phenotype, this class will be referred to here as “Axonal transport” defects. However, for an individual mutant further experiments are required to define the underlying cell biological mechanism.
Other: Hits that did not fall into any of the above phenotypic classes were grouped together into class six. This class consists of abnormal bouton morphology, lower Brp intensity, abnormal glutamate receptor staining, or disorganized active zones. Since these phenotypes are diverse, they were excluded from further analysis. However, they include interesting candidates that may participate in the proper formation of the NMJ and so are included in the table S1.
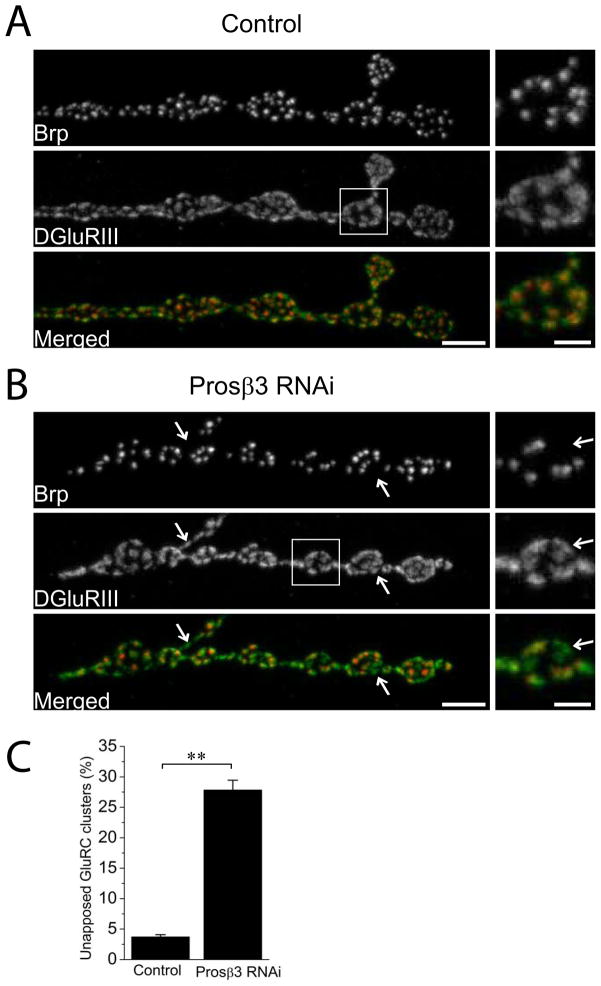
Figure 1. Neuron-driven RNAi targeting of a proteasomal subunit, Prosβ3, disrupts synaptic apposition.
Sample confocal images of muscle 4 NMJs of (A) Control (UAS-DCR2/+; Elav-Gal4/+) and (B) Prosβ3 RNAi knockdown (UAS-DCR2/+; UAS- Prosβ3 RNAi/+; Elav-Gal4/+ ) third instar larvae stained for the presynaptic active zone protein Brp (red) and the postsynaptic glutamate receptor DGluRIII (green). Inset shows a magnified image of boxed region to highlight the apposition of active zones and glutamate receptor clusters. Muscle 4 Type Ib NMJs from the genotypes in (A) and (B) were quantified for the percentage of unopposed DGluRIII clusters in C. Note that neuronal knockdown of Prosβ3 leads to significant increase in unapposed glutamate receptors (arrows); ** p<0.001; n = 16 for both genotypes. Error bars represent SEM. Scale bars represent 5 μm in lower magnification images and 2 μm in higher magnification inset.
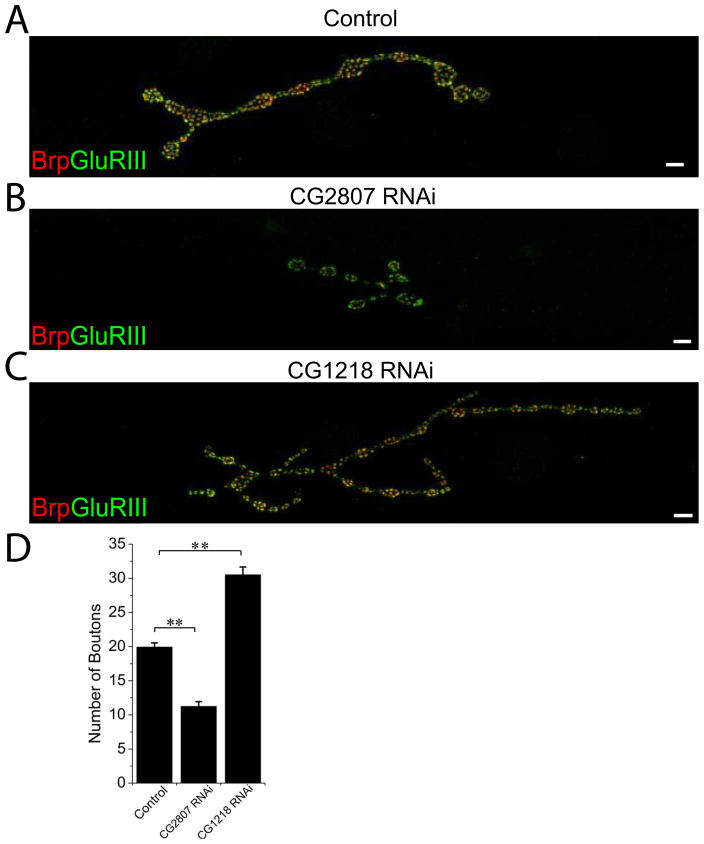
Figure 2. Neuronal RNAi knockdown of gene expression can positively or negatively affect synaptic growth.
Sample confocal images of muscle 4 NMJs of (A) Control (UAS-DCR2/+; Elav-Gal4/+) (B) CG2807 RNAi (UAS-DCR2/+; UAS-CG2807 RNAi/+; Elav-Gal4/+) and (C) CG1218 RNAi (UAS-DCR2/+; UAS-CG1218 RNAi/+; Elav-Gal4/+) third instar larvae stained for the presynaptic active zone protein Brp (red) and the postsynaptic glutamate receptor DGluRIII (green). Knockdown of spliceosome factor encoded by CG2807 results in severe NMJ undergrowth while depletion of zinc finger domain-containing uncharacterized protein encoded by CG1218 causes an increase in NMJ size. Number of boutons at Type Ib muscle 4 NMJs were quantified for the genotypes in (A)–(C) in D; n=31 for control, n=19 for CG2807 RNAi and n=19 for CG1218 RNAi; **p<0.001 both of the genotypes compared to control; error bars represent SEM. Note that the overgrown NMJ contains more boutons that are smaller in size. Scale bars represent 10 μm.
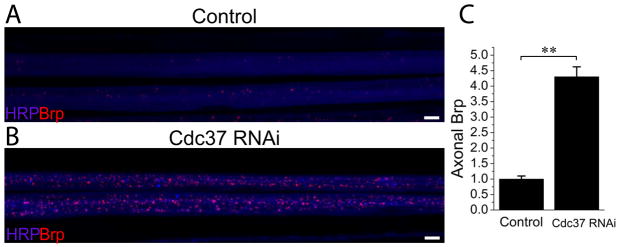
Figure 4. Neuronal RNAi knockdown of Cdc37 expression causes accumulations of synaptic protein Brp in axons.
Sample confocal images of two axonal bundles of (A) Control (UAS-DCR2/+; Elav-Gal4/+) and (B) Cdc37 RNAi (UAS-DCR2/+; UAS-Cdc37 RNAi/+; Elav-Gal4/+) third instar larvae stained for the presynaptic active zone protein Brp (red) and the neuronal membranes with HRP (blue). BRP intensity per axon area was quantified for both of the genotypes and normalized to control in C; n=10 for both genotypes, **p<0.001; error bars represent SEM. Note the presence of more numerous and larger Brp puncta in Cdc37 RNAi knockdown case. Scale bars represent 5 μm.
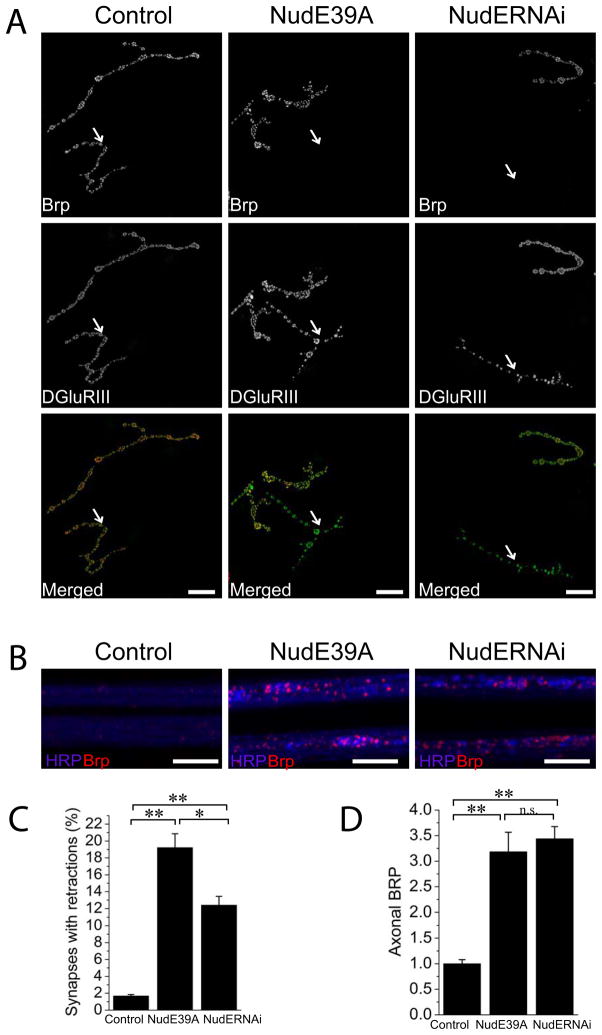
Figure 5. NudE39A genetic excision mutant replicates synaptic apposition and axonal transport defects seen in neuronal RNAi knockdown of NudE.
(A) Sample confocal images of muscle 4 NMJs of control (UAS-DCR2/+; Elav-Gal4/+), NudE RNAi (UAS-DCR2/+; UAS-NudE RNAi/+; Elav-Gal4/+) and in NudE39A excision mutant third instar larvae stained for the presynaptic active zone protein Brp (red) and the postsynaptic receptor DGluRIII (green). Chosen muscle 4 NMJs contain both Type Is and Type Ib synapses. Type Is synapses are labeled with arrows and are retracted in both NudE RNAi knockdown and NudE39A mutant animals. Scale bars represent 10 μm.
(B) Sample confocal image of two axonal bundles in control (UAS-DCR2/+; Elav-Gal4/+), neuron-driven RNAi against NudE and in NudE39A excision mutant third instar larvae stained for the presynaptic active zone protein Brp (red) and the neuronal membranes with HRP (blue). Note Brp accumulations in NudE RNAi knock down and in NudE39A mutant nerves. Scale bars represent 10 μm.
(C) Histogram showing quantification of the percentage of the NMJs with retractions per animal in genotypes from (A); n=14 larvae for all genotypes; *p<0.01, ** p<0.001.
(D) Histogram showing quantification of the BRP intensity per axon area in genotypes from (A) normalized to the BRP intensity in control axons; n= 13 for all genotypes; **p<0.001, n.s. – not significant; error bars represent SEM.
Gene set enrichment analysis of the phenotypic classes
We performed gene ontology analysis using the DAVID bioinformatics resources tool (Huang et al. 2009) to test for a link between each of the phenotypic classes and genes with a shared molecular function. DAVID analysis allows categorization of hits not only by their gene ontology (GO term) process but also incorporates KEGG pathway analysis and returns a list of enriched annotation terms. Using gene ontology (GO) annotations, our candidate hits were classified according to their predicted biological processes (BP), molecular functions (MF), and cellular components (CC). We performed this analysis on the genes belonging to each phenotypic class. Because many genes have multiple gene ontology (GO) process terms assigned to them, the genes in each phenotypic class were grouped into multiple GO process terms, many of which are parents or daughters of other terms. A summary of some of the major GO terms is presented in Table 1 and we concentrate only on the most significant ones for each phenotypic group.
To identify significant enrichment, a binomial P value was obtained based on the probability of observing the number of positively selected genes in each family compared to their representation within the collection of screened genes. Since this value is generated multiple times during the analysis, we used Bonferroni correction. Many of the generated categories are not independent because of the hierarchical nature of the GO terms, so the Bonferroni test is overly stringent for this analysis. Nonetheless, some of the identified categories are highly significant for all 3 parameters – binomial P value, Bonferroni P value and the fold enrichment.
Different genes are likely involved in different aspects of NMJ development. We analyzed each phenotypic class individually to identify functional annotation term enrichment. First, we analyzed the synaptic apposition phenotypic class. This analysis yielded a group of 17 genes involved in proteasomal degradation process (p=7.0×10−20), Table S2. These proteasome subunits have a 22-fold enrichment compared to the background list (the genes screened). An example of the synaptic apposition phenotype due to RNAi knockdown of a proteasome subunit Prosβ3 is presented in Figure 1. Quantification of the percentage of unapposed DGluRIII puncta in the Prosβ3 knockdown animals on muscle 4 showed a 9-fold increase in the unapposed DGluRIII clusters (p<0.001; n = 16 NMJs from at least 6 animals for each genotype). The identification of so many proteasome subunits strongly suggests that it is inhibition of the proteasome complex that promotes synaptic misapposition, rather than off-target effects or novel functions for the genes. Proteasomal degradation is a fundamental cellular process and its inhibition in other systems causes pleiotropic effects (Smalle et al. 2003; Lambertson et al 1999), so we were surprised that most of the proteasome subunits found in the screen fall into a single phenotypic class – synaptic apposition. This suggests that within the motoneuron a particular cell biological process, or potentially a single substrate, requires a functional proteasome system to form or maintain a normal active zone. This phenotype is different than that described for loss of the ubiquitin ligase highwire or components of the APC ligase complex (DiAntonio et al. 2001; Tian et al. 2011; van Roessel et al. 2004), indicating that this method of inhibiting the ubiquitin proteasome system identifies a unique role for this process. In mammalian systems, proteasome subunits influence synaptic function and are localized to synapses, with their best-described role in the post-synaptic maintenance of long-term plasticity (Bingol and Sheng 2011). Our findings highlight the potential importance of proteasome function for active zone development and maintenance in the presynaptic compartment.
Within the synaptic apposition group, another highly significant (p=2.6 × 10−9) GO term category was mitotic spindle organization, with 14 genes (Table 1) and 8-fold enrichment. These genes are involved in organization of the mitotic spindle and classical mutants for these genes are often lethal (Table S2). Our study involves post-mitotic knock down of these genes, suggesting that following cell division, this group of molecules is used again to promote the formation or maintenance of normal active zones opposite postsynaptic receptors. Organization of the mitotic spindle involves regulation of microtubules. Hence, the proper alignment of Brp-positive active zones opposite glutamate receptor clusters also likely relies on the organization of microtubules.
The second phenotypic class analyzed was NMJ undergrowth. Functional annotation of the genes in this group is diverse. Among them, we identified a splicing factor encoded by CG2807 which had decreased number of boutons (Figure 2B, D, p<0.001; n=19 NMJs from at least 6 animals for each genotype). For NMJ undergrowth, a group of ten genes involved in cytoskeletal and mitotic spindle organization was significantly enriched (p=1.5×10−5), Table S3. This group has some overlap with those identified in the synaptic apposition group, suggesting that microtubule organization may participate in both the growth of the entire synaptic structure as well as the organization of the many individual synapses within the terminal. A few microtubule-regulating proteins were previously demonstrated to promote synaptic terminal growth (Pennetta et al. 2002; Roos et al. 2000; Ruiz-Canada et al. 2004). Our analysis demonstrates that there are many genes with similar function that are involved in this process.
For the NMJ overgrowth phenotype, there was significant enrichment of a group of eight genes annotated as cell differentiation genes (p = 6.8×10−5). Some of these genes are known to affect the development of the NMJ, such as ether-a-go-go (Eag) (Budnik et al. 1990), while others are new targets (Table S4). Cell differentiation genes help the cell establish its identity, but the molecular functions of such genes are diverse and so do not immediately suggest a unifying cell biological process required to restrain synaptic terminal growth. As an example of the genes in this category, in Figure 2C, D we present the phenotype of RNAi knockdown of a zinc finger domain-containing uncharacterized protein encoded by CG1218 that has an ~50% increase in the number of boutons compared to control (p<0.001; n=19 NMJs from 6 animals).
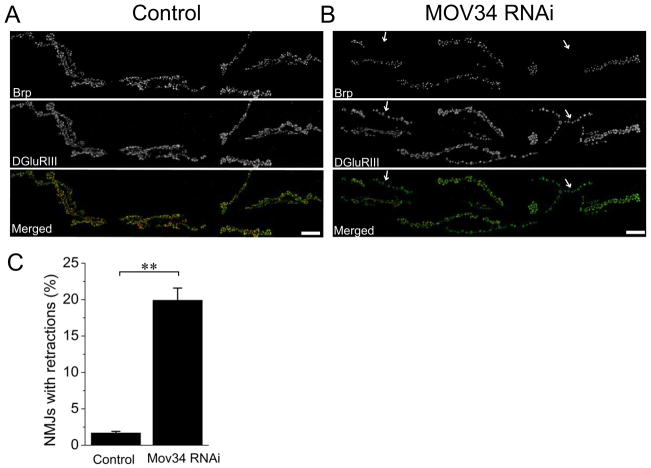
For synaptic retractions, there are two enriched groups including six proteasome subunits (p=4.3×10−4) and a group of five genes (p=4.5×10−3) annotated with mitotic spindle organization (Table S5). The profile is very similar to that found for synaptic apposition defects where the proteasome subunits and mitotic spindle organizers were identified. However, the gene number in each annotation term is much smaller for the synaptic retraction class. Mov34, one of such genes has retractions on 20% of its NMJs (Figure 3, p<0.001; n=15 animals for control and n=13 animals for Mov34 RNAi, over 100 NMJs were analyzed for each animal). This gene is annotated as being involved in microtubule organization but its mammalian homolog is a proteasome subunit and it has been identified as having both synaptic retractions and synaptic apposition defects when depleted by RNAi (Table S2 and S5). These similar gene enrichment profiles suggest that the retraction phenotype may be a subclass of the apposition phenotype. For example, severe apposition defects could trigger synaptic retractions. However, there are a number of genes with synaptic retraction defects that do not also have synaptic apposition phenotypes. Hence, apposition defects are not a necessary prerequisite for retraction of the synaptic terminal. These findings suggest that both apposition-dependent and apposition-independent mechanisms can promote synaptic terminal retractions.
Figure 3. Neuron-driven RNAi against Mov34 disrupts synaptic maintenance.
Sample confocal images of muscle 6/7 NMJs of (A) Control (UAS-DCR2/+; Elav-Gal4/+) and (B) Mov34 RNAi (UAS-DCR2/+; UAS-Mov34 RNAi/+; Elav-Gal4/+) third instar larvae stained for the presynaptic active zone protein Brp (red) and the postsynaptic glutamate receptor DGluRIII (green). In control NMJ, all branches labeled with DGluRIII are aligned with neuronal compartment labeled with Brp, while neuronal knockdown of Mov34 results in retracted branches (arrows). Percentage of NMJs with retractions per animal for both genotypes were quantified in C; n=15 for control and n=12 for Mov34 RNAi ; ** p<0.001; error bars represent SEM. Scale bars represent 20 μm.
We took advantage of an RNAi line with both synaptic retraction and apposition phenotypes to investigate the dose dependence of the RNAi system. For this, we took advantage of the temperature sensitivity of Gal4 expression (Duffy 2002) and compared knockdown of the proteasomal subunit prosα7 raised at 18°C (weak Gal4 expression) to the same line targeting prosα7 raised at 25°C (stronger Gal4 expression). We saw a decrease in both the apposition phenotype (pros α7 at 25°C: α7.2 ± 2%; pros α7 at 18°C: 29.4 ± 1.6%, p<0.01, n=10 NMJs from at least 4 animals for each genotype) as well as in the frequency of severe retractions (pros α7 at 25°C: 6.3 ± 0.7%; pros α7 at 18°C: 3.4 ± 1.1%, p<0.05, n=4 animals at 18°C and n=10 animals at 25°C, over 100 NMJs were analyzed per animal) at 18°C. These findings highlight that the severity of RNAi phenotypes are dose dependent and can be modified by the activity of the Gal4 driver.
The last phenotypic class analyzed was Axonal Transport Defect. The genes in this phenotypic group included a cell division cycle 37 (Cdc37) homolog with a 4-fold increase in axonal Brp accumulation (figure 4, p<0.001, n=9 animals for control and n=10 animals for Cdc37 RNAi, multiple nerves were analyzed for each animal). However, the most significant gene enrichment cluster (p=0.0041) was “cytoskeletal part,” which includes genes involved in maintaining the cytoskeleton, Table S6. Many of these proteins have microtubule binding properties, consistent with the movement of molecular motors along microtubules during axonal transport (Pack-Chung et al. 2007; Martin et al. 1999). Five of the six members of this group also have a synaptic retractions phenotype, consistent with previous findings that impaired axonal transport can lead to retractions (Eaton et al. 2002). However, our data indicate that impaired axonal transport is not always associated with a retraction phenotype (11 out of 32 RNAi lines with axonal transport defects did not have synaptic retractions (Table S1)). Conversely, retractions occurred in the absence of an axon transport defects 46 out of 67 times (Table S1). These findings suggest that 1) impaired axonal transport can impair synaptic stability, however 2) axon transport-independent mechanisms can also disrupt synaptic stability and 3) there is some unexplained selectivity of axon transport mutants such that only a subset lead to synaptic retractions.
Some genes belong to more than one phenotypic group, and some of the clusters that are enriched for different phenotypic classes may include overlapping genes. This is consistent with multiple functions for these genes, or for an overlap within our phenotypic groups. For example, severe synaptic apposition defects or defects in axonal transport could trigger retractions. While this may be true in some cases, in no case is one phenotypic group exclusively a subset of another. That is, every phenotype can occur in the absence of each of the other, suggesting that at least some distinct mechanisms are at play for each phenotypic class.
A role for NudE in synaptic maintenance
A large-scale RNAi screen is useful for identifying functional groups of genes that participate in a process of interest. However for any individual hit, RNAi is not sufficient to demonstrate a functional role for the gene because the phenotype may be due to an off-target effect. However, the list of hits does generate interesting candidates that can be validated by analysis of traditional loss-of-function mutants. We have tested the utility of this approach for a single hit from the screen, NudE.
RNAi knock down of NudE produces synaptic retraction and axonal transport phenotypes (Table S1). NudE encodes a Dynein adaptor protein (Kardon and Vale 2009). Mutations in other components of the dynein complex disrupt axon transport in the fly (Martin et al. 1999). To test whether the phenotypes are due to knock down of NudE rather than an off-target effect, we analyzed a traditional genetic mutant generated via imprecise excision of a P element, NudE39A(Wainman et al. 2009). This mutation displays an axonal transport defect that is very similar to that seen with RNAi knockdown (figure 5D, p=0.57 between traditional and RNAi mutants; n=13 animals per genotype, multiple nerves were analyzed for each animal). The excision mutant also displays a retraction phenotype, however, the frequency of retractions is higher than in the RNAi knockdown (figure 5C, p<0.001; n=14 animals per genotype), Figure 5. These findings are consistent with previous work demonstrating that the dynein complex is required for normal axonal transport and that the dynactin complex, which activates dynein, is required for stability of the Drosophila NMJ (Eaton et al. 2002).
The findings with NudE demonstrate that candidates identified in the RNAi screen can be validated with traditional mutants. However, we suggest that an equally important benefit of tissue- and temporally-selective RNAi screens is that it allows us to expand our understanding of the functions of essential genes. Traditional genetics limits our ability to probe the function of genes required early in development or in dividing cells. By restricting our genetic perturbation to postmitotic neurons we have identified important roles for the proteasome and for microtubule spindle organizers in synaptic development and maintenance.
Supplementary Material
Highlights.
In vivo screen in Drosophila identifies novel players in synaptic development and maintenance
Gene ontology analysis identifies overrepresented GO terms for different phenotypes
Proteasome subunits and mitotic spindle organizers are overrepresented in synaptic apposition phenotype group
NudE knock down causes axonal transport and retractions and the phenotype is replicated by a classical genetic mutant.
Acknowledgments
We would like to thank the members of the DiAntonio lab for helpful discussions as well as Xiaolu Sun and Emily Royse for their assistance during screening. We also thank Dr. Li-Wei Chang and Devjanee Swain Lenz for their help with data analysis and Dr. Michael Goldberg for providing NudE39A mutant flies. We also thank the Bloomington Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank for fly strains and antibodies. This work was supported by a grant from the NIH (NS043171 and ARRA supplement) to A.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banovic Daniel, Khorramshahi Omid, Owald David, Wichmann Carolin, Riedt Tamara, Fouquet Wernher, Tian Rui, Sigrist Stephan J, Aberle Hermann. Drosophila neuroligin 1 promotes growth and postsynaptic differentiation at glutamatergic neuromuscular junctions. Neuron. 2010 Jun 10;66(5):724–738. doi: 10.1016/j.neuron.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Bingol Baris, Sheng Morgan. Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron. 2011 Jan 13;69(1):22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Budnik V, Zhong Y, Wu CF. Morphological plasticity of motor axons in Drosophila mutants with altered excitability. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 1990 Nov;10(11):3754–3768. doi: 10.1523/JNEUROSCI.10-11-03754.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Ling, Locke Cody, Davis Graeme W. S6 kinase localizes to the presynaptic active zone and functions with PDK1 to control synapse development. The Journal of Cell Biology. 2011 Sep 19;194(6):921–935. doi: 10.1083/jcb.201101042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins Catherine A, DiAntonio Aaron. Synaptic development: insights from Drosophila. Current Opinion in Neurobiology. 2007 Feb;17(1):35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Cronin Shane JF, Nehme Nadine T, Limmer Stefanie, Liegeois Samuel, Andrew Pospisilik J, Schramek Daniel, Leibbrandt Andreas, de Matos Simoes Ricardo, Gruber Susanne, Puc Urszula, Ebersberger Ingo, Zoranovic Tamara, Gregory Neely G, von Haeseler Arndt, Ferrandon Dominique, Penninger Josef M. In vivo genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science (New York, NY) 2009 Jul 17;325(5938):340–343. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001 Jul 26;412(6845):449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- Dickman Dion K, Davis Graeme W. The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science (New York, NY) 2009 Nov 20;326(5956):1127–1130. doi: 10.1126/science.1179685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl Georg, Chen Doris, Schnorrer Frank, Su Kuan-Chung, Barinova Yulia, Fellner Michaela, Gasser Beate, Kinsey Kaolin, Oppel Silvia, Scheiblauer Susanne, Couto Africa, Marra Vincent, Keleman Krystyna, Dickson Barry J. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007 Jul 12;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Duffy Joseph B. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis (New York, NY: 2000) 2002 Oct;34(1–2):1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Eaton Benjamin A, Fetter Richard D, Davis Graeme W. Dynactin is necessary for synapse stabilization. Neuron. 2002 May 30;34(5):729–741. doi: 10.1016/s0896-6273(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Fouquet Wernher, Owald David, Wichmann Carolin, Mertel Sara, Depner Harald, Dyba Marcus, Hallermann Stefan, Kittel Robert J, Eimer Stefan, Sigrist Stephan J. Maturation of active zone assembly by Drosophila Bruchpilot. The Journal of Cell Biology. 2009 Jul 13;186(1):129–145. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf Ethan R, Heerssen Heather M, Wright Christina M, Davis Graeme W, DiAntonio Aaron. Stathmin is Required for Stability of the Drosophila Neuromuscular Junction. The Journal of Neuroscience. 2011 Oct 19;31(42):15026–15034. doi: 10.1523/JNEUROSCI.2024-11.2011. 10.1523/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf Ethan R, Daniels Richard W, Burgess Robert W, Schwarz Thomas L, DiAntonio Aaron. Rab3 dynamically controls protein composition at active zones. Neuron. 2009 Dec 10;64(5):663–677. doi: 10.1016/j.neuron.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Da Wei, Sherman Brad T, Lempicki Richard A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996 Nov;144(3):1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Ervin L, Fetter Richard D, Davis Graeme W. Negative Regulation of Active Zone Assembly by a Newly Identified SR Protein Kinase. PLoS Biol. 2009;7(9):e1000193. doi: 10.1371/journal.pbio.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon Julia R, Vale Ronald D. Regulators of the cytoplasmic dynein motor. Nature Reviews Molecular Cell Biology. 2009 Dec;10(12):854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann N, DeProto J, Ranjan R, Wan H, Van Vactor D. Drosophila liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis. Neuron. 2002 Mar 28;34(1):27–38. doi: 10.1016/s0896-6273(02)00643-8. [DOI] [PubMed] [Google Scholar]
- Kim Sungsu, Wairkar Yogesh P, Daniels Richard W, DiAntonio Aaron. The novel endosomal membrane protein Ema interacts with the class C Vps-HOPS complex to promote endosomal maturation. J Cell Biol. 2010 Mar 8;188(5):717–734. doi: 10.1083/jcb.200911126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R, Menon K, Zinn K. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Current Biology: CB. 2001 Mar 20;11(6):417–430. doi: 10.1016/s0960-9822(01)00124-5. [DOI] [PubMed] [Google Scholar]
- Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics. 1999 Sep;153(1):69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999 Mar;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Liebl Faith LW, Werner Kristen M, Sheng Qi, Karr Julie E, McCabe Brian D, Featherstone David E. Genome-wide P-element screen for Drosophila synaptogenesis mutants. Journal of Neurobiology. 2006 Mar;66(4):332–347. doi: 10.1002/neu.20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrus Scott B, DiAntonio Aaron. Preferential localization of glutamate receptors opposite sites of high presynaptic release. Current Biology: CB. 2004 Jun 8;14(11):924–931. doi: 10.1016/j.cub.2004.05.047. [DOI] [PubMed] [Google Scholar]
- Marrus Scott B, Portman Scott L, Allen Marcus J, Moffat Kevin G, DiAntonio Aaron. Differential Localization of Glutamate Receptor Subunits at the Drosophila Neuromuscular Junction. The Journal of Neuroscience. 2004 Feb 11;24(6):1406–1415. doi: 10.1523/JNEUROSCI.1575-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Iyadurai SJ, Gassman A, Gindhart JG, Jr, Hays TS, Saxton WM. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Molecular Biology of the Cell. 1999 Nov;10(11):3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccabe B. The BMP Homolog Gbb Provides a Retrograde Signal that Regulates Synaptic Growth at the Drosophila Neuromuscular Junction. Neuron. 2003 Jul;39(2):241–254. doi: 10.1016/s0896-6273(03)00426-4. [DOI] [PubMed] [Google Scholar]
- Gregory Neely G, Kubasend Keiji, Cammarato Anthony, Isobe Kazuya, Amann Sabine, Zhang Liyong, Murata Mitsushige, Elmén Lisa, Gupta Vaijayanti, Arora Suchir, Sarangi Rinku, Dan Debasis, Fujisawa Susumu, Usami Takako, Xia Cui-ping, Keene Alex C, Alayari Nakissa N, Yamakawa Hiroyuki, Elling Ulrich, Berger Christian, Novatchkova Maria, Koglgruber Rubina, Fukuda Keiichi, Nishina Hiroshi, Isobe Mitsuaki, Andrew Pospisilik J, Imai Yumiko, Pfeufer Arne, Hicks Andrew A, Pramstaller Peter P, Subramaniam Sai, Kimura Akinori, Ocorr Karen, Bodmersend Rolf, Penninger Josef M. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010a Apr 2;141(1):142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory Neely G, Hess Andreas, Costigan Michael, Keene Alex C, Goulas Spyros, Langeslag Michiel, Griffin Robert S, Belfer Inna, Dai Feng, Smith Shad B, Diatchenko Luda, Gupta Vaijayanti, Xia Cui-ping, Amann Sabina, Kreitz Silke, Heindl-Erdmann Cornelia, Wolz Susanne, Ly Cindy V, Arora Suchir, Sarangi Rinku, Dan Debasis, Novatchkova Maria, Rosenzweig Mark, Gibson Dustin G, Truong Darwin, Schramek Daniel, Zoranovic Tamara, Cronin Shane JF, Angjeli Belinda, Brune Kay, Dietzl Georg, Maixner William, Meixner Arabella, Thomas Winston, Andrew Pospisilik J, Alenius Mattias, Kress Michaela, Subramaniam Sai, Garrity Paul A, Bellen Hugo J, Woolfsend Clifford J, Penninger Josef M. A Genome-wide Drosophila Screen for Heat Nociception Identifies α2δ3 as an Evolutionarily Conserved Pain Gene. Cell. 2010b Nov 12;143(4):628–638. doi: 10.1016/j.cell.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieratschker Vanessa, Schubert Alice, Jauch Mandy, Bock Nicole, Bucher Daniel, Dippacher Sonja, Krohne Georg, Asan Esther, Buchner Sigrid, Buchner Erich. Bruchpilot in ribbon-like axonal agglomerates, behavioral defects, and early death in SRPK79D kinase mutants of Drosophila. PLoS Genetics. 2009 Oct;5(10):e1000700. doi: 10.1371/journal.pgen.1000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owald David, Fouquet Wernher, Schmidt Manuela, Wichmann Carolin, Mertel Sara, Depner Harald, Christiansen Frauke, Zube Christina, Quentin Christine, Körner Jorg, Urlaub Henning, Mechtler Karl, Sigrist Stephan J. A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. The Journal of Cell Biology. 2010 Feb 22;188(4):565–579. doi: 10.1083/jcb.200908055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack-Chung Eunju, Kurshan Peri T, Dickman Dion K, Schwarz Thomas L. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat Neurosci. 2007;10(8):980–989. doi: 10.1038/nn1936. [DOI] [PubMed] [Google Scholar]
- Pennetta Giuseppa, Hiesinger Peter Robin, Fabian-Fine Ruth, Meinertzhagen Ian A, Bellen Hugo J. Drosophila VAP-33A directs bouton formation at neuromuscular junctions in a dosage-dependent manner. Neuron. 2002 Jul 18;35(2):291–306. doi: 10.1016/s0896-6273(02)00769-9. [DOI] [PubMed] [Google Scholar]
- Pielage Jan, Bulat Victoria, Bradley Zuchero J, Fetter Richard D, Davis Graeme W. Hts/Adducin controls synaptic elaboration and elimination. Neuron. 2011 Mar 24;69(6):1114–1131. doi: 10.1016/j.neuron.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage Jan, Fetter Richard D, Davis Graeme W. Presynaptic spectrin is essential for synapse stabilization. Current Biology: CB. 2005 May 24;15(10):918–928. doi: 10.1016/j.cub.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Reeve Simon P, Lin Xinda, Sahin Bahar H, Jiang Fangfang, Yao Aiyu, Liu Zhihua, Zhi Hui, Broadie Kendal, Li Wei, Giangrande Angela, Hassan Bassem A, Zhang Yong Q. Mutational Analysis Establishes a Critical Role for the N Terminus of Fragile X Mental Retardation Protein FMRP. The Journal of Neuroscience. 2008 Mar 19;28(12):3221–3226. doi: 10.1523/JNEUROSCI.5528-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roessel, Peter David A, Elliott Iain M, Robinson Andreas Prokop, Brand Andrea H. Independent Regulation of Synaptic Size and Activity by the Anaphase-Promoting Complex. Cell. 2004 Nov 24;119(5):707–718. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Rohrbough Jeffrey, Rushton Emma, Woodruff Elvin, III, Fergestad Tim, Vigneswaran Krishanthan, Broadie Kendal. Presynaptic establishment of the synaptic cleft extracellular matrix is required for post-synaptic differentiation. Genes and Development. 2007 Oct 15;21(20):2607–28. doi: 10.1101/gad.1574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos Jack, Hummel Thomas, Ng Norman, Klambt Christian, Davis Graeme W. Drosophila Futsch Regulates Synaptic Microtubule Organization and Is Necessary for Synaptic Growth. Neuron. 2000 May;26(2):371–382. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada Catalina, Ashley James, Moeckel-Cole Stephanie, Drier Eric, Yin Jerry, Budnik Vivian. New synaptic bouton formation is disrupted by misregulation of microtubule stability in aPKC mutants. Neuron. 2004 May 27;42(4):567–580. doi: 10.1016/s0896-6273(04)00255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Smalle Jan, Kurepa Jasmina, Yang Peizhen, Emborg Thomas J, Babiychuk Elena, Kushnir Sergei, Vierstra Richard D. The Pleiotropic Role of the 26S Proteasome Subunit RPN10 in Arabidopsis Growth and Development Supports a Substrate-Specific Function in Abscisic Acid Signaling. The Plant Cell. 2003 Apr;15(4):965–980. doi: 10.1105/tpc.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Xiaolin, Li Jing, Valakh Vera, DiAntonio Aaron, Wu Chunlai. Drosophila Rae1 controls the abundance of the ubiquitin ligase Highwire in post-mitotic neurons. Nature Neuroscience. 2011 doi: 10.1038/nn.2922. advance online publication. http://dx.doi.org/10.1038/nn.2922. [DOI] [PMC free article] [PubMed]
- Viquez Natasha M, Füger Petra, Valakh Vera, Daniels Richard W, Rasse Tobias M, DiAntonio Aaron. PP2A and GSK-3beta act antagonistically to regulate active zone development. The Journal of Neuroscience. 2009 Sep 16;29(37):11484–11494. doi: 10.1523/JNEUROSCI.5584-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh Dhananjay A, Rasse Tobias M, Asan Esther, Hofbauer Alois, Schwenkert Isabell, Darrbeck Heike, Buchner Sigrid, Dabauvalle Marie-Christine, Schmidt Manuela, Qin Gang, Wichmann Carolin, Kittel Robert, Sigrist Stephan J, Buchner Erich. Bruchpilot, a Protein with Homology to ELKS/CAST, Is Required for Structural Integrity and Function of Synaptic Active Zones in Drosophila. Neuron. 2006 Mar 16;49(6):833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Wainman Alan, Creque Jacklyn, Williams Byron, Williams Erika V, Bonaccorsi Silvia, Gatti Maurizio, Goldberg Michael L. Roles of the Drosophila NudE protein in kinetochore function and centrosome migration. 2009 Jun 1;122(11):1747–1758. doi: 10.1242/jcs.041798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wairkar Yogesh P, Toda Hirofumi, Mochizuki Hiroaki, Furukubo-Tokunaga Katsuo, Tomoda Toshifumi, DiAntonio Aaron. Unc-51 controls active zone density and protein composition by downregulating ERK signaling. The Journal of Neuroscience. 2009 Jan 14;29(2):517–528. doi: 10.1523/JNEUROSCI.3848-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao KM, White K. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. Journal of Neurochemistry. 1994 Jul;63(1):41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.