Dicer is an RNase III family endoribonuclease and haploinsufficient tumor suppressor that processes mature miRNAs from the 5′ (5p) or 3′ (3p) arm of hairpin precursors. In murine Dicer knockout fibroblasts, the authors expressed human Dicer with point mutations in the RNase III, helicase, and PAZ domains and characterized miRNA expression by Northern blot and massively parallel sequencing of small RNAs. They show that inactivation of the RNase IIIA domain results in complete loss of 3p-derived mature miRNAs, but only partial reduction in 5p-derived mature miRNAs. Conversely, inactivation of the RNase IIIB domain by mutation of D1709, a residue mutated in a subset of nonepithelial ovarian cancers, results in complete loss of 5p-derived mature miRNAs, including the tumor-suppressive let-7 family, but only partial reduction in 3p-derived mature miRNAs. These results suggest that mutation of the clinically relevant residue D1709 within the RNase IIIB results in a uniquely miRNA-haploinsufficient state in which the let-7 family of tumor suppressor miRNAs is lost, while a complement of 3p-derived miRNAs remains expressed.
Keywords: Dicer, miRNA, RNase III, helicase, PAZ, cancer
Abstract
Dicer is an RNase III family endoribonuclease and haploinsufficient tumor suppressor that processes mature miRNAs from the 5′ (5p) or 3′ (3p) arm of hairpin precursors. In murine Dicer knockout fibroblasts, we expressed human Dicer with point mutations in the RNase III, helicase, and PAZ domains and characterized miRNA expression by Northern blot and massively parallel sequencing of small RNAs. We report that inactivation of the RNase IIIA domain results in complete loss of 3p-derived mature miRNAs, but only partial reduction in 5p-derived mature miRNAs. Conversely, inactivation of the RNase IIIB domain by mutation of D1709, a residue mutated in a subset of nonepithelial ovarian cancers, results in complete loss of 5p-derived mature miRNAs, including the tumor-suppressive let-7 family, but only partial reduction in 3p-derived mature miRNAs. Mutation of the PAZ domain results in global reduction of miRNA processing, while mutation of the Walker A motif in the helicase domain of Dicer does not alter miRNA processing. These results provide insight into the biochemical activity of human Dicer in vivo and, furthermore, suggest that mutation of the clinically relevant residue D1709 within the RNase IIIB results in a uniquely miRNA-haploinsufficient state in which the let-7 family of tumor suppressor miRNAs is lost while a complement of 3p-derived miRNAs remains expressed.
INTRODUCTION
MicroRNAs are a class of 22-nt RNAs that regulate diverse biological processes (Ghildiyal and Zamore 2009). They are transcribed initially as long primary transcripts (pri-miRNA) and then processed in the nucleus by Drosha to yield hairpin precursors (pre-miRNA) (Krol et al. 2010). Each pre-miRNA is transported into the cytoplasm and processed further by Dicer to produce a double-stranded species consisting of a miRNA guide strand, required for mRNA targeting, and a passenger strand that is degraded (Krol et al. 2010). The guide strand is incorporated into an RNA-induced silencing complex (RISC) consisting of Argonaute proteins. The RISC then targets transcripts through complementarity of positions 2–7, named the seed region, of the loaded miRNA to the 3′ untranslated region (3′ UTR) of its target mRNA (Bartel 2009). This binding subsequently results in translational inhibition and/or degradation of the target transcript (Ghildiyal and Zamore 2009).
The enzymes required for the biogenesis of miRNAs have been characterized biochemically. Dicer possesses multiple domains, including two universally conserved RNase III domains, a PAZ domain, and a less-conserved ATPase/DExD helicase (Carmell and Hannon 2004; Park et al. 2011). These domains have been characterized both in vitro and, to a lesser extent, in vivo (Welker et al. 2010; Park et al. 2011; Ohishi and Nakano 2012). The RNase III domains of Dicer are essential for miRNA maturation. Mutations within the RNase IIIA and IIIB domains of Dicer abrogate in vitro processing of the 3′ (3p) and 5′ (5p) arms of pre-miRNAs, respectively (Zhang et al. 2004; Heravi-Moussavi et al. 2012). The PAZ domain binds the free ends of pre-miRNAs and positions the RNase III domains of Dicer to cleave pre-miRNAs at a fixed distance from the 3p end, in the case of Giardia (MacRae et al. 2007), or the 5p end, in the case of mammalian Dicer (Park et al. 2011), of the pre-miRNA. The function of the helicase domain remains unclear, but its conservation correlates positively with the existence of endogenous small interfering RNAs (endo-siRNAs) processed from long double-stranded RNAs (dsRNAs) (Welker et al. 2010). In vitro, mutation of K70 in the ATP-binding Walker A motif within the helicase domain of human Dicer (hsDicer) does not impair processing of dsRNA, leading to the prediction that this residue is not required for processing of pre-miRNAs (Zhang et al. 2002). However, in vivo, in Caenorhabditis elegans, mutation of this motif, equivalent to a K70A mutation in hsDicer, results in loss of a subpopulation of endo-siRNAs but does not affect miRNA levels (Welker et al. 2010). Based on these observations, current models predict that the helicase is required for the unwinding and processive cleavage of dsRNAs (Welker et al. 2011).
In addition to biochemical characterization, the RNAi pathway has also been characterized genetically in various species, including fission yeast, nematode, fruit fly, mouse, and humans. Homozygous loss of mammalian Dicer, encoded from a single gene, often leads to profound defects, particularly during development, and can lead to proliferative delays and increased apoptosis (Harfe et al. 2005; Kanellopoulou et al. 2005; Ambros 2011; Mendell and Olson 2012), while heterozygous loss can contribute to pathological states (Lu et al. 2005; Kumar et al. 2007, 2009; Hill et al. 2009). In mouse models, deletion of a single copy of Dicer leads to enhanced tumor formation, demonstrating a role as a haploinsufficient tumor suppressor (Kumar et al. 2007, 2009). In humans, changes in expression levels of Dicer and miRNAs have been associated with tumor development (Garzon et al. 2010). Patients with germline mutations in Dicer develop familial pediatric pleuropulmonary blastoma (Hill et al. 2009). Recently, exome sequencing revealed a specific somatic mutation of D1709 within the RNase IIIB domain of Dicer in nonepithelial ovarian tumors that are haploid for this allele (Heravi-Moussavi et al. 2012).
Many questions remain about the mechanism underlying the pathological states promoted by alterations in Dicer. In particular, the effects of tumor-associated missense mutations within Dicer on miRNA-processing activity in vivo are unknown. This study uses Dicer knockout murine mesenchymal stem cells (MSCs), a fibroblast cell type, to carry out structure–function analysis of hsDicer in cell culture. Through point mutagenesis, we demonstrate that the RNase IIIA and IIIB domains of hsDicer process the 3p and 5p arms of miRNAs, respectively, in vivo. In particular, mutation of residue D1709, which is associated with human tumors, results in loss of maturation of the let-7 family of tumor suppressor miRNAs, as well as other miRNAs, derived from the 5p arm of precursors, but only partially impairs processing of miRNAs derived from the 3p arm. We also report that mutation within the PAZ domain results in global reduction of miRNA processing and that the ATP-binding activity of the Walker A motif within the helicase is dispensable for miRNA processing. These findings provide insight into the in vivo biochemical activity of Dicer.
RESULTS AND DISCUSSION
To perform structure–function analysis of human Dicer (hsDicer), we mutagenized residues in several functional domains (schematized in Fig. 1A). Within the RNase III domains, we targeted residues that coordinate metal ions to catalyze the cleavage of pre-miRNA. In the RNase IIIA domain, D1320 or E1564 was mutated to alanine, and in the RNase IIIB domain, D1709, which is altered in a subset of nonepithelial ovarian cancers (Heravi-Moussavi et al. 2012), was mutated to alanine. Within the PAZ domain, Y971 and Y972 are conserved residues required for optimal RNA binding by the PAZ domain of Ago proteins (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003; Zhang et al. 2004). Mutation of these aromatic residues within hsDicer was shown previously to reduce hsDicer activity in vitro (Zhang et al. 2004). As in this previous study, we replaced Y971/Y972 with alanines. Within the PAZ, we also mutated E1036 to alanine based on the previous observation that this residue is required for full hsDicer activity in vitro (Zhang et al. 2004). Within the ATPase Walker A motif of the helicase, we mutated K70 to alanine. This residue was shown previously, in C. elegans, to be required for the generation of a subset of endogenous siRNAs but dispensable for production of miRNAs (Welker et al. 2010).
FIGURE 1.
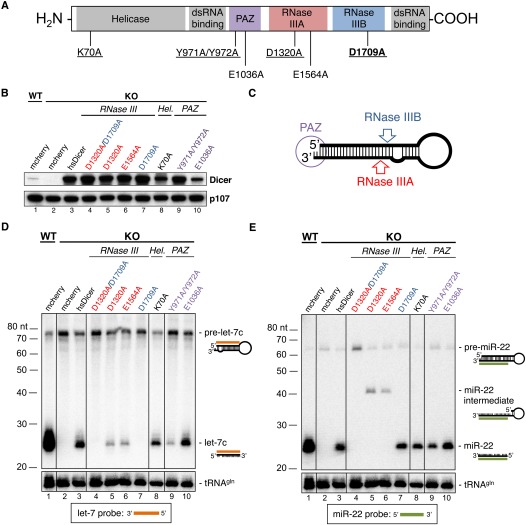
Mutational analysis of human Dicer (hsDicer). (A) Schematic of hsDicer. Domains are shaded. Mutations characterized in this study are indicated, with mutations characterized by small RNA-sequencing underlined. The residue mutated in human nonepithelial ovarian cancers is highlighted in bold. (B) Western blot of Dicer. Differences in signal intensity may reflect differences in immunoreactivity against murine and human Dicer. p107 was used as a loading control. (C) Schematic of the cleavage sites of the RNase IIIA and RNase IIIB domains, and the binding site of the PAZ domain relative to the precursor miRNA hairpin. The placement of the domains is based on findings reported here as well as published findings (Zhang et al. 2004; Park et al. 2011). (D) Northern blot of let-7c. (E) Northern blot of miR-22. The probes and visualized species are indicated to the right of each blot. Migration of a DNA ladder is shown to the left of each blot. tRNAgln was used as a loading control.
To assess the activity of each mutant, we transfected wild-type and mutant hsDicer constructs into Dicer−/− (Dicer KO) mesenchymal stem cells (MSCs), described in detail elsewhere (A Ravi, AM Gurtan, MS Kumar, A Bhutkar, C Chin, V Lu, JA Lees, T Jacks, and PA Sharp, unpubl.). These cells were derived from an adult Dicerf/f mouse (Harfe et al. 2005), immortalized with SV40 large T antigen, and infected with Adeno-Cre-GFP to delete both conditional alleles. As controls, we transfected Dicerf/f (Dicer WT) and Dicer KO MSCs with a fluorescent reporter plasmid encoding mCherry. Endogenous murine Dicer protein was expressed in Dicer WT cells but not in Dicer KO cells (Fig. 1B, lanes 1,2). Following transfection, wild-type and mutant hsDicer constructs were expressed at similar levels in Dicer KO cells, indicating that the mutations did not destabilize the protein.
To characterize the activity of each mutant, we performed Northern blotting on let-7c and miR-22, two abundant miRNAs derived from the 5p and 3p arms, respectively, of hairpin precursors. Mature miRNAs were undetectable in Dicer KO cells, confirming loss of Dicer activity (Fig. 1D,E). Following transfection of wild-type hsDicer into Dicer KO cells, we observed an increase in levels of mature miRNAs (Fig. 1D,E, cf. lanes 1–3) with a concomitant decrease in levels of detectable precursors (Fig. 1D), indicating that hsDicer is active in murine cells.
Simultaneous mutation of the RNase IIIA and IIIB domains (D1320A/D1709A) resulted in complete loss of miRNA-processing activity and failed to generate either mature let-7c or miR-22 (Fig. 1D,E, lane 4). The D1320A/D1709A mutant stabilized, but did not cleave, the miR-22 pre-miRNA. This stabilization suggests that the RNase III A/B double mutant is competing with a degradation pathway that targets pre-miR-22. Lin-28 is the best-characterized regulator of miRNA stability, modulating processing and degradation of let-7 (Heo et al. 2008; Piskounova et al. 2011). However, in both Dicer WT and KO MSCs, neither Lin-28a nor Lin-28b is expressed at the RNA level (data not shown). Therefore, an alternative pathway may be involved in degrading pre-miR-22.
In contrast to the RNase IIIA/B double mutant, constructs with a mutation in a single RNase III domain retained partial activity of the second, intact RNase III domain. Constructs with either D1320A or E1564A point mutations in the RNase IIIA domain (schematized in Fig. 1C) partially processed pre-let-7c to generate mature let-7c albeit at reduced levels relative to wild-type hsDicer. These mutants also generated a partially processed, stable pre-miR-22 intermediate but failed to generate mature miR-22. These results indicate that the RNase IIIB domain processes the 5p arm of pre-miRNAs, although at lower efficiency than wild-type hsDicer, in the absence of a functional RNase IIIA domain.
Mutation of D1709 blocked maturation of let-7c but did not affect maturation of miR-22 (Fig. 1D,E, lane 7). These data suggest that the RNase IIIB domain is required to process miRNAs from the 5p arm and that the RNase IIIA domain, in the absence of a functional RNase IIIB domain, can process the 3p arm, but not the 5p arm, of pre-miRNAs.
Mutation of the Walker A motif mutant (K70A), predicted to abrogate the putative helicase activity of hsDicer, has been reported in C. elegans to affect processing of endo-siRNAs but not miRNAs (Welker et al. 2010). In vitro, point mutation of K70 in hsDicer does not affect processing of dsRNA, suggesting that processing of pre-miRNAs would also be independent of ATP (Zhang et al. 2002). Deletion of the helicase domain of Dicer results in enhanced miRNA processing in vitro (Ma et al. 2008). In our experiments in vivo, the K70A mutant processed both let-7c and miR-22 at levels comparable to wild-type hsDicer, consistent with previous observations in vitro with hsDicer and in vivo with nematode Dicer.
The PAZ domain of hsDicer binds the free ends of the precursor miRNA. Mutation of Y971/Y972 and E1036 within the PAZ were reported previously to reduce processing of pre-miRNAs by hsDicer in vitro (Zhang et al. 2004). In our analyses, the Y971A/Y972A mutant displayed elevated levels of pre-let-7c relative to wild-type hsDicer, and reduced levels of both mature let-7c and miR-22, indicating a general reduction in activity, likely due to reduced binding of the precursors. In contrast to previous in vitro observations (Zhang et al. 2004), the activity of the E1036A mutant was comparable to that of wild-type protein. The presence of interacting partners in vivo, such as TRBP, may help stabilize the E1036A mutant and reduce the functional deficit that is observed in vitro using purified protein in the absence of such partners.
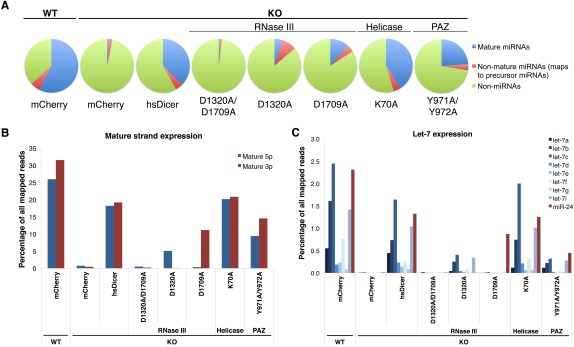
To confirm the results obtained by Northern blot and to extend our findings to all miRNAs transcribed in these fibroblasts, we performed massively parallel sequencing, in biological duplicate, of small RNAs (small RNA-seq), 15–50 nt in length, from Dicer WT control cells transfected with mCherry as well as Dicer KO cells transfected with either mCherry negative control or a subset of hsDicer point mutants characterized by Northern blot. Results from individual replicates are provided in Supplemental Data 1–3. Mature miRNAs comprised 57% of all reads in Dicer WT cells transfected with mCherry, but only 1% of all reads in Dicer KO cells transfected with mCherry (Fig. 2A; Supplemental Data 1). Comparison of mCherry-transfected Dicer WT and KO libraries did not reveal any experimentally reproducible Dicer-independent miRNAs expressed in MSCs. Mature miRNAs were restored comparably well with either transfection of wild-type hsDicer (37% of all reads) or the K70A helicase mutant (41%), further supporting the observation that the helicase is not required for miRNA processing. No recovery of miRNAs was observed upon transfection of the D1320A/D1709A RNase IIIA/B double mutant (1%), and partial recovery was observed with transfection of the D1320A RNase IIIA single mutant (5%), the D1709A RNase IIIB mutant (11%), and the Y971A/Y972A PAZ mutant (24%) (Fig. 2A). In cells expressing D1320A, reads mapping to pre-miRNAs were more abundant than reads mapping to mature miRNAs, in contrast to the other libraries from cells expressing fully or partially active hsDicer (Fig. 2A; Supplemental Data 1). This result may be due to increased accumulation, in D1320A-expressing cells, of partially processed pre-miRNAs such as the miR-22 intermediate observed by Northern blot (Fig. 1E). In cells expressing D1320A or D1709A, we observed an increased ratio of miRNA star to mature strands relative to cells expressing wild-type hsDicer, although in all libraries star strands were less abundant than mature strands (data not shown).
FIGURE 2.
Small RNA-sequencing from Dicer WT and KO cells transfected with hsDicer constructs. (A) Summary of total library reads mapped to mature miRNAs (blue), pre-miRNAs (orange), or non-miRNA loci (green). One mismatch was allowed for mapping. Mature miRNAs were identified as reads with an exact 5′ end and a ±1-nt length variation at the 3′ end compared with previously published annotations (Chiang et al. 2010). (B) Reads mapping to mature 5p or 3p arms of precursors. (C) Expression of the let-7 family of miRNAs. (B,C) miRNA composition was calculated as a percentage of total reads mapped, with 0 or 1 mismatch, to the mouse genome (mm9). The sequencing was performed in biological duplicate. The results shown are the average of the duplicate libraries.
Consistent with Northern blot analysis, the RNase III single mutants exhibited a marked bias toward processing of a specific arm of the pre-miRNA. RNase IIIA domain mutant D1320A, but not RNase IIIB mutant D1709A, restored let-7c expression to 23% of levels observed with transfection of wild-type hsDicer (Table 1). Conversely, D1709A, but not D1320A, restored miR-22 expression to 73% of levels relative to wild-type hsDicer (Table 1). Globally, the D1320A RNase IIIA mutant showed partial recovery (28% relative to wild-type hsDicer) of 5p miRNAs, and no recovery of 3p miRNAs (Fig. 2B, cf. D1320A to hsDicer; raw data provided in Supplemental Data 3). Conversely, the D1709A RNase IIIB mutant showed partial recovery (58% relative to wild-type hsDicer) of 3p miRNAs and no recovery of 5p miRNAs (Fig. 2B, cf. D1709A to hsDicer; raw data provided in Supplemental Data 3). Expression of all members of the let-7 family was partially restored in the D1320A RNase IIIA mutant library but was absent in the D1709A RNase IIIB mutant library (Fig. 2C). 3p-derived miRNA miR-24 was absent in cells transfected with the D1320A RNase IIIA mutant but was detected in the D1709A RNase IIIB mutant. Within each library from Dicer KO cells expressing hsDicer, K70A, and Y971A/Y972A, 5p- and 3p-derived miRNAs were comparably represented, indicating that both RNase III domains were comparably active in these constructs (Fig. 2B,C; Supplemental Data 2). In the Y971A/Y972A sequencing libraries, although 5p-derived miRNAs appeared to be slightly underrepresented relative to 3p-derived miRNAs (Fig. 2B), this result was not consistent between biological replicates (Supplemental Data 2). Fidelity of miRNA processing was not affected in cells expressing Y971A/Y972A mutations within the PAZ (data not shown), a domain that positions pre-miRNAs for cleavage at fixed distances from the precursor termini (MacRae et al. 2007; Park et al. 2011).
TABLE 1.
Expression of miRNAs relative to wild-type hsDicer (%)
Our results mark the first characterization in cell culture of the RNase III domains of human Dicer. Furthermore, our helicase and PAZ mutants add to, and are consistent with, the in vivo structure–function studies performed recently analyzing the mechanism by which worm and human Dicer process miRNAs (Welker et al. 2010; Park et al. 2011). Our findings indicate that each RNase III domain can retain partial activity in vivo when the other RNase III domain is inactivated. Confirming conclusions from in vitro experiments (Zhang et al. 2004), loading of pre-miRNA into hsDicer in vivo is directional, with the RNase IIIA and IIIB domains responsible for processing of the 3p and 5p arms of precursors, respectively (Fig. 1C). Consistent with our results, a recently published forward genetic screen in Chinese hamster ovary cells identified a role for the RNase IIIA domain in processing 3p miRNAs (Ohishi and Nakano 2012).
Missense mutations of a haploid allele of Dicer have been reported in human nonepithelial ovarian cancers, notably in the RNase IIIB domain (Heravi-Moussavi et al. 2012), but the consequences of these mutations on Dicer activity in vivo have been unclear. Our results indicate that inactivation of a single RNase III domain within Dicer dramatically alters the miRNA expression profile of a cell. Importantly, catalytically inactive mutations in the RNase IIIB domain solely generate miRNAs from the 3p arm of the hairpin precursor. Specifically, we conclude that patient tumors expressing hsDicer with a mutation at D1709 in the RNase IIIB are likely deficient in 5p miRNAs while competent for expression of 3p miRNAs. Thus, this mutation represents a unique, miRNA-haploinsufficient state in which an entire family of tumor suppressors, the 5p-derived let-7 family, is absent while a complement of 3p-derived miRNAs, including the ubiquitously abundant miR-22, remains expressed. This miRNA expression profile may allow cells to selectively inactivate regulation of tumor-suppressive networks by let-7 family members while maintaining regulation of cellular pathways by miRNAs from the 3p arm. Since cells null for Dicer and thus all miRNAs are sensitive to many types of stress (Leung et al. 2011a), it is possible that retention of miRNAs from the 3p arm might be critical for tumor survival. In total, these and previous results strongly indicate that alterations in regulation by miRNAs can be critical in human cancer.
MATERIALS AND METHODS
Cell culture
Murine mesenchymal stem cell (MSC) cultures were passaged in α-MEM supplemented with pen/strep and 10% FBS. Cells were transfected with Lipofectamine 2000 (Invitrogen) and either pCAGGS-mCherry, wild-type pCAGGS-Flag-hsDicer, or mutant pCAGGS-Flag-hsDicer constructs. Forty-eight hours after transfection, MSCs were harvested for protein or total RNA, as indicated below.
Western blot
Cells were lysed with 2× sample buffer (Bio-Rad) containing 5% β-mercaptoethanol, boiled for 5 min, and subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, proteins were transferred to PVDF membranes using a submerged transfer apparatus (Bio-Rad). After blocking with 5% nonfat dried milk in TBS-T (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.1% Tween 20), the membrane was incubated with the primary antibody diluted in TBST, washed extensively, and incubated with the appropriate horseradish peroxidase–linked secondary antibody (Amersham). Chemiluminescence was used for detection. Western blots were performed with anti-Dicer (Bethyl Laboratories), for detection of murine and human Dicer, or anti-p107 as a loading control (Santa Cruz Biotechnology).
Northern blot
RNA was prepared using Qiazol Reagent according to the manufacturer's protocol (QIAGEN). Total RNA was mixed with an equal volume of formamide loading buffer, denatured for 5 min at 95°C, and run for 1 h at 35 W on a 12% denaturing polyacrylamide gel (Sequagel, National Diagnostics) after 30 min of pre-running. A semi-dry transfer apparatus set to 18 V was used to transfer the RNA to a Hybond-N+ nylon membrane (GE Healthcare Life Sciences) for 1.5 h at 4°C. RNA was then UV cross-linked at 1.2 × 106 μJ in a Crosslinker 2400 (Stratagene) on top of Whatman paper. The membrane was prehybridized with Ultrahyb Oligo Buffer (Ambion) for 1 h and then probed overnight at 37°C with a 5′-end-labeled DNA oligo probe complementary to mature let-7c, mature miR-22, or glutamine tRNA. The membrane was washed twice for 30 min in 2× SSC/0.1% SDS buffer, exposed to a phosphorimager screen, and then imaged on a Storm scanner (Molecular Dynamics). Northern blots were carried out in biological duplicate.
Small RNA cloning
Small RNA sequencing from mesenchymal stem cell lines was performed, in biological duplicate, as described previously but with bar-coded samples for multiplex sequencing (Leung et al. 2011b). (The GEO Accession Number for this data set is GSE36978.) A detailed protocol is available upon request. Multiplexed Illumina sequence reads were bucketed by sample bar code and processed using the FASTX-Toolkit (from the laboratory of Gregory Hannon) to strip adaptors, drop resulting reads <15 bp, and aggregate identical sequences in each individual library. Processed reads were mapped to the mouse genome (build mm9 in UCSC Genome Browser) using the Bowtie short read alignment tool (Langmead et al. 2009), allowing for up to a single base pair mismatch per alignment. All unique matches to the genome were first processed to identify sense-strand alignments to miRNA annotation (Chiang et al. 2010; Kozomara and Griffiths-Jones 2011). Any repeat matches to the mouse genome were further processed to identify possible matches to miRNA annotation.
Using custom software, reads mapping to full-length mature miRNA annotations with exact ends and allowance for a 1-bp slip at the 3′ end were categorized as mature miRNAs. These were subsequently used to quantify the levels of mature miRNAs in various data sets under study. Mature miRNA matches were also classified according to 5′ or 3′ pre-miRNA arm annotation in each library. Precision of mature miRNA processing was assessed in each library within a ±10-bp window of the annotated 5′ and 3′ ends of mature miRNAs. Levels of miRNA precursor intermediates were quantified in each library, and length distributions were calculated using read alignments anchored at the location of the Dicer cut with allowance for a ±1-bp slip, for each of the annotated miRNAs on either the 5′ or 3′ arm of the corresponding pre-miRNA.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Mauro Calabrese for generating the pCAGGS-Flag-hsDicer construct. We also thank members of the Sharp laboratory for helpful discussions. Massively parallel Illumina sequencing was performed by the Biopolymers & Proteomics Core Facility in the Swanson Biotechnology Center at the David H. Koch Institute for Integrative Cancer Research at M.I.T. Bioinformatic analysis was performed in the Bioinformatics & Computing division of the Swanson Biotechnology Center. This work was supported by United States Public Health Service grants RO1-GM34277 from the National Institutes of Health, PO1-CA42063 from the National Cancer Institute to P.A.S., and partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. A.M.G. acknowledges support from a Leukemia and Lymphoma Society grant (5198-09).
Author contributions: A.M.G. designed the experiments. A.M.G. and V.L. performed the experiments and analyzed data. A.B. performed the informatics for the sequencing data. A.M.G. and V.L. prepared the figures. A.M.G. and P.A.S. wrote the paper. P.A.S. provided supervision and assisted with manuscript preparation. All authors reviewed and approved the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032680.112.
REFERENCES
- Ambros V 2011. MicroRNAs and developmental timing. Curr Opin Genet Dev 21: 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ 2004. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol 11: 214–218 [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM 2010. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov 9: 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ 2005. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci 102: 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, Fejes AP, Chow C, Tone A, Kalloger SE, et al. 2012. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 366: 234–242 [DOI] [PubMed] [Google Scholar]
- Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, et al. 2009. DICER1 mutations in familial pleuropulmonary blastoma. Science 325: 965 doi: 10.1126/science.1174334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S 2011. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39: 673–677 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 23: 2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P 2011a. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA 2011b. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 18: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA 2008. Auto-inhibition of human Dicer by its internal helicase domain. J Mol Biol 380: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA 2007. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 14: 934–940 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Olson EN 2012. MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Nakano T 2012. A forward genetic screen to study mammalian RNA interference—essential role of RNase IIIa domain of Dicer1 in 3′ strand cleavage of dsRNA in vivo. FEBS J 279: 832–843 [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI 2011. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147: 1066–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10: 1026–1032 [DOI] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL 2010. Dicer's helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL 2011. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell 41: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426: 468–474 [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]