In maize, thylakoid assembly 8 (tha8) causes defects in the biogenesis of chloroplast thylakoid membranes. THA8 contains more than 10 pentatricopeptide repeats (PPR). THA8 is the first member of this subfamily with a defined molecular function. THA8 localizes to chloroplasts where it associates specifically with the ycf3-2 and trnA group II introns.
Keywords: pentatricopeptide repeat, plastid, group II intron, chloroplast, mitochondria
Abstract
A maize gene designated thylakoid assembly 8 (tha8) emerged from a screen for nuclear mutations that cause defects in the biogenesis of chloroplast thylakoid membranes. The tha8 gene encodes an unusual member of the pentatricopeptide repeat (PPR) family, a family of helical repeat proteins that participate in various aspects of organellar RNA metabolism. THA8 localizes to chloroplasts, where it associates specifically with the ycf3-2 and trnA group II introns. The splicing of ycf3-2 is eliminated in tha8 mutants, and trnA splicing is strongly compromised. Reverse-genetic analysis of the tha8 ortholog in Arabidopsis thaliana showed that these molecular functions are conserved, although null alleles are embryo lethal in Arabidopsis and seedling lethal in maize. Whereas most PPR proteins have more than 10 PPR motifs, THA8 belongs to a subfamily of plant PPR proteins with only four PPR motifs and little else. THA8 is the first member of this subfamily with a defined molecular function, and illustrates that even small PPR proteins have the potential to mediate specific intermolecular interactions in vivo.
INTRODUCTION
Chloroplasts are organelles of photosynthetic eukaryotes that originate from a cyanobacterial ancestor. Chloroplasts in land plants have retained ∼100 genes, most of which encode components of the basal chloroplast gene-expression machinery or subunits of photosynthetic complexes. The expression of the small chloroplast genome requires hundreds of nuclear gene products; some of these were acquired from the cyanobacterial ancestor, but many evolved in the eukaryotic host (for review, see Barkan 2011a).
Post-transcriptional aspects of gene expression in land plant chloroplasts are particularly complex, and include RNA editing, segmental mRNA stabilization, and the splicing of ∼20 group II introns. Group II introns are large ribozymes that splice via the same chemical steps as nuclear pre-mRNA introns. Although group II introns are considered to be “self-splicing” (Lambowitz and Zimmerly 2011), self-splicing has not been reported for any land plant chloroplast introns. In fact, 14 nuclear genes have been identified that are required for the splicing of group II introns in angiosperm chloroplasts (for review, see Falcon de Longevialle et al. 2010; Chateigner-Boutin et al. 2011; Watkins et al. 2011). Each is required for the splicing of distinct, but overlapping intron subsets, and each intron has been shown to require multiple proteins. These proteins are not related to nuclear splicing factors, nor to the intron-encoded “maturases” that are typical of group II introns in bacteria (Lambowitz and Zimmerly 2011). Instead, these proteins are drawn from unusual families of RNA-binding proteins whose members function predominantly in mitochondria and chloroplasts (for review, see Barkan 2011a). For example, several chloroplast splicing factors are members of the pentatricopeptide repeat (PPR) family, a family of helical repeat proteins that bind RNA and that are defined by degenerate 35 amino acid tandem repeats (Small and Peeters 2000). PPR proteins are found in all eukaryotes, but the family is remarkable for its size in terrestrial plants, with over 450 members (Lurin et al. 2004). All of the PPR proteins that have been thoroughly characterized promote various aspects of organellar RNA metabolism and interact with one or several organellar RNAs (for review, see Schmitz-Linneweber and Small 2008). The mechanism behind their target specificity is not understood, but is anticipated to involve a series of interactions between helical repeating units in the protein and individual nucleotides in the RNA (Fujii et al. 2011; Prikryl et al. 2011).
In this work we describe a new PPR-containing chloroplast splicing factor, which we named THA8 (thylakoid assembly 8). THA8 was discovered during a phenotype-driven screen for maize mutants with defects in the translocation of proteins across the thylakoid membrane. Chloroplast protein defects in tha8 mutants phenocopy those of hcf106 and tha4 mutants (Voelker and Barkan 1995; Walker et al. 1999), which have mutations in genes encoding components of the “twin arginine translocase” (Tat) for transporting proteins across the thylakoid membrane (for review, see Cline and Dabney-Smith 2008). However, our results show that THA8's direct function is in the splicing of two group II introns, and imply that the Tat pathway defects in tha8 mutants are a secondary effect of bioenergetic deficits. We show that THA8 is required for the splicing of the ycf3-2 and trnA introns, and that it associates specifically with these two introns in stable ribonucleoprotein particles (RNPs) in vivo. We show further that these functions are conserved in Arabidopsis. THA8 is an unusually small PPR protein, with only four PPR motifs and little else. In that context, THA8's specificity for two introns is intriguing. Recombinant THA8 binds RNA in vitro, albeit weakly and with no detectable specificity for the ycf3-2 intron. Furthermore, recombinant THA8 fails to bind in vitro to the other known ycf3-2 splicing factors, APO1 (Watkins et al. 2011) and OTP51 (Falcon de Longevialle et al. 2008). Thus, the basis for the specific association between this small but essential splicing factor and its cognate introns remains to be determined.
RESULTS
tha8 mutants have decreased levels of thylakoid protein complexes and a defect in the Tat thylakoid transport pathway
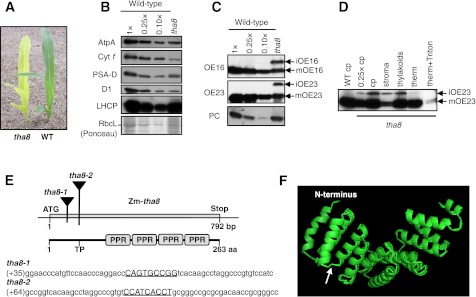
Proteins are translocated from the chloroplast stroma to the lumen of the thylakoid membrane system via one of two machineries, the Sec and Tat machineries (Cline and Dabney-Smith 2008). The thylakoid assembly 8-1 (tha8-1) mutant was identified during a screen for maize mutants with defects in these mechanisms. tha8-1 mutants have pale green leaves (Fig. 1A) and die after the development of approximately three leaves, as is typical of nonphotosynthetic maize mutants. tha8-1 mutants have substantially reduced levels of core subunits of the thylakoid ATP synthase, cytochrome b6f, photosystem I (PSI), and photosystem II (PSII) complexes, but only a modest decrease in the accumulation of the stromal protein RbcL (Fig. 1B). Whereas the Sec substrate plastocyanin (PC) accumulates normally in tha8 mutants, two Tat substrates, OE16 and OE23, accumulate to reduced levels (Fig. 1C). Furthermore, tha8 mutants accumulate precursors of OE16 and OE23 (iOE16 and iOE23), which lack chloroplast import sequences, but have retained the lumenal targeting signals (Fig. 1C). These thylakoid protein deficiencies are similar to those observed previously in maize mutants with defects in the chloroplast Tat machinery (Voelker and Barkan 1995; Walker et al. 1999). Fractionation of chloroplasts isolated from tha8-1 mutants, followed by protease treatments of the isolated thylakoids, confirmed that the overaccumulating iOE23 is found outside of the thylakoid membrane (Fig. 1D). These results indicate that tha8-1 mutants have a defect in Tat transport.
FIGURE 1.
Chloroplast protein and pigment defects in tha8 mutants. (A) Visual phenotype of a tha8 mutant and normal sibling (WT) grown for 8 d in soil. (B) Western blot analysis of thylakoid proteins in tha8 mutants. Proteins were extracted from leaves of wild-type and tha8 seedlings and analyzed on replicate immunoblots by probing with antibodies to AtpA (α subunit of ATP synthase), Cyt f (subunit of cytochrome b6f complex), D1 (reaction center protein of photosystem II), PSA-D (core subunit of photosystem I), or LHCP (major light harvesting chlorophyll binding protein of PSII). An example of a Ponceau S stained filter is shown below, with RbcL (large subunit of Rubisco) marked. (C) Aberrant metabolism of Tat substrates in tha8 mutants. Mature (m) and incompletely processed stromal intermediates (i) are marked. (D) iOE23 and iOE16 in tha8 mutants accumulate outside of the thylakoid membrane. Mutant chloroplasts (cp) were separated into stroma and thylakoids. The thylakoids were treated with thermolysin (therm) with or without the addition of Triton X-100. Analogous results were obtained with iOE16 (data not shown). (E) Positions of the Mu insertions in tha8 mutants. The tha8 gene lacks introns. The approximate position of the transit peptide (TP) cleavage site is marked. The sequences flanking the insertions are shown below, with the 9-bp target site duplications underlined. (F) Predicted structure of THA8. This structure prediction was made via the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) using the sequence of mature maize THA8 (i.e., lacking the inferred transit peptide). The arrow marks the beginning of the canonical PPR tract.
Identification of the tha8 locus
The tha8-1 mutant arose in maize lines with active Mu transposons, a high copy transposon system. To identify the causal mutation, Mu insertions that cosegregate with tha8 were sought by Southern blot display of Mu elements in homozygous mutant and related wild-type seedlings (data not shown). This analysis revealed a Mu1 insertion that is tightly linked to tha8. Molecular cloning of this insertion site and corresponding cDNA showed it to map ∼70 bp downstream from the predicted start codon of an unannotated, intronless gene (Fig. 1E).
To determine whether the linked insertion was responsible for the tha8-1 phenotype, we recovered an independent Mu insertion in the same gene through a PCR-based reverse-genetic screen of our large collection of nonphotosynthetic maize mutants. The second allele, tha8-2, has a Mu insertion ∼100 bp downstream from the predicted start codon, and has a visible phenotype similar to that of tha8-1 (Fig. 1E; data not shown). A complementation cross between plants that were heterozygous for each of the two alleles produced progeny segregating ∼25% chlorophyll-deficient plants, confirming this genomic region to be the tha8 locus. The thylakoid protein deficiencies were similar between tha8-1, tha8-2, and tha8-1/8-2 heteroallelic plants (data not shown). The experiments described below were performed with either tha8-1 homozygotes (protein work) or tha8-1/tha8-2 material (RNA work). These will be referred to hereafter as “tha8” without distinguishing between the alleles, as they were phenotypically indistinguishable.
THA8 is the founding member of a subfamily of PPR proteins in land plants that consist of four PPR motifs and little else
The tha8 locus encodes a 263-amino acid polypeptide with four pentatricopeptide repeats (PPR) and no other apparent functional motifs (Fig. 1E). PPRs are degenerate repeats of ∼35 amino acids found predominantly in proteins involved in organellar gene expression (Small and Peeters 2000; Schmitz-Linneweber and Small 2008). PPR motifs are usually found in tandem arrays of between ∼10 and 25 copies, and PPR tracts with only four copies are rare (Lurin et al. 2004). BLAST searches identified three closely related THA8 paralogs in maize, and orthologs for each of the four THA8-like proteins in rice and Arabidopsis. Only one THA8-like protein is encoded in the genome of the moss Physcomitrella patens. An alignment and phylogenetic tree of THA8 and THA8-like proteins are presented in Supplemental Figure S1. All THA8-like proteins are similar in size, and most THA8-like proteins begin with a predicted mitochondria or chloroplast targeting sequence. No proteins with strong similarity to THA8 were detected in the unicellular alga Chlamydomonas reinhardtii.
THA8-like proteins comprise a subfamily of PPR proteins that are defined by a conserved segment of ∼40 amino acids, followed by four PPR motifs. Structural models of THA8 computed by the I-TASSER algorithm (Roy et al. 2010) predict that the conserved N-terminal region adopts a helical hairpin structure that continues the α-solenoid architecture beyond the four canonical PPR motifs (Fig. 1F). Thus, similar to what was observed for the chloroplast protein PPR10 (Pfalz et al. 2009), virtually all of mature THA8 is predicted to consist of PPR-like modules, even though several of these modules are not predicted to be PPR motifs. Short PPR tracts are appended to catalytic domains in human mitochondrial RNA polymerase and plant organellar ribonuclease P (Gobert et al. 2010; Ringel et al. 2011). However, functions have not been assigned to any proteins in the THA8-subfamily or to any other protein with fewer than 10 PPR motifs and no other functional domains.
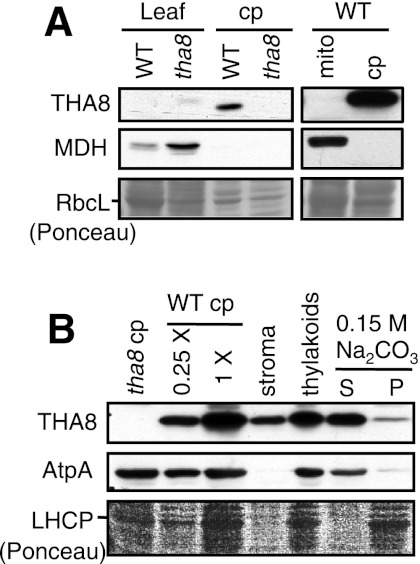
We anticipated that THA8 localizes to chloroplasts because it begins with a predicted organelle targeting sequence and it influences chloroplast biogenesis. To test this prediction, we used a polyclonal antiserum raised to recombinant THA8 to probe immunoblots of leaf, chloroplast, and mitochondrial extracts (Fig. 2A). The mitochondrial protein malate dehydrogenase (MDH) served as a marker for mitochondrial proteins. The THA8 antibody did not detect THA8 in total leaf homogenates. However, a protein whose size is consistent with that of mature THA8 (∼25 kDa) was detected in wild-type chloroplasts and was absent in tha8 mutant chloroplasts (Fig. 2A). THA8 was not detected in mitochondria purified from wild-type leaves (Fig. 2A). Immunoblot analyses of fractionated chloroplasts detected THA8 in the stromal and thylakoid fractions (Fig. 2B). Treatment of thylakoids with Na2CO3 released ∼80% of THA8 from the thylakoid membrane, suggesting that THA8 is extrinsically associated with the stromal face of the membrane. The extrinsic thylakoid protein AtpA exhibited a comparable release from the membrane after this treatment (Fig. 2B). These results show that THA8 partitions between the chloroplast stroma and the stromal face of thylakoid membranes. THA8 was recently found to be enriched in chloroplast nucleoid preparations (Majeran et al. 2012). As nucleoids can associate with thylakoid membranes (Liu and Rose 1992), THA8's nucleoid association might account for its partial cofractionation with thylakoid membranes.
FIGURE 2.
THA8 partitions between the chloroplast stroma and the extrinsic face of thylakoid membranes. (A) THA8 is enriched in isolated chloroplasts. Immunoblots of leaf, chloroplast (cp), and mitochondrial (mito) extracts from wild-type (WT) and tha8 mutant seedlings probed with antibodies to THA8 or the mitochondrial protein malate dehydrogenase (MDH). A portion of each Ponceau S-stained membrane is shown to illustrate sample loading. (B) Intra-chloroplast location of THA8. Purified chloroplasts were separated into stromal and thylakoid fractions. The thylakoids were washed with Na2CO3 to remove extrinsic proteins, repelleted (P) and the supernatant (S) removed. Equivalent proportions of each fraction were analyzed by immunoblotting with antibodies to THA8 and AtpA, a membrane-extrinsic subunit of the ATP synthase. Unfractionated tha8 and WT chloroplasts are shown to the left.
THA8 is required for splicing of the ycf3-2 and trnA group II introns
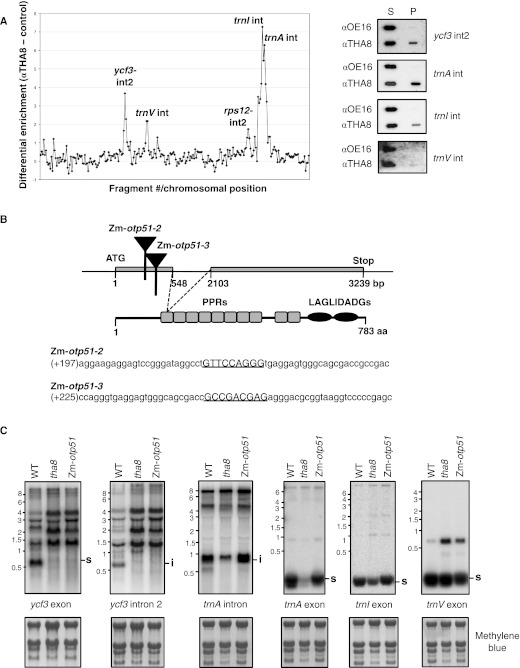
Our attention was initially drawn to THA8 due to the Tat-transport defect in tha8 mutants. However, we were unable to detect an association between THA8 and the Tat machinery by native gel electrophoresis or coimmunoprecipitation, and neither the abundance nor membrane integration of the Tat subunits THA4 and HCF106 were altered in tha8 mutants (data not shown). Because most characterized PPR proteins influence organellar RNA metabolism, the presence of PPR motifs suggested that THA8 might instead influence chloroplast gene expression, with the Tat-transport defect arising as a secondary effect. We therefore determined whether THA8 is associated with specific chloroplast RNAs by performing genome-wide RNA coimmunoprecipitation assays (RIP-Chip) from native chloroplast extract (Schmitz-Linneweber et al. 2005). Antibodies against THA8 were used for a coimmunoprecipitation from chloroplast stromal extract. RNA was isolated from the supernatant and pellet, differentially labeled with a green or a red fluorescing dye, and hybridized to a tiling microarray of the maize chloroplast genome. A negative control experiment used antibodies to the lumenal protein OE16, which is not expected to bind RNA. RNA from three chromosomal regions was strongly enriched in the THA8 coimmunoprecipitation: the second intron in ycf3 (ycf3-2), the trnA intron, and the trnI intron (Fig. 3A). RNA derived from the trnV intron and rps12-intron 2 also appeared to coimmunoprecipitate with THA8, but to a lesser extent. Slot-blot hybridization of RNAs from the same immunoprecipitations confirmed that the ycf3-2 and trnA introns are strongly enriched in THA8 coimmunoprecipitations (Fig. 3A, right). The trnI intron is also enriched, but less so, and the trnV intron was not detectably enriched. The trnA and trnI introns are found in adjacent genes in the same polycistronic transcription unit; the weaker coimmunoprecipitation of the trnI intron suggested that THA8 might associate only with the trnA intron, with the trnI intron coimmunoprecipitating to a lesser extent due to its presence on some of the same RNA molecules.
FIGURE 3.
THA8 is found in intron-containing complexes and is required for the splicing of two chloroplast introns. (A) RIP-chip experiment using THA8 antiserum. The log2-transformed enrichment ratios (ratio of signal in the immunoprecipitation pellet versus supernatant) are plotted according to position on the chloroplast genome after subtracting values obtained in a negative control immunoprecipitation with antibody to OE16 (left). Coimmunoprecipitation of RNAs represented by the prominent peaks was further tested by slot-blot hybridization of RNA purified from immunoprecipitation supernatants (S) and pellets (P) (right). (B) Zm-otp51 mutants. Zm-otp51 corresponds to maize locus GRMZM2G325019. The positions of the Mu insertions are marked with triangles. The mutant progeny of a complementation cross between Zm-otp51-2 and Zm-otp51-3 was used for the RNA analyses. The sequences flanking the insertion sites are shown below, with the 9-bp target site duplications underlined. (C) RNA gel-blot hybridizations demonstrating chloroplast splicing defects in a tha8 mutant. Gels included ∼5 μg of seedling leaf RNA from plants of the indicated genotypes. The rRNAs on the same filters, detected by staining with methylene blue, are shown below. (s) Mature spliced product, (i) excised intron.
The association of THA8 with specific chloroplast introns suggested that THA8 might influence intron RNA metabolism. We therefore analyzed transcripts from all intron-containing chloroplast genes in tha8 mutants. The Arabidopsis PPR protein OTP51 was shown previously to be required for the splicing of the ycf3-2 intron (Falcon de Longevialle et al. 2008). We recovered mutant alleles of the orthologous maize gene, Zm-otp51, in a reverse genetic screen of our mutant collection (Fig. 3B), and analyzed tha8 and Zm-otp51 RNAs in parallel for comparative purposes.
RNA gel-blot hybridizations showed that tha8 mutants have strong defects in the splicing of the ycf3-2 and trnA introns (Fig. 3C), the two introns that were strongly enriched in the RNA coimmunoprecipitation assays. In both instances, the loss of the spliced RNAs is accompanied by a decrease in the level of the excised intron, confirming these to be splicing defects. Zm-otp51 mutants also exhibit a strong defect in ycf3-2 splicing, showing this function to be conserved between Arabidopsis and maize. However, the functions of Zm-otp51 and tha8 do not overlap completely, as Zm-otp51 mutants exhibit normal trnA splicing. The abundance of spliced trnI is reduced slightly in tha8 mutants and the abundance of spliced trnV is unaltered, suggesting no more than a minimal contribution of THA8 to the splicing of these introns. The splicing of all other chloroplast introns was assayed (Supplemental Fig. S2), but no additional splicing defects were detected in tha8 mutants. Overall, there was an excellent correlation between those introns that strongly coimmunoprecipitate with THA8 and those whose splicing is THA8 dependent, implying that THA8 functions directly in the splicing of the ycf3-2 and trnA introns. Furthermore, these two splicing defects are sufficient to account for the chloroplast protein deficiencies in tha8 mutants, as Ycf3 is required for the accumulation of photosystem I (Ruf et al. 1997) and trnA is required for chloroplast translation, and thus for the accumulation of all photosynthetic enzyme complexes harboring plastid-encoded subunits.
Arabidopsis OTP51 mutants have mild defects in atpF and trnV splicing in addition to their strong defect in ycf3-2 splicing (Falcon de Longevialle et al. 2008). Zm-otp51 mutants have similar splicing defects (Supplemental Fig. S2A). However, the atpF splicing defect is much less severe than those caused by mutations in the maize genes crs1, wtf1, and rnc1 (Jenkins et al. 1997; Watkins et al. 2007; Kroeger et al. 2009), indicating that OTP51 plays only an accessory role. The splicing of the petB intron is also compromised slightly in the Zm-otp51 mutant (Supplemental Fig. S2B); an analogous defect was not reported for the Arabidopsis mutant. Nonetheless, the defects associated with loss of OTP51 function in maize and Arabidopsis are very similar overall.
THA8 is found in large complexes and coimmunoprecipitates with known trnA splicing factors
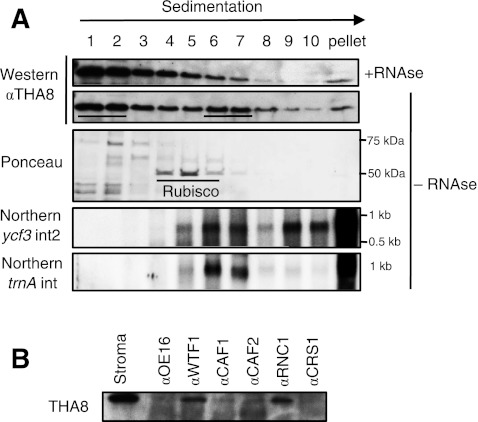
Previously characterized chloroplast splicing factors are found in large ribonucleoprotein complexes in the chloroplast stroma (Jenkins and Barkan 2001; Till et al. 2001; Asakura and Barkan 2007; Watkins et al. 2007; Asakura et al. 2008; Kroeger et al. 2009). To determine whether THA8 behaves similarly, stroma was fractionated by sedimentation through a sucrose gradient and fractions were analyzed by probing immunoblots with THA8 antibody (Fig. 4A). THA8 peaked in two regions of the gradient: a peak at the top of the gradient contains small, possibly monomeric particles; a peak in fractions 6 and 7 is estimated to contain particles of ∼600–700 kDa, based on its position relative to that of Rubisco (∼550 kDa). Treatment of the extract with RNAse A prior to sucrose gradient centrifugation reduced the representation of THA8 in the high molecular weight peak, indicating that these particles contain RNA (Fig. 4A). RNA gel-blot hybridization of RNA purified from gradient fractions showed that THA8's primary RNA targets, the ycf3-2 and trnA introns, cosediment with THA8 in fractions 6 and 7 (Fig. 4A). Intron RNA was also detected in higher molecular weight fractions, possibly due to their tethering to the chloroplast nucleoid complex, fragments of which contaminate stromal preparations.
FIGURE 4.
THA8 is found in large ribonucleoprotein particles. (A) Sucrose gradient fractionation of untreated (−RNAse) and RNAse A-treated chloroplast stroma. An equivalent aliquot of each fraction was used for Western blot analysis with THA8 antibody. The underlined fractions in the untreated sample contain the two major THA8 peaks. Rubisco (∼550 kDa) serves as a size marker. This sedimentation pattern was observed in two independent experiments (data not shown). RNA extracted from fractions of the control gradient (−RNAse) was analyzed by RNA gel-blot hybridization with probes specific for the ycf3-2 and trnA introns. The RNAs detected have the size expected for excised intron RNAs. (B) THA8 coimmunoprecipitates with the trnA splicing factors WTF1 and RNC1. Chloroplast stroma was used for coimmunoprecipitations with antibodies against various chloroplast splicing factors, as indicated at top. An immunoprecipitation with an antibody to OE16 served as a negative control. The immunoprecipitation pellets were analyzed by probing immunoblots with antibodies to THA8.
THA8 shares intron targets with four previously described splicing factors: RNC1, WTF1, OTP51, and APO1. An RNC1/WTF1 homodimer binds the trnA intron and is required for its splicing (Watkins et al. 2007; Kroeger et al. 2009). When WTF1 and RNC1 were immunoprecipitated from stromal extract, THA8 coimmunoprecipitated (Fig. 4B). These results suggest that THA8, WTF1, and RNC1 associate simultaneously with the trnA intron, their only shared intron target. An analogous experiment could not be performed with OTP51 and APO1, because antibodies to these proteins are not available. THA8 did not significantly coimmunoprecipitate with the chloroplast splicing factors CAF1, CAF2, or CRS1 (Fig. 4B), with which it does not share intron targets (Till et al. 2001; Ostheimer et al. 2003).
THA8 function is conserved between dicots and monocots
To learn whether the function of THA8 is conserved between maize (a monocot) and Arabidopsis (a dicot), we analyzed Arabidopsis mutants with insertions in the Arabidopsis tha8 ortholog, At-tha8 (At3g27750). At-tha8-1 carries a T-DNA insertion ∼70 bp upstream of the stop codon, and At-tha8-2 carries a Ds3 transposon insertion near the middle of the protein-coding region (Fig. 5A). Homozygous At-tha8-1 mutants exhibit retarded growth and greening, which in mature plants is mostly confined to the middle of the rosette (Fig. 5B). At-tha8-2 homozygotes are albino as seeds (Fig. 5C) and fail to germinate, consistent with the annotation of At-THA8 as embryo-essential in Arabidopsis (Bryant et al. 2011). A cross between At-tha8-1/tha8-1 and At-tha8-2/+ plants yielded heteroallelic progeny with an intermediate phenotype, confirming that the visible phenotypes result from disruption of At-tha8. At-tha8-1/At-tha8-2 are seedling-lethal when grown in soil but can survive if transferred from sucrose-containing agar medium to soil after ∼1 mo (Fig. 5D).
FIGURE 5.
Arabidopsis tha8 mutants. (A) At-THA8 (At3g27750) encodes a 222-amino acid protein that is predicted to contain three PPR motifs, but with a fourth motif inferred by comparison with other THA8 orthologs. At-tha8-1 and At-tha8-2 have T-DNA and Ds insertions, respectively, in the At-THA8 locus. (B) Visual phenotype of homozygous At-tha8-1 mutants. Homozygous At-tha8-1 plants exhibit stunted growth and delayed greening with a mild chlorophyll deficiency in the middle of the rosette. (C) Defective seeds segregating in siliques of self-pollinated At-tha8-2+ plants. (D) Heteroallelic progeny of a complementation cross between At-tha8-1/+ and At-tha8-2/+ plants.
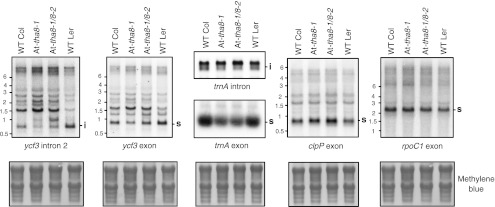
THA8's primary intron targets in maize are the ycf3-2 and trnA introns. The splicing of the orthologous introns was assayed by RNA gel-blot analysis of leaf RNA from 4-wk-old At-tha8-1 and At-tha8-1/tha8-2 mutants grown on sucrose-supplemented agar (Fig. 6). It was impractical to assay chloroplast splicing in strong mutant alleles (i.e., At-tha8-2 homozygotes) because of their embryo-lethal phenotype. However, the abundance of both the excised intron and the spliced product from ycf3-2 and trnA were reduced even in plants with the weak alleles, demonstrating a conserved role for THA8 orthologs in the splicing of these introns. The less severe ycf3 splicing defect in At-tha8-1/tha8-2 mutants compared with At-tha8-1 homozygotes might be explained by differences in the genetic background of the two alleles: At-tha8-2 was recovered from a Landsberg erecta (Ler) background, in which ycf3-2 appears to be more efficiently spliced than in the Columbia-0 (Col) background from which At-tha8-1 was recovered (Fig. 6).
FIGURE 6.
At-THA8 and maize THA8 promote the splicing of orthologous introns. Leaf RNA was analyzed by RNA gel-blot hybridization using probes specific for the indicated sequences. The methylene blue stained filters below illustrate equal loading of the samples. WT Col and WT Ler are the ecotypes in which At-tha8-1 and At-tha8-2 arose, respectively.
Arabidopsis chloroplast genomes encode three group II introns that are not found in maize: two in the clpP gene and one in the rpoC1 gene. At-THA8 does not appear to contribute to the splicing of these introns, as the spliced products from both genes accumulate normally in At-tha8-1 and At-tha8-1/8-2 mutants (Fig. 6).
Assaying the RNA and protein-binding activities of recombinant THA8 and Zm-OTP51
The long PPR tracts typical of most PPR proteins are believed to provide an extensive surface for RNA interaction, possibly via a modular 1 PPR motif-1 nucleotide mechanism (Fujii et al. 2011; Prikryl et al. 2011; Filipovska and Rackham 2012). According to this view, the longer the PPR tract, the greater the potential for high-affinity, sequence-specific interactions with RNA. To find out whether THA8, with only four PPR motifs and little else, is capable of binding its target RNA, we assayed the RNA-binding activity of recombinant THA8 (rTHA8). An MBP-THA8 fusion protein was expressed in E. coli, purified by amylose affinity chromatography, and then cleaved to separate MBP from THA8. This material was fractionated by size-exclusion chromatography, and contaminating MBP was reduced by incubation of THA8 fractions with amylose resin (Fig. 7A). The two other known ycf3-2 splicing factors, Zm-OTP51 and Zm-APO1, were also expressed as recombinant proteins (with StrepII and MBP tags, respectively), as they are potential interaction partners of THA8 (Figs. 7A, 8). Recombinant APO1 has been shown to bind RNA (Watkins et al. 2011). OTP51's RNA binding activity has not been reported, although it was expected to bind RNA via its long PPR tract and LAGLIDADG domain (Falcon de Longevialle et al. 2008).
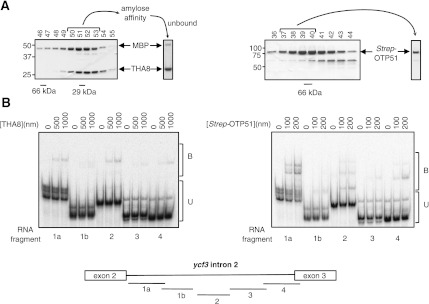
FIGURE 7.
RNA-binding activities of rTHA8 and rZm-OTP51. (A) Purification of rTHA8 (left) and rZm-OTP51 (right). Proteins were purified from E. coli via successive affinity and size-exclusion chromatography steps, as described in the Materials and Methods. Aliquots of fractions from the size-exclusion column were analyzed by SDS-PAGE; the elution positions of globular size standards are shown below. The bracketed fractions were pooled for a second amylose-affinity step to remove contaminating MBP (THA8) or for direct use in RNA-binding assays (Strep-OTP51). Coomassie-stained gels of the final preparations are shown to the right. Residual MBP remained in the THA8 preparation, but MBP does not bind RNA (Kroeger et al. 2009). (B) Gel mobility-shift assays using rTHA8 (left) and Strep-OTP51 (right). Radiolabeled RNAs of ∼150 nt were derived from regions of the ycf3-2 intron, as diagrammed below. (B) Bound RNA; (U) unbound RNA.
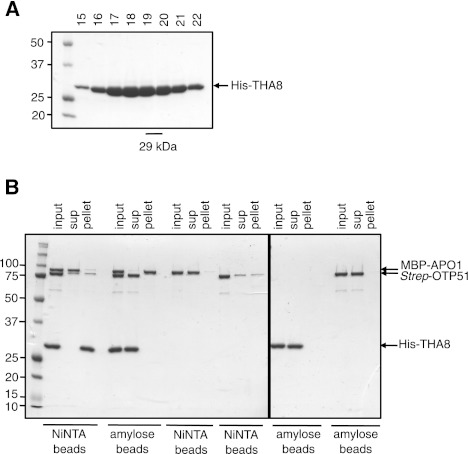
FIGURE 8.
In vitro pulldown assays for interactions between the three characterized ycf3-intron 2 splicing factors. (A) SDS-PAGE showing elution of His-THA8 from a gel filtration column. (B) Equal amounts of MBP-APO1, Strep-OTP51, and His-THA8 were combined (input), and then subjected to affinity chromatography on either NiNTA or amylose-coupled agarose beads. Each protein alone was also incubated with the beads to assess nonspecific interactions. Equal proportions of material eluted from the washed beads (pellet) and the corresponding supernatants (sup) are shown.
Gel mobility-shift assays were used to assess the ability of rZm-OTP51 and rTHA8 to bind five overlapping fragments of ycf3-2 (Fig. 7B). rTHA8 bound detectably but weakly to RNA, with binding detected only at high protein concentrations (500 μM). Zm-OTP51 bound RNA with higher affinity, with possible preference for RNA 1a, representing the first 197 nt of the intron. Previous results with APO1 likewise suggested specificity for this intron fragment (Watkins et al. 2011). However, definition of the RNA sites bound by each protein will require more detailed experimentation and is beyond the scope of the current study.
The very weak RNA-binding activity of THA8 is consistent with its short PPR tract. However, it seems unlikely that this RNA-binding activity can account for THA8's specific and stable association with the ycf3-2 and trnA introns in vivo. We therefore considered the possibility that THA8 might be recruited to the ycf3-2 intron by interacting with the other known ycf3-2 splicing factors APO1 and OTP51. To test this hypothesis, in vitro pulldown assays were performed with recombinant MBP-APO1, 6xHistidine-tagged THA8 (His-THA8), and Strep-OTP51. All three proteins were combined, and the mixture was then subjected to affinity chromatography on either NiNTA or amylose beads. The NiNTA beads pulled down His-THA8 as expected, but did not enrich either MBP-APO1 or Strep-OTP51 (Fig. 8). Likewise, the amylose beads pulled down MBP-APO1, but did not enrich either His-THA8 or Strep-OTP51 (Fig. 8). These results suggest that THA8 does not interact directly with either APO1 or OTP51, but we cannot eliminate the possibility that the recombinant proteins are misfolded and not fully functional.
DISCUSSION
In this study we describe a new component of the complex network of nucleus-encoded splicing factors in angiosperm chloroplasts (see summary in Fig. 9). THA8 is required for ycf3-2 splicing and strongly enhances trnA splicing. THA8 promotes trnA splicing in conjunction with the nucleus-encoded proteins WTF1 and RNC1, and, most likely, with the chloroplast gene product MatK (Zoschke et al. 2010). THA8 is also the third nucleus-encoded splicing factor discovered for ycf3-2, following OTP51 (Falcon de Longevialle et al. 2008) and APO1, a member of a plant-specific protein family with a novel zinc-dependent RNA-binding domain (Watkins et al. 2011). Like THA8, OTP51 and APO1 are absolutely required for ycf3-2 splicing, while also enhancing the splicing of several other introns. However, the “secondary” intron targets of these proteins do not overlap. THA8 is found in stromal ribonucleoprotein particles of ∼600–700 kDa (Fig. 4A), as are WTF1 and RNC1 (Watkins et al. 2007; Kroeger et al. 2009). Furthermore, THA8, WTF1, and RNC1 coimmunoprecipitate from chloroplast stroma, providing strong evidence that these three proteins are bound simultaneously to the trnA intron.
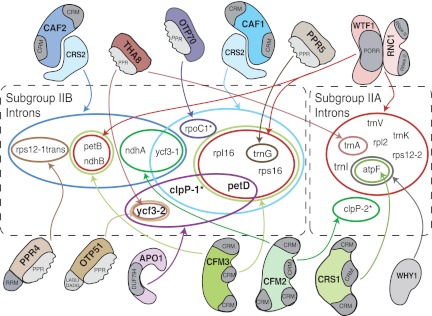
FIGURE 9.
Summary of nucleus-encoded proteins that promote the splicing of group II introns in angiosperm chloroplasts. Mutations in genes encoding the indicated proteins in maize (CAF1, CAF2, CRS1, CRS2, OTP51, PPR4, PPR5, RNC1, THA8, WHY1, WTF1) and/or Arabidopsis (CRS1, CAF1, CAF2, THA8, OTP51, OTP70, CFM2, CFM3) disrupt the splicing of the indicated introns. Where orthologous proteins have been analyzed, intron targets are conserved. All of the proteins analyzed in maize have been shown to coimmunoprecipitate with those introns whose splicing they promote. OTP51 has several minor targets (trnK, trnV, atpF, petB) not diagrammed (Falcon de Longevialle et al. 2008; Supplemental Fig. S2B). The MatK protein encoded in the trnK intron associates with all chloroplast subgroup IIA introns and is likely to be required for their splicing (Zoschke et al. 2010). This information is summarized (Jenkins et al. 1997; Vogel et al. 1999; Jenkins and Barkan 2001; Till et al. 2001; Ostheimer et al. 2003; Asakura and Barkan 2006, 2007; Schmitz-Linneweber et al. 2006; Watkins et al. 2007; Asakura et al. 2008; Beick et al. 2008; Falcon de Longevialle et al. 2008; Prikryl et al. 2008; Kroeger et al. 2009; Chateigner-Boutin et al. 2011; Watkins et al. 2011).
The group II introns in land plant chloroplasts fall into subgroups IIA and IIB, which represent an ancient division (Michel et al. 1989). The known splicing factors for these introns are summarized in Figure 9. The splicing-factor requirements within each intron subgroup overlap considerably, with less sharing of splicing factors between subgroups. THA8 acts on one intron in each subgroup. The ycf3-2 intron is unusual in that (1) each of its three known splicing factors has few other intron targets; and (2) it is the only subgroup IIB intron that does not require the peptidyl-tRNA hydrolase homolog CRS2 or any characterized member of the CRM domain family. It seems likely that these distinct features of the ycf3-2 intron and its cognate splicing factors reflect differences in splicing mechanism and/or evolutionary origin.
The results in this study add to the evidence that the machinery for chloroplast RNA splicing is highly conserved among monocot and dicot plants. We showed previously that the CRM domain proteins CAF1, CAF2, CRS1, CFM2, and CFM3 act on orthologous introns in maize and Arabidopsis (Till et al. 2001; Ostheimer et al. 2003; Asakura and Barkan 2006, 2007; Asakura et al. 2008). Here we demonstrate that THA8 orthologs in maize and Arabidopsis target orthologous introns, and that maize OTP51 functions in ycf3-2 splicing, as does its Arabidopsis ortholog (Falcon de Longevialle et al. 2008). Given this strong precedent, it seems very likely that the functions of those chloroplast splicing factors that have been studied in just one angiosperm species have conserved functions throughout the angiosperms. In fact, conservation of chloroplast splicing factors probably extends throughout the land plants, both vascular and nonvascular, as their chloroplast intron content is largely conserved (Turmel et al. 2006), and putative orthologs of THA8 and most other angiosperm chloroplast splicing factors can be found even in the moss Physcomitrella patens (data not shown). In contrast, chloroplast introns and their cognate splicing factors in green algae (e.g., Chlamydomonas reinhardtii) appear to have evolved independently of those in land plants (Turmel et al. 2006; Goldschmidt-Clermont 2009).
Although the molecular functions of THA8 orthologs in maize and Arabidopsis are conserved, the macroscopic phenotypes of tha8 mutants in the two species differ. Null alleles of maize tha8 condition a pale green, seedling-lethal phenotype, whereas Arabidopsis THA8 is essential for embryogenesis. We were nonetheless able to discern that THA8 function in Arabidopsis is similar to that in maize through analysis of weak alleles. THA8's role in trnA splicing likely accounts for its requirement during Arabidopsis embryogenesis: the deficiency for trnA will compromise chloroplast translation, which is known to be required for embryogenesis in Arabidopsis (Bryant et al. 2011). In contrast, numerous maize mutants with severe plastid translation defects have been shown to develop normally through the first few weeks of seedling growth, at which time they die due to their photosynthetic defect. In fact, the chloroplast splicing factors CAF2, WTF1, PPR4, and PPR5 are likewise embryo essential in Arabidopsis, but not in maize (Ostheimer et al. 2003; Asakura and Barkan 2006; Schmitz-Linneweber et al. 2006; Beick et al. 2008; Kroeger et al. 2009). The basis for this difference between the species is likely due to the presence of an essential gene, accD, in Arabidopsis chloroplasts, whose function in fatty acid synthesis is provided by a nuclear gene in maize (Bryant et al. 2011).
THA8 belongs to a subfamily of PPR proteins that have four PPR motifs preceded by a short conserved N-terminal region that is also predicted to adopt a helical hairpin structure. The THA8-like subfamily includes four members in angiosperms and one in the moss Physcomitrella patens. THA8 is the only member of this group to be functionally characterized. THA8 stands out among the PPR proteins studied so far, in that it has few PPR motifs and yet it associates with specific RNAs in vivo. As a point of comparison, seven PPR proteins have been shown previously to promote the splicing of group II introns (PPR4, PPR5, OTP51, and OTP70 in chloroplasts [Schmitz-Linneweber et al. 2006; Beick et al. 2008; Falcon de Longevialle et al. 2008; Chateigner-Boutin et al. 2011]; OTP43, BIR6, and ABO5 in mitochondria [Falcon de Longevialle et al. 2007; Koprivova et al. 2010; Liu et al. 2010]), and each of these has at least 10 PPR motifs. If PPR tracts bind RNA via the modular mechanism that is currently predicted (Fujii et al. 2011; Prikryl et al. 2011; Filipovska and Rackham 2012), it seems unlikely that THA8's four PPR motifs can account for its stable and specific association in vivo with just two introns. We have so far been unable to detect strong interactions between recombinant THA8 and either intron RNA or other splicing factors that could account for the in vivo findings. This suggests the possibility that additional proteins may be involved in recruiting THA8 to specific introns.
Our attention was originally drawn to tha8 because the thylakoid protein defects of tha8 mutants resemble those of hcf106 and tha4 mutants, which lack components of the Tat thylakoid transport machinery. However, we were unable to detect any defect in the assembly of the Tat machinery in tha8 mutants, nor any association between THA8 and Tat components. These negative results, together with our finding that THA8 is a chloroplast splicing factor, make a direct role for THA8 in the Tat pathway very unlikely. Presumably, the Tat pathway defect in tha8 mutants is a secondary effect of compromised thylakoid energetics that result from the chloroplast gene expression defects. However, it is rare that nonphotosynthetic mutants exhibit Tat-pathway defects; for example, mutants such as crp1, ppr10, pet2, and cps1 are pale green, seedling lethal, and lack various components of the thylakoid electron transport chain, but accumulate normal levels of mature OE16 and OE23 (data not shown). Thus, specific aspects of the energetic deficits in tha8 mutants likely underlie their Tat defect, but the relevant deficits remain unclear. This history is reminiscent of the evolving view of APO1, originally suggested to be an Fe-S cluster assembly factor (Amann et al. 2004), but now known to be a chloroplast splicing factor (Watkins et al. 2011), and highlights the challenge of distinguishing direct from indirect effects during mutant analysis.
MATERIALS AND METHODS
Cloning the tha8 gene
A Mu1 insertion that cosegregates with tha8-1 was discovered by using Southern blot hybridization to display Mu elements in plants of known genotype (Voelker et al. 1997). This insertion was cloned and its position determined by inverse PCR and DNA sequencing. To validate the identification of the tha8 locus, an independent insertion in the same gene (tha8-2) was isolated in a reverse-genetic screen of the PML (http://pml.uoregon.edu/photosyntheticml.html) collection. The two mutations failed to complement when heterozygous plants were crossed. The tha8 locus is designated as AC217965.2_FG012 at UniProt. Its position is coincident with gene model GRMZM2G466032 in the current maize genome release. However, the direction of transcription shown for GRMZM2G466032 is incorrect, so the inferred protein product is not correctly annotated.
Zm-otp51 mutants
Two insertion alleles in the maize ortholog of Arabidopsis OTP51 were recovered in a reverse-genetic screen of our maize mutant collection, using Mu-specific and gene-specific primers as described previously (Ostheimer et al. 2003). The gene-specific primers were designed and tested by John Chapman Robbins in Maureen Hanson's laboratory (Cornell University). The mutants analyzed here are the heteroallelic progeny of a complementation cross between Zm-otp51-2 and Zm-otp51-3. Genotypes were confirmed by PCR with primers pl1_3_1032 and EOMuMix (see Supplemental Table S1 in the Supplemental Material).
Arabidopsis tha8 mutants
Insertions in the Arabidopsis tha8 ortholog, At-THA8 (locus At3g27750) were obtained from the SALK collection (At-tha8-1:SALK_140334, Col background) (Alonso et al. 2003) and from the Cold Spring Harbor Genetrap collection (At-tha8-2: CSHL_GT16880, Ler background) (Sundaresan et al. 1995). The plants were genotyped using insertion-specific primers LBb1.3 (for SALK_140334) or Ds3-1 (for CSHL_GT16880), and a gene-specific primer Attha8_rev (see Supplemental Table S1; Supplemental Fig. S3).
Plant growth
Maize seedlings used for phenotypic analyses were germinated in soil and grown in a growth chamber under the following conditions: 16-h days, 27°C, ∼500 μmol photons m−2 s−1; 8 h nights, 25°C. Leaf material was harvested after 7–8 d, at which time seedlings had between two and three leaves. Arabidopsis plants were germinated and grown on Murashige and Skoog medium supplemented with 2% sucrose in 16-h days at 21°C, 50–70 μmol photons m−2 sec−1 for ∼4 wk. Then, the plants were transferred to soil and grown until maturity in 16-h days at 21°C, 50–70 μmol photons m−2 sec−1.
Production of THA8 antiserum
A NotI–SalI fragment of the tha8 cDNA was cloned into pET-28a(+) (Novagen), such that the encoded protein carries an N-terminal 6× histidine tag followed by 43 vector-encoded amino acids and THA8's C-terminal 219 amino acids. The protein was expressed in E.coli BL21(DE3) cells (Stratagene), purified by nickel-affinity chromatography (Ni-NTA, Qiagen) under native conditions, and used for antibody production in rabbits at the University of Oregon Polyclonal Antibody Facility. The antisera were affinity-purified using THA8 antigen immobilized on NHS HiTrap columns (GE Healthcare). The antibody was eluted in 0.1 M glycine (pH 2.6), and immediately neutralized by addition of 1 M Tris base. The glycine buffer was substituted with PBS using a Centricon filter unit (Millipore). BSA and sodium azide were added to concentrations of 0.5% and 0.05% (w/v), respectively. Antibodies were divided into aliquots and stored at −80°C.
Analysis of leaf and chloroplast proteins
Leaf proteins were isolated and analyzed by immunoblotting as described previously (Barkan 1998). Chloroplast isolation, chloroplast fractionation, and the antibodies used to detect thylakoid proteins were as described (Voelker and Barkan 1995). Mitochondria were purified from etiolated maize seedlings as described by Asakura et al. (2008). Antibodies to the mitochondrial protein MDH were a generous gift of Kathy Newton. Thylakoids were treated with proteases or chaotropic agents as in Walker et al. (1999). Coimmunoprecipitation of THA8 with other splicing factors from chloroplast stroma was assayed using the antibodies and procedure described previously (Till et al. 2001; Ostheimer et al. 2003; Asakura and Barkan 2007, 2008; Watkins et al. 2007; Prikryl et al. 2008; Kroeger et al. 2009).
Sucrose gradient fractionation of chloroplast stroma
Chloroplast stroma was prepared as described (Jenkins and Barkan 2001). A total of 400 μL of stroma (∼10 mg protein/mL) was mixed with 1.5 μL of 10 mg/mL RNAse A (RNAse treatment) or with water (control) and left on ice for 30 min. Thereafter, stroma was fractionated by sedimentation through 5-mL 10%–30% sucrose gradients as described previously (Watkins et al. 2007). Ten 500-μL fractions plus an additional fraction containing pelleted material were collected and used for RNA isolation and SDS-PAGE.
RNA immunoprecipitation and microarray analysis (RIP–chip)
The RIP–chip assay was performed with maize chloroplast stroma and a tiling microarray of the maize chloroplast genome, as described previously (Schmitz-Linneweber et al. 2005; Barkan 2009). The same RNA samples were used for the slot-blot hybridizations, using the indicated intron-specific probes and the method described previously (Barkan 2009).
Northern blotting and poisoned primer extension (PPE)
Total leaf RNA was isolated with TriReagent (MRC). RNA gel-blot hybridizations were performed as described (Barkan 1998), using 5 μg of total seedling leaf RNA per lane. Probes were PCR products labeled by random-hexamer priming with [α-32P]dCTP. The PCR primers are listed in Supplemental Table S1. The ndhB probe corresponds to position 89551–90139 nt of the chloroplast genome. Poisoned primer extension assays were performed as described (Asakura and Barkan 2006; Barkan 2011b), with the primers listed in Supplemental Table S1. Radioactive gels and blots were exposed to a PhosphorImager screen (Molecular Dynamics) and analyzed using ImageQuant software (GE Healthcare).
Expression of recombinant proteins
MBP-ZmAPO1 expression and purification were described previously (Watkins et al. 2011). Mature Zm-OTP51 (amino acids 79–783 of the protein encoded by locus GRMZM2G325019) was expressed from a cDNA constructed by separate amplification of each of the two exons, followed by fusion via overlapping PCR. An N-terminal Strep-tag II (WSHPQFEK) (Schmidt and Skerra 2007) was introduced through one of the primers. The product was cloned into pGEM-T Easy (Promega) and then subcloned into pET-28a(+) using the NcoI and BamHI sites. Sequences encoding mature Zm-tha8 (amino acids 33–263) were amplified from maize leaf cDNA. Mature THA8 with an N-terminal 6× histidine tag was expressed from a clone containing this cDNA inserted into the NdeI/HindIII sites of pET-28c(+) (Novagen). The MBP-THA8 fusion was generated by cloning this cDNA into pMAL-TEV using BamHI and PstI sites. Proteins were expressed in Rosetta 2 cells (MBP-THA8) or Rosetta 2 (DE3) cells (Strep-OTP51 and His-THA8) (Novagen). The cells were grown at 37°C until OD600∼0.6, and then moved to 20°C. After 30 min at 20°C, protein expression was induced with 0.5 mM IPTG (final concentration) and grown for 4–5 h before harvesting. The cell pellet was resuspended in ice-cold lysis buffer (50 mM Tris at pH 7.5, 300–400 mM NaCl, 0.06% β-mercaptoethanol) and sonicated. The lysate was cleared by centrifugation at 13,000g for 20 min. The proteins were purified by affinity chromatography using NiNTA agarose beads (Qiagen), Strep-Tactin Sepharose (IBA) or amylose-coupled agarose beads (New England Biolabs) as appropriate, according to manufacturer's instructions. MBP-THA8 was treated with TEV protease overnight to cleave off MBP. Prior to purification on a gel-filtration column (1 cm × 30 cm Superdex 200 equilibrated in lysis buffer), particulates were removed by centrifugation at 13,000g for 10 min and filtration through Amicon Ultrafree-MC (0.45 μm). Column fractions were dialyzed against lysis buffer supplemented with 50% glycerol using a Slide-A-Lyzer dialysis cassette (Pierce) and stored at −20°C.
RNA-binding assays
Gel mobility-shift assays were performed as described in Watkins et al. (2011), using the same fragments of the ycf3-2 intron. Reactions included 40 pM RNA, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 4 mM DTT, 0.08 units/μL RNAsin, 0.04 mg/mL BSA, and 100 ng/μL heparin. Gels were dried and exposed to a PhosphorImager screen.
In vitro protein–protein pulldown assays
Twelve micrograms each of MBP-ZmAPO1, Strep-ZmOTP51 and His-ZmTHA8 were combined in 100-μL reactions containing 50 mM Tris-HCl (pH 7.5), 60 mM NaCl, 10 mM β-mercaptoethanol, 0.01% Nonidet P-40, 10 mM imidazole, and 10% glycerol. The controls contained 12 μg of each protein on its own in 100-μL reactions. An aliquot (8 μL) was removed from each reaction as the “input.” The reactions were incubated at RT for 10 min and then for 45 min on ice. Sixty microliters of either NiNTA (Qiagen) or amylose (New England Biolabs) agarose beads, pre-rinsed in the reaction buffer, were added to samples and the incubation continued at 4°C on a rotator for 60 min. The beads were pelleted by centrifugation, and an aliquot (10 μL) of the supernatant was removed for later analysis. The beads were washed four times with 700 μL of reaction buffer supplemented with 20 mM imidazol and lacking glycerol. The proteins bound to the beads (“pellet” fraction) were eluted in 90 μL of SDS-PAGE loading buffer, and 12 μL of the elution was analyzed by SDS-PAGE alongside the input and supernatant fractions.
DATA DEPOSITION
The sequence of the tha8 locus and cDNA have been deposited in GenBank under accession numbers AY530729 and AY530730, respectively.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are especially grateful to Susan Belcher for help with tha8 and otp51 complementation crosses, Roz Williams-Carrier for identifying the Mu1 insertion that cosegregates with tha8, and Ken Cline (University of Florida) for assistance with analyzing connections between THA8 and Tat components. We also thank Steve Theg (University of California-Davis) and David Kramer (Washington State University) for contributing to the analysis of thylakoid energetics in tha8 mutants, John Robbins (Cornell University) for designing the primers used to screen for Zm-otp51 mutants, and Kathy Newton (University of Missouri) for providing antibodies to the mitochondrial protein MDH. This work was supported by National Science Foundation grants IOS-0922560 and MCB-0744960 to A.B., a postdoctoral fellowship from the National Institutes of Health to R.M., and a postdoctoral fellowship from The Danish Council for Independent Research (Natural Sciences) to A.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032623.112.
REFERENCES
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amann K, Lezhneva L, Wanner G, Herrmann RG, Meurer J 2004. ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A 2006. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A 2007. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell 19: 3864–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Bayraktar O, Barkan A 2008. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA 14: 2319–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A 1998. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57 [Google Scholar]
- Barkan A. 2009. Genome-wide analysis of RNA-protein interactions in plants. In Plant systems biology (ed. D Belostotsky), Vol. 553, pp. 13–37. Humana Press, New York. [Google Scholar]
- Barkan A 2011a. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. 2011b. Studying the structure and processing of chloroplast transcripts. In Chloroplast research in Arabidopsis: Methods and protocols (ed. P Jarvis), Vol. 774, pp. 183–197. Humana Press, New York. [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A 2008. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D 2011. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis thaliana. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Colas des Francs-Small C, Delannoy E, Kahlau S, Tanz SK, Falcon de Longevialle A, Fujii S, Small I 2011. OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J 65: 532–542 [DOI] [PubMed] [Google Scholar]
- Cline K, Dabney-Smith C 2008. Plastid protein import and sorting: different paths to the same compartments. Curr Opin Plant Biol 11: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon de Longevialle A, Meyer EH, Andrès C, Taylor NL, Lurin C, Millar AH, Small ID 2007. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon de Longevialle A, Hendrickson L, Taylor N, Delannoy E, Lurin C, Badger M, Millar AH, Small I 2008. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Falcon de Longevialle AF, Small ID, Lurin C 2010. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3: 691–705 [DOI] [PubMed] [Google Scholar]
- Filipovska A, Rackham O 2012. Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol Biosyst 8: 699–708 [DOI] [PubMed] [Google Scholar]
- Fujii S, Bond CS, Small ID 2011. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc Natl Acad Sci 108: 1723–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giege P 2010. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17: 740–744 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. 2009. Chloroplast RNA splicing. In The Chlamydomonas sourcebook: Organellar and metabolic processes (ed. D Stern, E Harris), pp. 915–936. Academic Press, Oxford, UK. [Google Scholar]
- Jenkins B, Barkan A 2001. Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J 20: 872–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B, Kulhanek D, Barkan A 1997. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Colas des Francs-Small C, Calder G, Mugford ST, Tanz S, Lee BR, Zechmann B, Small I, Kopriva S 2010. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J Biol Chem 285: 32192–32199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger T, Watkins K, Friso G, Wijk Kv, Barkan A 2009. A plant-specific RNA binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2011. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616 doi: 10.1101/cshperspect.a003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JW, Rose RJ 1992. The spinach chloroplast chromosome is bound to the thylakoid membrane in the region of the inverted repeat. Biochem Biophys Res Commun 184: 993–1000 [DOI] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J 63: 749–765 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ 2012. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Ostheimer G, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A 2003. Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J 22: 3919–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar O, Prikryl J, Barkan A 2009. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, Wijk KJ, Barkan A 2008. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Rojas M, Schuster G, Barkan A 2011. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D 2011. Structure of human mitochondrial RNA polymerase. Nature 478: 269–273 [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Kossel H, Bock R 1997. Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J Cell Biol 139: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TG, Skerra A 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc 2: 1528–1535 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A 2005. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A 2006. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 Pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N 2000. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A 2001. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C 2006. The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol 23: 1324–1338 [DOI] [PubMed] [Google Scholar]
- Voelker R, Barkan A 1995. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J 14: 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R, Mendel-Hartvig J, Barkan A 1997. Transposon-disruption of a maize nuclear gene, tha1, encoding a chloroplast SecA homologue: in vivo role of cp-SecA in thylakoid protein targeting. Genetics 145: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Boerner T, Hess W 1999. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res 27: 3866–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Roy L, Coleman E, Voelker R, Barkan A 1999. The maize tha4 gene functions in sec-independent protein transport in chloroplasts and is related to hcf106, tatA, and tatB. J Cell Biol 147: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K, Kroeger T, Cooke A, Williams-Carrier R, Friso G, Belcher S, Wijk Kv, Barkan A 2007. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K, Rojas M, Friso G, Wijk Kv, Meurer J, Barkan A 2011. APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Borner T, Schmitz-Linneweber C 2010. An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]