Translation termination in eukaryotes is carried out by the class 1 release factor eRF1. A cavity model for stop codon recognition argues that three distinct binding pockets on the surface of eRF1 are important in recognition of the three specific nucleotides of stop codons. The binding pocket sequence motifs are typically divergent in organisms that depend on an altered code. Here, evidence supporting the cavity model is provided through a mutational analysis performed on reporter proteins in Saccharomyces cerevisiae. Moreover, the identified key residues extend beyond the previously identified TASNIKS and YCF motifs.
Keywords: eRF1, translation termination, stop codon, protein synthesis, translation, ciliates
Abstract
The initiation and elongation stages of translation are directed by codon–anticodon interactions. In contrast, a release factor protein mediates stop codon recognition prior to polypeptide chain release. Previous studies have identified specific regions of eukaryotic release factor one (eRF1) that are important for decoding each stop codon. The cavity model for eukaryotic stop codon recognition suggests that three binding pockets/cavities located on the surface of eRF1's domain one are key elements in stop codon recognition. Thus, the model predicts that amino acid changes in or near these cavities should influence termination in a stop codon-dependent manner. Previous studies have suggested that the TASNIKS and YCF motifs within eRF1 domain one play important roles in stop codon recognition. These motifs are highly conserved in standard code organisms that use UAA, UAG, and UGA as stop codons, but are more divergent in variant code organisms that have reassigned a subset of stop codons to sense codons. In the current study, we separately introduced TASNIKS and YCF motifs from six variant code organisms into eRF1 of Saccharomyces cerevisiae to determine their effect on stop codon recognition in vivo. We also examined the consequences of additional changes at residues located between the TASNIKS and YCF motifs. Overall, our results indicate that changes near cavities two and three frequently mediated significant effects on stop codon selectivity. In particular, changes in the YCF motif, rather than the TASNIKS motif, correlated most consistently with variant code stop codon selectivity.
INTRODUCTION
In prokaryotes, translation termination occurs when a Class I release factor (RF) recognizes a stop codon located in the ribosomal A site. The two Class I release factors, RF1 and RF2, recognize UAA/UAG and UAA/UGA codons, respectively, and then facilitate polypeptide chain release (Ito et al. 2000). The Class II release factor, RF3, is a GTPase that disassociates the Class I RF from the ribosome following polypeptide release (Zavialov et al. 2001). In eukaryotes, this process differs in several ways. Polypeptide release occurs after the Class I release factor, eRF1, recognizes any one of three termination codons (UAA, UAG, and UGA) (Kisselev et al. 2003). The eukaryotic Class II release factor, eRF3, is a GTPase that facilitates recognition of some termination signals, and also greatly enhances the rate of polypeptide chain release mediated by eRF1 (Frolova et al. 1996; Salas-Marco and Bedwell 2004; Alkalaeva et al. 2006). Thus, eRF3 plays a much more direct role in the termination process than the prokaryotic Class II factor, RF3.
eRF1 has a modular organization with three functional domains (Song et al. 2000). Domain one mediates stop codon recognition and contains the conserved TASNIKS and YxCxxxF (YCF) motifs (Bertram et al. 2000; Song et al. 2000; Frolova et al. 2002). Domain two contains the universally conserved GGQ motif that promotes polypeptide chain release (Frolova et al. 1999; Song et al. 2000). Domain three of eRF1 interacts with eRF3 (Ito et al. 1998; Merkulova et al. 1999).
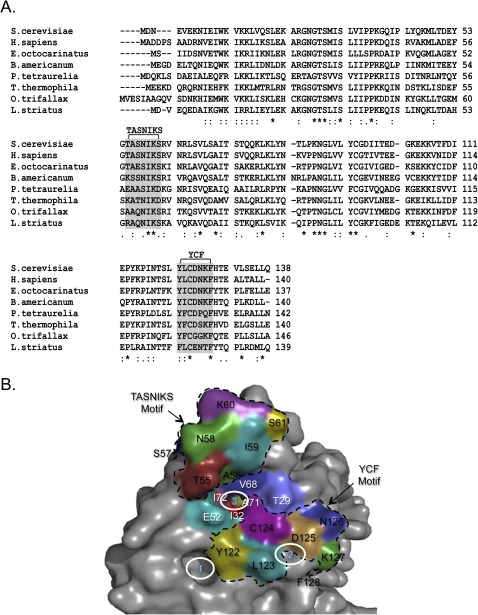
A number of organisms do not recognize all three stop codons utilized in the standard genetic code. Such “variant code” species are particularly common among the ciliated protozoa, a group of unicellular eukaryotes (Kim et al. 2005). In general, these species either utilize UGA as a stop codon (with UAA and UAG recoded to glutamine codons) (Horowitz and Gorovsky 1985; Lozupone et al. 2001; Kim et al. 2005) or UAA/UAG as stop codons (with UGA recoded to cysteine or tryptophan codons) (Meyer et al. 1991; Lozupone et al. 2001). Past studies found that fusion of eRF1 domain one from a variant code organism to eRF1 domains two and three from a standard code organism often conferred variant stop codon recognition (Kervestin et al. 2001; Salas-Marco et al. 2006; Lekomtsev et al. 2007a,b; Fan-Minogue et al. 2008; Eliseev et al. 2011). These results demonstrated that eRF1 domain one plays a key role in stop codon recognition. The primary amino acid sequences of domain one from standard and variant code organisms have also been examined to identify more defined regions involved in stop codon recognition. Two sequence elements within domain one, the TASNIKS and YCF motifs, were found to be highly conserved in standard code organisms, but more divergent in variant code species (Fig. 1A; Knight et al. 2000; Song et al. 2000; Lozupone et al. 2001; Kim et al. 2005), suggesting that these sequence elements play a role in stop codon recognition. Mutagenesis studies examined the effects of changing amino acids within the TASNIKS and YCF motifs of eRF1 (Bertram et al. 2000; Frolova et al. 2002; Seit-Nebi et al. 2002; Kolosov et al. 2005; Fan-Minogue et al. 2008; Cheng et al. 2009). Those studies confirmed the general importance of both motifs in stop codon recognition. However, the relative importance of specific amino acids examined in different studies has been difficult to compare directly because of the different experimental systems and strategies used.
FIGURE 1.
Domain 1 of eRF1 mediates stop codon recognition. (A) Protein sequence alignment of eRF1 domain one from multiple species. The star symbol indicates fully conserved residues; the colon indicates strong conservation; and the period indicates weak conservation. (B) Location of eRF1 residues in the context of the human eRF1 structure (Song et al. 2000). YCF and TASNIKS residues are in black type and bordered by dotted lines. Residues between the TASNIKS and YCF motifs are in white type. The three solid white circles indicate the cavities proposed to mediate stop codon recognition (Bertram et al. 2000).
Several models of eukaryotic stop codon recognition have been proposed. An early model proposed that residues located in the TASNIKS motif, T55/A56/S57 (Saccharomyces cerevisiae numbering) act as a linear “peptide anticodon” that interacts directly with the stop codon (Nakamura and Ito 2002). A second model suggested that residues in or near the TASNIKS motif serve as key determinants in decoding stop codons. These residues, located in a helical region of the protein, were proposed to assume either a relaxed or tight conformation dependent upon the stop codon recognized. Moreover, it was suggested that residues located on the helix, G54/T55 and S57/N58 (S. cerevisiae numbering), recognize the second and third nucleotide of the stop codon, respectively (Muramatsu et al. 2001). Further supporting a role for the TASNIKS motif in stop codon recognition, another study found that an mRNA containing 4-thiouracil in the first position of the stop codon could be cross-linked to residues in (or near) the TASNIKS motif of human eRF1, particularly the K60 residue (S. cerevisiae numbering; underlined) (Chavatte et al. 2002).
Additional mutagenesis studies have shown that mutations in other regions of domain one also influence stop codon recognition. This led to the proposal of a “nonlinear” model where stop codons are recognized by positive and negative determinants (Frolova et al. 2002; Seit-Nebi et al. 2002). Another group used a genetic approach to identify mutations in domain one that altered the recognition of specific stop codons in vivo. Those results, in conjunction with the crystal structure of human eRF1 (Song et al. 2000), led to a model in which three cavities (or pockets) located on the surface of domain one were proposed to recognize each of the three nucleotides of the stop codon (Fig. 1B; Bertram et al. 2000). Consistent with the cavity model, a crystallization study of an eRF1/eRF3 complex found that a component of the precipitant solution, ATP, was stably bound to one of the proposed cavities (Cheng et al. 2009).
In the current study, we used a previously established yeast system to determine the effects of different eRF1 mutations on stop codon selectivity (Keeling et al. 2004; Salas-Marco and Bedwell 2004; Kallmeyer et al. 2006; Salas-Marco et al. 2006; Fan-Minogue et al. 2008). In particular, we compared the relative importance of the TASNIKS and YCF motifs in stop codon recognition. To do this, we introduced amino acid changes found in six divergent TASNIKS and YCF motifs from a series of variant code species, both separately and together, into the eRF1 protein of the standard code organism Saccharomyces cerevisiae. We then assayed stop codon recognition of each mutant protein expressed as the sole source of eRF1 in vivo. We found that many of these changes altered stop codon recognition in a manner consistent with a previously proposed cavity model of stop codon recognition (Bertram et al. 2000). Additional missense mutations located between the TASNIKS and YCF motifs near cavity three also modified stop codon discrimination in distinct ways. When taken together, our results suggest that key determinants for eRF1 stop codon recognition map to a surface of domain one that includes three cavities previously proposed to recognize the three nucleotides of stop codons. Importantly, this surface includes, but extends beyond, the TASNIKS or YCF motifs. Moreover, changes in the YCF motif correlated most consistently with variant code stop codon selectivity.
RESULTS
To examine the relative importance of residues within the TASNIKS and YCF motifs for stop codon recognition, we introduced these motifs from six variant code species into eRF1 of the standard code organism Saccharomyces cerevisiae. eRF1 mutants that supported growth in the absence of wild-type eRF1 were assayed directly to determine their pattern of stop codon recognition. Mutants unable to support cell viability as the sole source of eRF1 were transformed into a sup45Δ strain that also expressed wild-type eRF1 under GAL1 promoter control. These strains were grown in galactose medium to express both wild-type and mutant eRF1, shifted to glucose medium to halt production of wild-type eRF1, and grown for six more generations to deplete wild-type eRF1. The efficiency of translation termination at each stop codon was measured using a well-established dual luciferase readthrough reporter system (Grentzmann et al. 1994; Howard et al. 2000; Keeling et al. 2004; Salas-Marco and Bedwell 2004, 2005; Salas-Marco et al. 2006; Kamenski et al. 2007; Fan-Minogue et al. 2008).
A single amino acid change at L123 from the Euplotes and Blepharisma YCF motifs results in strong UAA/UAG specificity
Euplotes species utilize UAA and UAG as stop codons, while Blepharisma species utilize UAA (and possibly UAG) (Meyer et al. 1991; Lozupone et al. 2001; Kim et al. 2005). We first examined the effect of introducing divergent amino acids from the TASNIKS and YCF motifs of Euplotes octocarinatus and Blepharisma americanum into S. cerevisiae eRF1. Expression of mutant eRF1 proteins containing the individual TASNIKS or YCF motifs all supported cell viability as the sole source of eRF1, while the double mutants containing both motifs from either organism were unable to sustain cell viability. The ability of the double mutants to recognize UAA, UAG, and UGA stop codons was thus assayed using the shift assay described above.
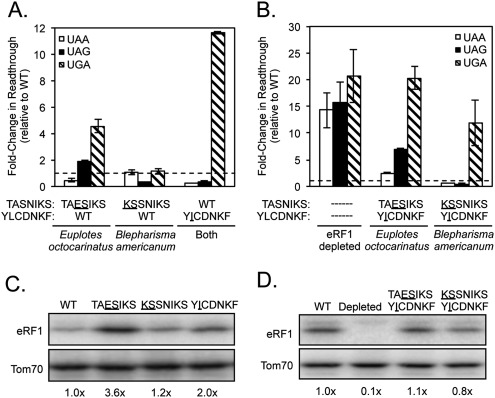
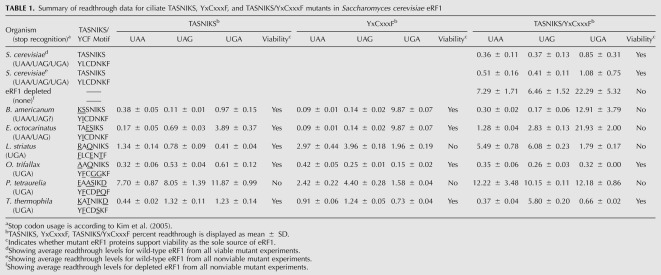
Both Euplotes octocarinatus and Blepharisma americanum eRF1 proteins contain only a single divergent isoleucine at position 123 in their YCF motifs (YICDNKF) instead of the leucine residue at the corresponding position of the S. cerevisiae YCF motif (YLCDNKF). Introduction of the L123I mutation reduced termination efficiency at the UGA codon >11-fold when compared with wild-type (S. cerevisiae) eRF1 (Fig. 2A; Table 1). In contrast, this mutant exhibited more efficient termination at UAA and UAG stop codons than wild-type eRF1. These results suggested that recognition of guanine (UGA) in the second position was inhibited, while adenine recognition in position 2 (UAA and UAG) was enhanced by this single amino acid substitution. The contribution of the TASNIKS motifs to the variant stop codon recognition used by these organisms was much less direct. A UAA/UAG-specific pattern of stop codon recognition was not observed in the mutant carrying the Blepharisma KSSNIKS motif. The Euplotes TAESIKS mutant showed a partial loss of UGA recognition (as well as a slight loss of UAG recognition) (Fig. 2A; Table 1). The double mutant carrying the Euplotes TAESIKS and YICDNKF motifs exhibited a greater loss of UGA recognition than either motif alone, but also showed defects in UAA and UAG recognition (Fig. 2B). Finally, the mutant carrying both the Blepharisma KSSNIKS and YICDNKF motifs exhibited the same strong UAA/UAG-specific readthrough phenotype as the YCF mutant alone (Fig. 2B), suggesting that Blepharisma americanum is a UAA/UAG-specific organism. Taken together, these results indicate that the isoleucine at residue 123 plays a key role in excluding guanine in the second position of the stop codon in these two UAA/UAG-specific variant-code organisms, consistent with the close proximity of L123 to cavity two in the Stansfield model (see Fig. 1B).
FIGURE 2.
The L123I mutation from the Euplotes and Blepharisma YCF motifs reduces eRF1 recognition of the UGA codon. (A,B) Fold change in readthrough by viable (A) and inviable (B) strains expressing the indicated mutant eRF1 proteins compared with wild-type S. cerevisiae eRF1. The data are expressed as mean values (relative to wild-type eRF1) ± standard deviation. The horizontal dashed lines represent wild-type S. cerevisiae eRF1 levels. (C,D) Western blots showing steady-state eRF1 proteins levels from strains assayed in A and B, respectively.
TABLE 1.
Summary of readthrough data for ciliate TASNIKS, YxCxxxF, and TASNIKS/YxCxxxF mutants in Saccharomyces cerevisiae eRF1
Previous findings have shown that a significant decrease in the level of eRF1 protein reduces the efficiency of translation termination at all three stop codons (Salas-Marco and Bedwell 2004). To exclude such nonspecific effects, we routinely included two control assays. First, we measured readthrough in a strain depleted of eRF1 protein. As expected, this strain exhibited a large (15- to 20-fold) decrease in termination efficiency at each of the three stop codons (Fig. 2B). This uniform pattern of reduced termination efficiency is distinct from the stop codon-specific results obtained with most eRF1 mutants discussed above. We also monitored the steady-state level of each mutant eRF1 protein by Western blot analysis. Nonviable eRF1 mutants were assayed after a galactose to glucose shift, followed by continued growth for six generations to deplete WT eRF1 (as done for the readthrough assays). None of the Euplotes or Blepharisma substitutions resulted in a significant reduction in eRF1 abundance (Fig. 2C,D). However, the YICDNKF and TAESIKS mutants displayed twofold and 3.6-fold increases in eRF1 abundance, respectively. Similar increases were previously noted for some eRF1 mutants (Fan-Minogue et al. 2008) and are thought to reflect a regulatory mechanism that couples eRF1 abundance to the efficiency of translation termination (S Conard, A Kallmeyer, and D Bedwell, unpubl.). When taken together, these results indicate that the codon-specific changes in termination efficiency described above were not simply due to inadequate levels of the mutant eRF1 proteins.
S. cerevisiae eRF1 displays UGA specificity when the Paramecium, Tetrahymena, or Oxytricha YCF motifs are introduced
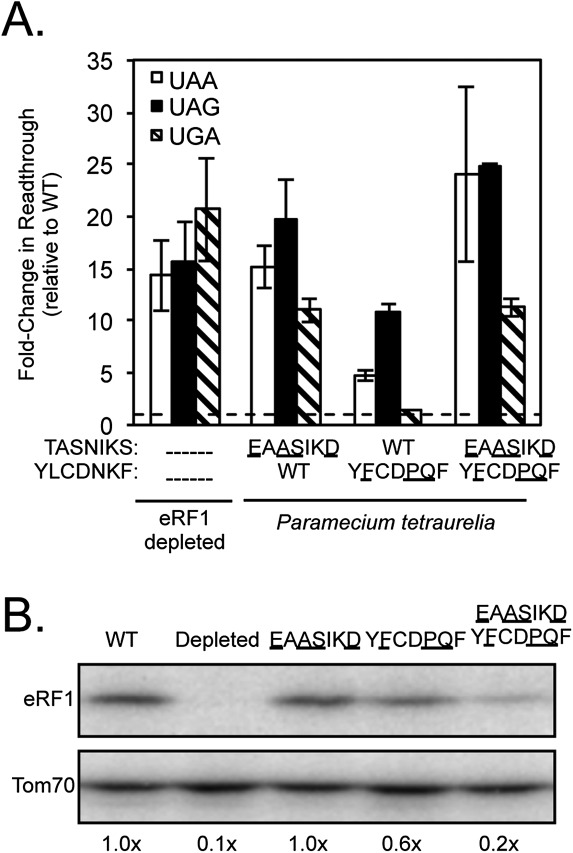
Paramecium species have reassigned UAA and UAG to glutamine codons, and use only UGA as a termination codon (Preer et al. 1985; Kim et al. 2005). To determine whether the TASNIKS and YCF motifs of Paramecium tetraurelia contain determinants for UGA specificity, we introduced the TASNIKS (EAASIKD), YCF (YFCDPQF), or both motifs from this organism into S. cerevisiae eRF1. The YCF mutant displayed strong UGA specificity, as five- and 10-fold decreases in termination efficiency were observed at UAA and UAG codons, respectively (Fig. 3A; Table 1). In contrast, termination at the UGA codon decreased only slightly (1.5-fold). These results indicate that the three amino acid changes in the YFCDPQF motif were sufficient to reduce recognition of adenine in the second position of UAA and UAG stop codons. The TASNIKS and double mutants showed strong decreases in termination efficiency at all three stop codons, suggesting that they compromised overall eRF1 protein function and/or abundance when introduced into S. cerevisiae eRF1. A large (fivefold) decrease in eRF1 abundance of the double mutant is also consistent with its general readthrough phenotype (Fig. 3B). These results show that the divergent YCF motif is sufficient to provide a strong bias toward UGA stop codon specificity as observed in Paramecium species.
FIGURE 3.
Introduction of the YCF motif of Paramecium tetraurelia into S. cerevisiae eRF1 results in UGA specificity. (A) Fold change in readthrough by strains expressing the indicated mutant eRF1 proteins compared with wild-type S. cerevisiae eRF1. The data are expressed as mean values (relative to wild-type eRF1) ± standard deviation. The horizontal dashed lines represent wild-type S. cerevisiae eRF1 levels. (B) Western blot showing steady-state eRF1 proteins levels from strains in A.
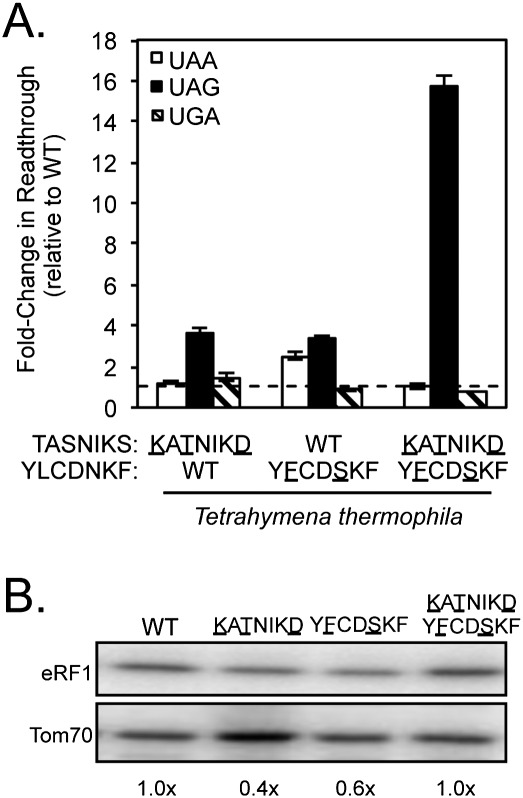
Like Paramecium tetraurelia, Tetrahymena species also mediate translation termination exclusively at UGA codons (Horowitz and Gorovsky 1985; Kim et al. 2005). To further explore how divergent TASNIKS and YCF sequences influence stop codon selectivity, we next tested the amino acid changes found in the TASNIKS and YCF motifs of Tetrahymena thermophila. Introduction of the Tetrahymena YCF motif (YFCDSKF) into S. cerevisiae eRF1 resulted in modest UGA specificity (Fig. 4A; Table 1). However, the increases in readthrough at UAA and UAG codons (2.5-fold and 3.4-fold, respectively) were much less than was observed with the Paramecium YCF motif. UGA-specific termination was not observed with strains expressing eRF1 proteins containing the Tetrahymena TASNIKS motif (KATNIKD) or the KATNIKD/ YFCDSKF double mutant. Instead, these mutants exhibited large (fourfold and 16-fold increases, respectively) in readthrough at the UAG termination codon. These results suggest that one or more of the changes in the TASNIKS motif decrease recognition of guanine in the third position (UAG). Western blot analysis was also carried out to determine the level of mutant eRF1 proteins. The YCF mutant (YFCDSKF) resulted in a 40% reduction in eRF1 abundance, while the TASNKS (KATNIKD) mutant displayed a 60% decrease in eRF1 protein (Fig. 4B). Since termination at one (or more) of the three stop codons remained at or near the normal level, we conclude that these modest decreases in eRF1 abundance were not sufficient to induce a global decrease in termination efficiency. When taken together, these results suggest that alterations within the Tetrahymena YCF motif reduce recognition of adenine in position two of stop codons, while changes in the Tetrahymena TASNIKS motif provide a bias against guanine in position three of the stop codon. While other residues in Tetrahymena eRF1 probably also contribute to its UGA specificity, the TASNIKS and YCF motifs clearly provide important contributions to its overall pattern of stop codon recognition.
FIGURE 4.
An eRF1 mutant carrying the Tetrahymena YCF motif displays UGA specificity. (A) Fold change in readthrough by strains expressing the indicated mutant eRF1 proteins compared with wild-type S. cerevisiae eRF1. The data are expressed as mean values (relative to wild-type eRF1) ± standard deviation. The horizontal dashed lines represent wild-type S. cerevisiae eRF1 levels. (B) Western blot showing steady-state eRF1 proteins levels from strains in A.
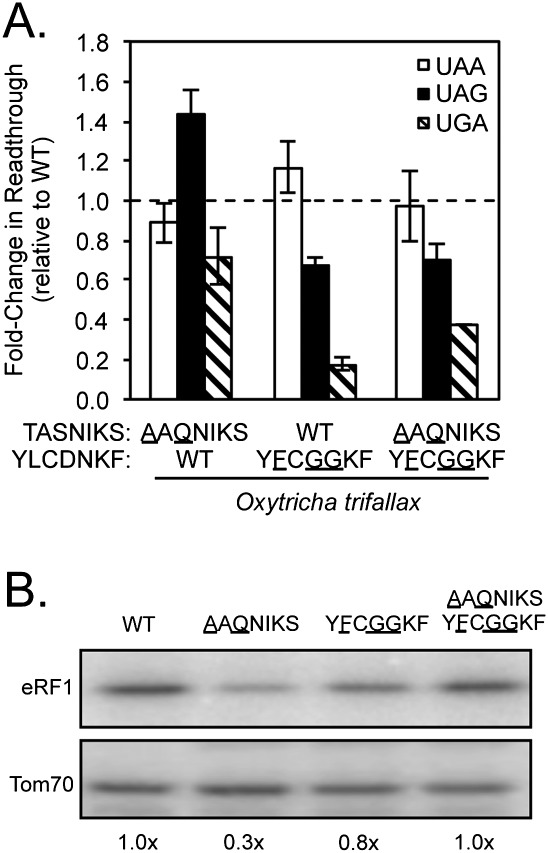
We next examined the effects of introducing the TASNIKS and YCF motifs from a third UGA-specific organism, Oxytricha trifallax, into wild-type S. cerevisiae eRF1. Interestingly, these changes produced distinct changes in the pattern of stop codon recognition as compared with the results obtained with the Paramecium or Tetrahymena motifs. Rather than a decrease in UAA or UAG specificity, we observed a sixfold increase in UGA termination efficiency in the YCF mutant relative to wild-type eRF1 (Fig. 5A; Table 1). This specific increase in UGA termination efficiency suggests that the Oxytricha YCF motif (YFCGGKF) primarily enhances recognition of guanine in the second position (UGA) without significantly reducing adenine recognition (UAA and UAG) at that position. In contrast, little or no change in the termination efficiency could be attributed to the Oxytricha TASNIKS motif (AAQNIKS), and termination of the TASNIKS/YCF double mutant was similar to the YCF mutant. The abundance of each mutant eRF1 protein remained similar to wild-type eRF1 with the exception of the TASNIKS mutant (Fig. 5B), which was reduced to 30% of the wild-type eRF1 protein level. In spite of that decrease, the termination efficiency was largely unchanged, indicating that a sufficient amount of mutant eRF1 was present for efficient termination. These results suggest that the Oxytricha YCF motif enhances UGA recognition, but other determinants in Oxytricha eRF1 beyond the TASNIKS and YCF motifs must act to reduce termination at UAA/UAG codons in order to confer UGA-specific termination.
FIGURE 5.
The Oxytricha YCF motif increases UGA recognition by S. cerevisiae eRF1. (A) Fold change in readthrough by strains expressing the indicated mutant eRF1 proteins compared with wild-type S. cerevisiae eRF1. The data are expressed as mean values (relative to wild-type eRF1) ± standard deviation. The horizontal dashed lines represent wild-type S. cerevisiae eRF1 levels. (B) Western blot showing steady-state eRF1 proteins levels from strains in A.
Some amino acid changes in variant TASNIKS and YCF motifs enhance termination efficiency at one or more stop codons
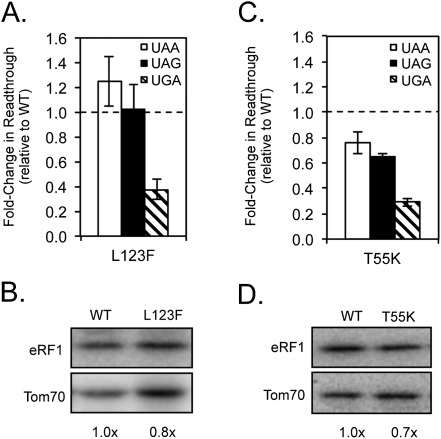
The results described above show that divergent residues in the Paramecium, Tetrahymena, and Oxytricha YCF motifs confer UGA specificity by reducing UAA/UAG termination or enhancing UGA recognition. These three motifs, YFCDPQF, YFCDSKF, and YFCGGKF, respectively, share the L123F alteration. We also showed that the sole amino acid change in the Euplotes and Blepharisma YCF motifs, L123I, conferred a strong bias against UGA termination. When taken together, these results suggest that the identity of the amino acid at residue 123 has a strong effect on the efficiency of UGA recognition. To explore this further, we introduced L123F alone into S. cerevisiae eRF1 and examined its effects on stop codon recognition (Fig. 6A; Table 2). The L123F mutation resulted in an approximately threefold enhancement of termination efficiency at the UGA codon, while termination at UAA/UAG remained similar to wild-type eRF1 levels. Little or no change was observed in the steady-state level of the L123F eRF1 protein relative to wild-type eRF1 (Fig. 6B). These results demonstrate that the L123F alteration acts to enhance UGA recognition, rather than reduce UAA/UAG recognition.
FIGURE 6.
The L123F and T55K mutations enhance UGA recognition by S. cerevisiae eRF1. (A,C) Fold change in readthrough by strains expressing the indicated mutant eRF1 proteins compared with wild-type S. cerevisiae eRF1. The data are expressed as mean values (relative to wild-type eRF1) ± standard deviation. The horizontal dashed lines represent wild-type S. cerevisiae eRF1 levels. (B,D) Western blot showing steady-state eRF1 proteins levels from strains in A,C.
TABLE 2.
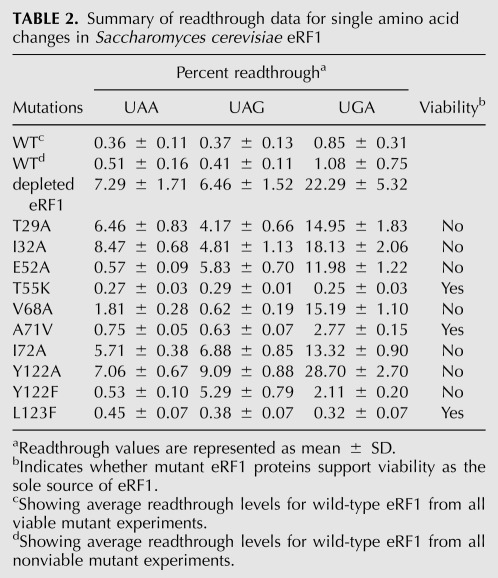
Summary of readthrough data for single amino acid changes in Saccharomyces cerevisiae eRF1
Our earlier results showed that the presence of the Tetrahymena TASNIKS motif (KATNIKD) resulted in a bias against efficient termination at the UAG codon when placed in the context of S. cerevisiae eRF1, suggesting that these changes together prevent guanine binding in position three (Fig. 4A). Of the three changes in this motif, T55 is closest to cavity three of the Bertram model. Accordingly, we also examined the consequences of introducing a T55K mutation into S. cerevisiae eRF1 (Fig. 6C; Table 2). Surprisingly, this mutant protein recognized each of the three stop codons more efficiently than WT eRF1 (ranging from 20% better at UAA to 70% better at UGA). The steady-state level of the T55K eRF1 protein was only slightly reduced relative to wild-type eRF1 (Fig. 6D). These results suggest that the T55K change in the first position of the TASNIKS motif may act to counterbalance other negative changes that contribute to variant stop codon selection in order to optimize UGA specificity in Tetrahymena species. When taken together, these results suggest that the L123F and T55K mutations enhance recognition of specific stop codons in variant-code species, presumably to fine-tune the overall termination efficiency.
The Loxodes TASNIKS and YCF motifs both contribute to UGA-specific stop codon recognition
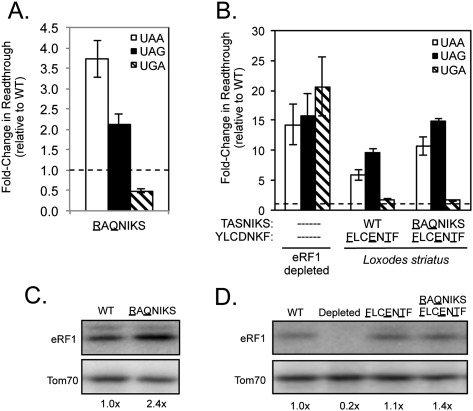
The results above suggest that the YCF motif generally plays a more prominent role than the TASNIKS motif in determining stop codon specificity for the five variant-code organisms examined above. To test this further, we introduced the TASNIKS, YCF, or TASNIKS/YCF motifs of the UGA-specific species Loxodes striatus into S. cerevisiae eRF1 (Fig. 7). Unlike the other UGA-specific ciliates above that carry the L123F change in their YCF motifs, this organism contains the standard code leucine at position 123 (Kim et al. 2005). We found that the Loxodes TASNIKS mutant (RAQNIKS) reduced UAA/UAG recognition by two to fourfold, while the YCF mutant (FLCENTF) reduced UAA/UAG recognition by five to 10-fold (Fig. 7A,B; Table 1). Thus, the introduction of either motif into S. cerevisiae eRF1 resulted in varying degrees of UGA specificity. The double mutant (RAQNIKS/FLCENTF) reduced termination at UAA and UAG codons (11-fold and 15-fold, respectively), significantly more than either the RAQNIKS or FLCENTF mutants alone. These results suggest that the Loxodes TASNIKS and YCF motifs act together to suppress termination at UAA and UAG codons and optimize UGA-specific termination. Once again, Western blot analysis indicated that the steady-state level of each eRF1 mutant protein remained similar to (or above) wild-type eRF1 levels (Fig. 7C,D).
FIGURE 7.
Introduction of the Loxodes TASNIKS and/or YCF motifs reduce UAA/UAG recognition by S. cerevisiae eRF1. (A,B) Fold change in readthrough by strains expressing the indicated mutant eRF1 proteins compared with wild-type S. cerevisiae eRF1. The data are expressed as mean values (relative to wild-type eRF1) ± standard deviation. The horizontal dashed lines represent wild-type S. cerevisiae eRF1 levels. (C,D) Western blot showing steady-state eRF1 proteins levels from strains in A,B.
Mutations between the TASNIKS and YCF motifs have distinct effects on stop codon recognition
In their model for stop codon recognition, Bertram et al. (2000) proposed that eRF1 cavity one binds the conserved uracil in position one of stop codons, while cavities two and three bind the purines in positions two and three, respectively. The YCF motif occupies most of the space between the three proposed cavities on the surface of eRF1, while the first residues of the TASNIKS motif are distal to cavity three (see Fig. 1B). The results described above showed that the introduction of several variant code TASNIKS and YCF elements into S. cerevisiae eRF1 altered stop codon recognition in a manner that is generally consistent with this cavity model. This led us to hypothesize that changes in other amino acids between the TASNIKS and YCF motifs may also influence the pattern of stop codon recognition.
Seven additional eRF1 mutants carrying single amino acid changes were generated. Introduction of T29A, I32A, or I72A mutations individually into S. cerevisiae eRF1 caused severe readthrough at all stop codons, indicating that these mutations each caused general defects in termination (Table 2). These results show that these residues play important, but poorly defined role(s) in the structure or function of eRF1. The A71V mutation showed a two- to threefold defect in termination at all three stop codons, and thus did not influence stop codon selectivity. Other mutations showed more specific roles on stop codon recognition. The V68A mutation exhibited a fourfold reduction in termination efficiency at UAA and a more severe (14-fold) reduction in termination at UGA, but no change in termination at the UAG codon. These results suggest that the V68A mutation caused a strong bias against stop codons with an adenine in position three, consistent with the location of this residue near cavity three.
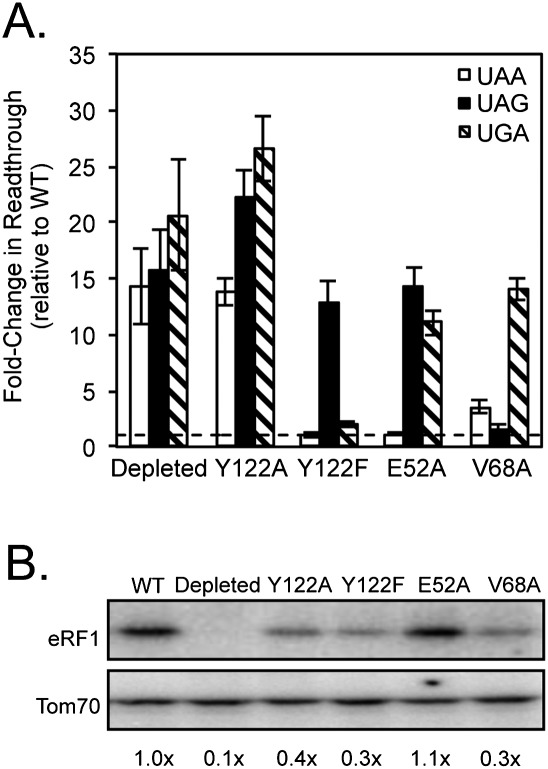
Based on structural studies, the Y122 residue in the YCF motif is adjacent to cavity one and is predicted to form a hydrogen bond with E52, which borders cavity three (Kolosov et al. 2005). To examine the importance of this interaction, we next mutated each of these residues in S. cerevisiae eRF1. The E52A mutation significantly reduced termination at UAG and UGA codons (14- and 11-fold, respectively), but not at the UAA codon, suggesting that this mutation reduced termination at codons with guanine in positions 2 or 3. The Y122A mutant exhibited high levels of readthrough at all three stop codons, indicating that this mutation resulted in a nonfunctional protein in vivo. To overcome this problem, we introduced a more conservative change, Y122F (which differs only by the loss of the hydroxyl group thought to participate in the hydrogen bond with E52). The Y122F mutant displayed efficient termination at UAA and UGA codons, but 13-fold less-efficient termination at the UAG codon (Fig. 8A; Table 2). This suggested that recognition of guanine in the third position of the stop codon was inhibited by the Y122F mutation. The Y122A, Y122F, and V68A mutants each displayed two- to threefold reductions in the abundance of eRF1 protein (Fig. 8B). Since the Y122F and V68A mutants each retained efficient termination at one or more stop codons, we conclude that these eRF1 levels were sufficient to facilitate efficient termination in vivo.
FIGURE 8.
Mutations in eRF1 residues near cavity three show altered stop codon recognition. (A) Fold change in readthrough by strains expressing the indicated mutant eRF1 proteins compared with wild-type S. cerevisiae eRF1. The data are expressed as mean values (relative to wild-type eRF1) ± standard deviation. The horizontal dashed lines represent wild-type S. cerevisiae eRF1 levels. (B) Western blot showing steady-state eRF1 proteins levels from strains in A.
DISCUSSION
The variant genetic codes of most ciliated protozoa appear to use either UAA/UAG or UGA as stop codons (Kim et al. 2005), and a number of studies have implicated amino acid changes within the TASNIKS and YCF motifs as important mediators of those two outcomes (Bertram et al. 2000; Knight et al. 2000; Song et al. 2000; Lozupone et al. 2001; Frolova et al. 2002; Seit-Nebi et al. 2002; Kim et al. 2005; Kolosov et al. 2005; Fan-Minogue et al. 2008; Cheng et al. 2009). In this study, we examined the relative importance of these motifs from six variant-code species in the context of S. cerevisiae eRF1. We found that the single amino acid change L123I found in both the Euplotes and Blepharisma YCF motifs reduced UGA-specific termination by 12-fold, resulting in the conversion of S. cerevisiae eRF1 to primarily a UAA/UAG pattern of stop codon recognition. Consistent with our results, Stansfield and colleagues found that an L123V mutation resulted in a fivefold decrease in UGA-specific termination (Bertram et al. 2000). We previously demonstrated that a C124S mutation in a Euplotes octocarinatus/S. cerevisiae hybrid eRF1 protein converted it from a UAA/UAG pattern of recognition to omnipotent recognition (UGA recognition was restored while UAA/UAG recognition was maintained) (Fan-Minogue et al. 2008). These results demonstrate that single amino acid changes at these two adjacent residues near cavity two either enhance (C124S) or inhibit (L123I) recognition of guanine in the second position of the stop codon (UGA) (Bertram et al. 2000; Fan-Minogue et al. 2008), and are entirely consistent with their location directly adjacent to cavity two of the Bertram model.
This study provides new insights into possible mechanisms of stop codon reassignment. It is generally assumed that recoding requires the acquisition of amino acid changes within eRF1 that reduce its ability to recognize some, but not all, stop codons. However, our results show that the acquisition of variant stop codon usage involves multiple amino acid changes with both negative and positive effects on stop codon recognition that may act cooperatively to optimize variant code utilization. For example, we found that incorporating the two or three amino acid changes from the Paramecium or Tetrahymena YCF motifs into S. cerevisiae eRF1 significantly reduced termination at UAA/UAG codons, while maintaining efficient UGA termination (see Figs. 3A, 4A). However, incorporation of the Oxytricha YCF or a single divergent residue shared by all three organisms (L123F) did not reduce UAA or UAG termination. Instead, it enhanced termination at the UGA codon by two- to fivefold (Fig. 5A). Similarly, we found that the T55K mutation from the Tetrahymena TASNIKS motif enhanced termination efficiency at all three stop codons by up to fourfold. These results demonstrate that some amino acid changes reduce the recognition of one or more stop codons, while others buffer that effect by enhancing the recognition of other stop codons. The net effect is to optimize recognition of one (or a subset) of stop codons.
To test the contribution of residues beyond the TASNIKS and YCF motifs, we altered a series of amino acids between the TASNIKS and YCF motifs into S. cerevisiae eRF1 to determine whether other patterns of stop codon recognition could be obtained (Fig. 8). Among these, mutant eRF1 proteins carrying the Y122F, E52A, or V68A mutations exhibited intriguing patterns of stop codon recognition. Y122F is thought to hydrogen bond to E52 (Kolosov et al. 2005), which borders cavity three in the Bertram cavity model. The Y122F mutant displayed a 13-fold reduction in termination efficiency at UAG, while termination at UAA and UGA codons was largely unaffected. This indicates that the Y122F mutation strongly and specifically inhibited recognition of guanine at the third position (as in UAG). These findings are in general agreement with a previous study that showed a threefold decrease in termination efficiency at UAG using an in vitro release assay using human eRF1 (Kolosov et al. 2005). In contrast to Y122F, we found that the E52A mutant exhibited efficient termination at UAA, but a strong defect in termination at UAG and UGA (reduced 14-fold and 11-fold, respectively). This indicates that E52A shows a strong bias against guanine residues at the second or third positions (UAG and UGA), a distinct phenotype from the Y122F mutant. The different nature of these phenotypes indicates that the potential hydrogen bonding of E52 and Y122 is not their only contribution to standard code stop codon recognition.
Residue V68 is also directly adjacent to cavity three. eRF1 carrying the V68A mutation mediated efficient termination at UAG, while termination at both UAA and UGA was decreased (fourfold and 14-fold, respectively). Thus, the V68A mutant inhibited recognition of adenine at the third position (as in UAA/UGA), a result that is consistent with cavity three serving to bind the third nucleotide of the stop codon. A recent crystallization study of an eRF1/eRF3 complex found ATP, a component of the precipitant solution, bound to this same pocket (Cheng et al. 2009). Importantly, V68 was identified as one of the hydrophobic residues that bound the adenine base, implicating the potential importance of this residue in decoding stop codons (Cheng et al. 2009). However, those investigators concluded that the bound ATP was located in a cavity that bound the second, rather than the third nucleotide of the stop codon. Our results demonstrate that the Y122F, E52A, and V68A mutations generally exhibit strong effects on selection of the third nucleotide of the stop codon. Furthermore, given the importance of L123 and C124 in recognition of the second nucleotide of the stop codon, we believe that the sum of our results are most consistent with the cavity model proposed by Bertram and coworkers (Bertram et al. 2000).
Unfortunately, there is a significant body of conflicting data in the literature regarding several of these key eRF1 mutations. A study by von der Haar and colleagues showed that a V68A mutation caused a bias against stop codons with an adenine in position three, consistent with our data and the assignment of this residue near cavity three (Merritt et al. 2010). However, another study reported that this mutation allowed termination to remain similar to wild-type eRF1 in S. cerevisiae cells (Bertram et al. 2000). The reason for this discrepancy is not known. Two other studies examined the E52A mutation (E55A in human eRF1) (Kolosov et al. 2005; Cheng et al. 2009). In our study, we found that this mutation allows termination at UAA, but largely excludes UAG or UGA codons. In contrast, Cheng et al. (2009) reported that an E52A mutant exhibited only subtle (1.5- to twofold) defects at all three stop codons in an S. cerevisiae eRF1 depletion strain expressing mutant derivatives of human eRF1. Another study characterized the E52A mutation in human eRF1 using an fmet release assay (a highly purified system) (Kolosov et al. 2005). They reported that the E52A mutant eRF1 showed a strong defect at the UAG codon, and more subtle (twofold) defects at UAA and UGA codons. Again, the reasons for these different results are not clear. However, the use of such diverse assay systems, eRF1 species, and experimental conditions has previously made it difficult to compare results from different studies in the context of a unified model for stop codon recognition.
There are several advantages to the experimental system used in the current study. First, results were obtained using a homologous in vivo S. cerevisiae system in which mutant eRF1 derivatives were maintained in the absence of selective pressure for suppressor mutations until the assays were carried out, thus minimizing the potential for second-site mutations that could influence the results obtained. Second, a relatively small number of amino acids were changed in the context of S. cerevisiae eRF1 in each experiment, and swaps of entire domains to produce hybrid eRF1 proteins that could complicate the interpretation of results was avoided. Third, control strains expressing either wild-type S. cerevisiae eRF1 or a strain completely depleted of eRF1 were included. These controls demonstrated that this system provides a broad dynamic range for both decreased termination efficiency (ranging from 17- to 26-fold; see Table 1) and increased termination efficiency (up to fivefold). Together, these results show that our system allows quantitation of termination efficiency over a 130-fold range. Finally, many other proteins have been shown to influence the efficiency of translation termination in vivo (von der Haar and Tuite 2007). Consequently, the use of in vivo termination measurements ensures that all data were obtained in a physiologically relevant environment. When taken together, our results provide a unified picture of the functional consequences of these mutations in the context of S. cerevisiae eRF1 in its natural in vivo environment.
Our current data reinforces previous findings that the TASNIKS and YCF motifs play important roles in stop codon recognition. However, it also clearly shows that changes in the YCF consistently result in stronger and more specific effects on the expected variant patterns of stop codon selection. It is informative to consider these results in the context of our recent study that examined a hybrid eRF1 containing domain one of Euplotes octocarinatus fused to domains two and three of Saccharomyces cerevisiae eRF1 (referred to as Eo/Sc eRF1) (Fan-Minogue et al. 2008). We found that Eo/Sc eRF1 showed a UAA/UAG pattern of stop codon recognition and did not efficiently recognize the UGA codon. When the Euplotes variant TAESIKS motif in Eo/Sc eRF1 was changed to the standard TASNIKS sequence (Eo/Sc eRF1 E57S/S58N) we found that omnipotent stop codon recognition was restored. Importantly, we also showed that termination at UAA/UAG codons no longer required eRF3 (either in vitro or in vivo), while UGA termination retained the usual eRF3 requirement. Those results led us to propose a conformational model in which eRF3 induced eRF1 to acquire a final position/conformation on the pretermination complex after stop codon recognition that was dependent on the bound stop codon (UAA/UAG vs. UGA). We reasoned that the Eo/Sc eRF1 E57S/S58N mutant maintained the UAA/UAG position/conformation constitutively, thus relieving the requirement for eRF3 to enhance termination at those stop codons. However, this mutant continued to require eRF3 in order to attain the alternate position/conformation required for termination at UGA. The implications of this model is that the primary role of the TASNIKS motif is not direct stop codon recognition; instead, it acts to couple stop codon recognition and GTP hydrolysis to the acquisition of alternate positions/conformations of eRF1 on the pretermination complex prior to polypeptide release. The data presented in the current study are generally consistent with the TASNIKS motif mediating such a coupling function, since the introduction of TASNIKS motifs from most variant code organisms showed only a limited ability to determine stop codon specificity. In contrast, the YCF motifs from those organisms were generally much more efficient at conferring the stop codon specificity of each organism, consistent with a role in direct molecular contacts with the residues of the stop codon as predicted by the cavity model (Bertram et al. 2000). Ultimately, structural studies on these distinct termination complexes will be required to confirm the validity of this model.
MATERIALS AND METHODS
Strain used
The S. cerevisiae strain used in this study was YDB447 (MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2- 80 suc2-Δ901 sup45:: HIS3 [psi-]) (Salas-Marco and Bedwell 2004).
Plasmids
pDB800, a centromere-based plasmid that expresses the wild-type SUP45 gene (which encodes S. cerevisiae eRF1) and contains a LEU2 selectable marker, was used as a control in all dual luciferase reporter readthrough assays. pDB967, a centromere-based plasmid that expresses wild-type eRF1 with an N-terminal HA-tag from the GAL1 promoter with a TRP1 selectable marker (Salas-Marco et al. 2006), was used for eRF1 deletion experiments during luciferase assays and Western blot analysis. pDB1047, a centromere-based plasmid containing a LEU2 selectable marker and expresses wild-type S. cerevisiae eRF1 with an N-terminal HA-tag under control of the SUP45 promoter, served as wild-type eRF1 control for Western blot analysis. All mutant forms of eRF1 were expressed from pDB1047. eRF1 mutations were made by two-stage site-directed mutagenesis of pDB1047 using the QuikChange mutagenesis kit (Stratagene). Each mutant was verified by sequencing the entire SUP45 ORF, as well as adjacent 5′ and 3′ regions. A complete list of plasmids generated for this study is listed in Supplemental Table 1.
Viability assay
Yeast strains expressing each mutant eRF1 protein as the sole source of eRF1 were initially tested for cell viability. The viability assay was performed as previously described (Salas-Marco et al. 2006). Briefly, LEU2 selectable plasmids expressing mutant eRF1 proteins were transformed into YDB447 (sup45Δ) containing pUKC802, which expresses wild-type eRF1 from a plasmid carrying a URA3 selectable marker. Each transformation was plated on synthetic minimal (SM) medium supplemented with 2% glucose with required amino acids (but lacking leucine). After initial growth, single colonies were streaked onto plates containing 5-fluoroorotic acid (5-FOA). Since 5-FOA inhibits growth of cells that express the URA3 gene, this led to colony formation from cells that had lost pUKC802 if the mutant eRF1 protein could support cell viability as the sole source of eRF1. If no colonies were obtained on 5-FOA plates, it was concluded that the mutant eRF1 protein was unable to support cell viability as the sole source of cellular eRF1.
eRF1 depletion experiments
To determine stop codon recognition of eRF1 mutants that were unable to support cell viability as the sole source of eRF1, relevant plasmids were transformed into YDB447/pDB967 (which expresses wild-type eRF1 under GAL1 promoter control). The resulting strains were maintained by growth on galactose-containing medium. To monitor stop codon recognition of each mutant eRF1 protein, cultures were grown in SM medium containing 2% galactose and the appropriate amino acids overnight. The next morning, cells were diluted back and allowed to grow for at least 5 h to mid-log (defined as an A600 reading of 0.5–1.0 OD/mL). Cells were harvested, washed three times with SM medium containing 2% glucose, and then resuspended in SM medium with 2% glucose and amino acids and grown overnight for six generations until they again reached mid-log. Readthrough and steady-state eRF1 protein levels were tested with YDB447/pDB967 (without a mutant plasmid) as a control to confirm depletion of eRF1. YDB447 carrying a plasmid encoding wild-type eRF1 (pDB800 or pDB1047) under SUP45 promoter control was included in each experiment as a positive control.
Dual-luciferase readthrough assays
Readthrough assays were performed as previously described (Grentzmann et al. 1998; Keeling et al. 2004; Salas-Marco and Bedwell 2004; Kallmeyer et al. 2006; Salas-Marco et al. 2006). The dual-luciferase reporter used contained the Renilla reniformis (Renilla) and Photinus pyralis (firefly) luciferase genes separated by a linker region containing a readthrough cassette with either a sense or stop codon. The efficiency of termination (expressed as readthrough) of either codon was monitored by firefly luminescence activity. Firefly activity was normalized to Renilla activity, which served as an internal normalization control. Readthrough levels of each mutant were determined in at least two independent experiments with each sample being assayed in quadruplicate. “Percent Readthrough” for wild-type S. cerevisiae eRF1 driven by the SUP45 promoter (pDB800) or GAL1 promoter control (pDB967) was expressed as the firefly/Renilla ratio obtained from the mutant (stop codon) reporter divided by the firefly/Renilla ratio from the wild-type (sense codon) reporter (x 100). The resulting values for each construct are expressed as the overall mean obtained using the same strains and conditions. For ease of comparison, the final readthrough data were expressed as the fold-change in readthrough (normalized to the wild-type control) ± standard deviation.
Western blots
Ten A600 units of cells were harvested and incubated in 10% trichloroacetic acid on ice for 30 min. Cells were washed with 100% acetone and dried. After resuspension in 120 μL of SDS lysis buffer (50 mM Tris-HCl at pH 7.5, 1 mM EDTA, 1% SDS), cells were lysed by mechanical disruption using glass beads. Samples were boiled for 3 min and cleared by centrifugation for 10 min in a microcentrifuge. Protein concentration was measured using the Lowery Method (Lowry et al. 1951). Total protein (20 μg) from crude extracts was resolved on an SDS–polyacrylamide gel, followed by transfer to an Immobilon-P transfer membrane (Millipore) and Western blotting. Cells from depletion experiments were collected after the galactose to glucose shift procedure as described above.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank the Heflin Center Genomics Core Facility for DNA sequencing of eRF1 mutant alleles. This work was supported by NIH grant GM068854 (to D.M.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031997.111.
REFERENCES
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I 2000. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6: 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Seit-Nebi A, Dubovaya V, Favre A 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J 21: 5302–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al. 2009. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev 23: 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseev B, Kryuchkova P, Alkalaeva E, Frolova L 2011. A single amino acid change of translation termination factor eRF1 switches between bipotent and omnipotent stop-codon specificity. Nucleic Acids Res 39: 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM 2008. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell 30: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341 [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Seit-Nebi A, Kisselev L 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham RH 1994. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci 91: 5848–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486 [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, Gorovsky MA 1985. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci 82: 2452–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Shirts BH, Petros LM, Flanigan KM, Gesteland RF, Atkins JF 2000. Sequence specificity of aminoglycoside-induced stop codon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann Neurol 48: 164–169 [PubMed] [Google Scholar]
- Ito K, Ebihara K, Nakamura Y 1998. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA 4: 958–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y 2000. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403: 680–684 [DOI] [PubMed] [Google Scholar]
- Kallmeyer AK, Keeling KM, Bedwell DM 2006. Eukaryotic release factor 1 phosphorylation by CK2 protein kinase is dynamic but has little effect on the efficiency of translation termination in Saccharomyces cerevisiae. Eukaryot Cell 5: 1378–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I 2007. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell 26: 625–637 [DOI] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Frolova L, Kisselev L, Jean-Jean O 2001. Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO Rep 2: 680–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OT, Yura K, Go N, Harumoto T 2005. Newly sequenced eRF1s from ciliates: the diversity of stop codon usage and the molecular surfaces that are important for stop codon interactions. Gene 346: 277–286 [DOI] [PubMed] [Google Scholar]
- Kisselev L, Ehrenberg M, Frolova L 2003. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J 22: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF 2000. The early evolution of the genetic code. Cell 101: 569–572 [DOI] [PubMed] [Google Scholar]
- Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Efimov A, Kisselev L 2005. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res 33: 6418–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekomtsev S, Kolosov P, Bidou L, Frolova L, Rousset JP, Kisselev L 2007a. Different modes of stop codon restriction by the Stylonychia and Paramecium eRF1 translation termination factors. Proc Natl Acad Sci 104: 10824–10829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekomtsev SA, Kolosov PM, Frolova LY, Bidou L, Rousset JP, Kisselev LL 2007b. How does Euplotes translation termination factor eRF1 fail to recognize the UGA stop codon? Mol Biol 41: 924–931 [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Lozupone CA, Knight RD, Landweber LF 2001. The molecular basis of nuclear genetic code change in ciliates. Curr Biol 11: 65–74 [DOI] [PubMed] [Google Scholar]
- Merkulova TI, Frolova LY, Lazar M, Camonis J, Kisselev LL 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett 443: 41–47 [DOI] [PubMed] [Google Scholar]
- Merritt GH, Naemi WR, Mugnier P, Webb HM, Tuite MF, von der Haar T 2010. Decoding accuracy in eRF1 mutants and its correlation with pleiotropic quantitative traits in yeast. Nucleic Acids Res 38: 5479–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Schmidt HJ, Plumper E, Hasilik A, Mersmann G, Meyer HE, Engstrom A, Heckmann K 1991. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc Natl Acad Sci 88: 3758–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T, Heckmann K, Kitanaka C, Kuchino Y 2001. Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett 488: 105–109 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K 2002. A tripeptide discriminator for stop codon recognition. FEBS Lett 514: 30–33 [DOI] [PubMed] [Google Scholar]
- Preer JR Jr, Preer LB, Rudman BM, Barnett AJ 1985. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature 314: 188–190 [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24: 7769–7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol 348: 801–815 [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Fan-Minogue H, Kallmeyer AK, Klobutcher LA, Farabaugh PJ, Bedwell DM 2006. Distinct paths to stop codon reassignment by the variant-code organisms Tetrahymena and Euplotes. Mol Cell Biol 26: 438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Kisselev L 2002. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep 3: 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D 2000. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- von der Haar T, Tuite MF 2007. Regulated translational bypass of stop codons in yeast. Trends Microbiol 15: 78–86 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124 [DOI] [PubMed] [Google Scholar]