Riboswitches are RNA elements that bind to effector ligands and control gene expression. S-Adenosyl Methionine (SAM) binds the aptamer domain of the SAM-I riboswitch and induces conformational changes in the expression domain to form an intrinsic terminator (transcription OFF state). Without SAM the riboswitch forms the transcription ON state, allowing read-through transcription. The mechanistic link between the SAM/aptamer recognition event and subsequent secondary structure rearrangement by the riboswitch is unclear. The authors present binding measurements and in-line probing that are consistent with the hypothesis that when SAM is present, stacking interactions with the AT helix stabilize a partially formed P1 helix in the hybrids. Molecular modeling indicates that continuous stacking between the P1 and the AT helices is plausible with SAM bound. These findings raise the possibility that conformational intermediates may play a role in ligand-induced aptamer folding.
Keywords: riboswitch, aptamer, S-adenosyl methionine, RNA folding, ligand binding
Abstract
Riboswitches are RNA elements that bind to effector ligands and control gene expression. Most consist of two domains. S-Adenosyl Methionine (SAM) binds the aptamer domain of the SAM-I riboswitch and induces conformational changes in the expression domain to form an intrinsic terminator (transcription OFF state). Without SAM the riboswitch forms the transcription ON state, allowing read-through transcription. The mechanistic link between the SAM/aptamer recognition event and subsequent secondary structure rearrangement by the riboswitch is unclear. We probed for those structural features of the Bacillus subtilis yitJ SAM-I riboswitch responsible for discrimination between the ON and OFF states by SAM. We designed SAM-I riboswitch RNA segments forming “hybrid” structures of the ON and OFF states. The choice of segment constrains the formation of a partial P1 helix, characteristic of the OFF state, together with a partial antiterminator (AT) helix, characteristic of the ON state. For most choices of P1 vs. AT helix lengths, SAM binds with micromolar affinity according to equilibrium dialysis. Mutational analysis and in-line probing confirm that the mode of SAM binding by hybrid structures is similar to that of the aptamer. Altogether, binding measurements and in-line probing are consistent with the hypothesis that when SAM is present, stacking interactions with the AT helix stabilize a partially formed P1 helix in the hybrids. Molecular modeling indicates that continuous stacking between the P1 and the AT helices is plausible with SAM bound. Our findings raise the possibility that conformational intermediates may play a role in ligand-induced aptamer folding.
INTRODUCTION
The ongoing discovery of riboswitches in the last one and a half decades has added to our understanding of the active role of RNA as a gene regulatory element. Riboswitches are folded RNA elements that sense and bind to certain cellular metabolites and regulate the expression of genes related to the metabolites (Auger et al. 2002; Mironov et al. 2002). Riboswitches are found near the 5′ untranslated region (5′ UTR) of mRNA in g-positive bacteria, a few plants, and in certain fungi (Sudarsan et al. 2003). Riboswitch gene control mechanisms include modulation of rho-independent transcription terminator formation, sequestering of ribosome binding sites, and control of alternative splicing by ribozyme cleavage (Nahvi et al. 2002; Cheah et al. 2007). Riboswitches appear capable of manipulating the expression of genes related to metabolite biosynthesis without direct interaction with proteins. Moreover, riboswitches have been suggested as potential drug targets (Blount and Breaker 2006; Blount et al. 2007; Kim et al. 2009; Lee et al. 2009; Cressina et al. 2011; Daldrop et al. 2011; Deigan and Ferré-D'Amaré 2011; Maciagiewicz et al. 2011) and for a number of potential engineering applications (Wieland and Hartig 2008; Link and Breaker 2009; Topp and Gallivan 2010), highlighting the importance of understanding mechanisms for specificity and recognition of riboswitches by small molecule ligands (Link and Breaker 2009; Haller et al. 2011).
The S-box system, also called the SAM-I riboswitch was identified by careful analysis (Grundy and Henkin 1998) of sequencing data from Bacillus subtilis (Médigue et al. 1995; Kunst and Ogasawara 1997). It controls the expression of genes involved in metabolism, synthesis, and transport of SAM and sulfur-containing amino acids using S-adenosyl methionine (SAM) as the effector molecule (Grundy and Henkin 1998; Epshtein et al. 2003; McDaniel et al. 2003; Winkler et al. 2003; Wang and Breaker 2008). The SAM-I riboswitch is one of five classes within the SAM riboswitch family. Each class has its own sequence conservation and gene control mechanism (Montange et al. 2010). In the SAM-I riboswitch, binding of SAM to the riboswitch causes structural rearrangement of the RNA leading to transcription modulation (McDaniel et al. 2003; Winkler et al. 2003). The small molecule SAM is the cue for bacterial sulfur metabolism. The remarkable ability of such a small molecule to alter the global structure of the macromolecule (RNA) motivates the search for an in-depth understanding of the regulatory mechanism. Such an understanding, in turn, will provide insights into the principles of metabolite recognition and discrimination by RNA.
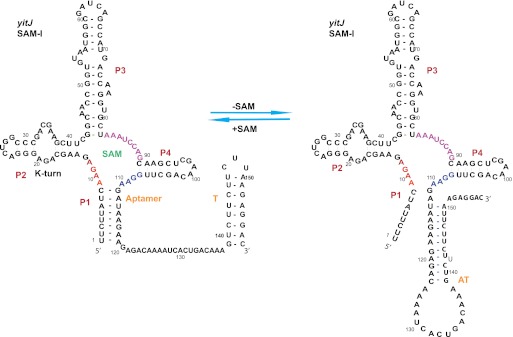
The SAM-I riboswitch is comprised of two functional domains (Fig. 1). The first domain, called the aptamer, binds to SAM at raised cellular SAM concentration. Under these conditions, the second domain, called the expression platform, undergoes structural rearrangement to allow the formation of a stable transcription terminator stem–loop. At low cellular SAM levels, the interplay between the two domains is altered to stabilize the antiterminator (AT). These structural alterations in the expression platform allow readthrough transcription (McDaniel et al. 2003; Winkler et al. 2003). The crystal structures of the SAM-I riboswitch in Thermoanearobacter tencongensis upstream of the metF-H2 gene and in B. subtilis upstream of the yitJ gene revealed that SAM binds to a pocket formed by four coaxially stacked helices (P1, P2, P3, P4) joined by junctions J1/2, J2/3, and J3/4 (Fig. 1; Montange and Batey 2006; Lu et al. 2010; Montange et al. 2010). Mutational analysis and biophysical studies show that a kink-turn-mediated pseudoknot formation assists the global folding of the riboswitch cooperatively with Mg2+ present (Winkler et al. 2001; McDaniel et al. 2005; Heppell and Lafontaine 2008; Lu et al. 2010).
FIGURE 1.
The secondary structure and sequence of the B. subtilis yitJ SAM-I riboswitch. The residues of the junction regions J1/2, J3/4, and J1/4 are colored in red, magenta, and blue, respectively. The aptamer, AT, and T are decision-making regions: denoting the aptamer element, antiterminator, and the terminator sequences, respectively.
Since the riboswitch is believed to undergo conformational changes from transcription “ON” state to “OFF” state in the presence of SAM, the question arises: How can a limited set of contacts by a small molecule ligand cause a large-scale rearrangement of secondary structure folding in the target macromolecule? The dynamic nature of the expression platform spanning the AT and the terminator poses a major challenge in understanding the gene control mechanism. In the absence of SAM, the P1 helix is dynamic even in the isolated aptamer (Stoddard et al. 2010; Hennelly and Sanbonmatsu 2011).
We examine the basis for discrimination by SAM between ON and OFF state riboswitch RNA, in terms of secondary and tertiary structure, using the B. subtilis yitJ SAM-I riboswitch as a model system. The base-pairing patterns in the P1 helix and the AT helix are similar in the B. subtilis yitJ SAM-I riboswitch. This observation led us to the design of RNA constructs to mimic aspects of an OFF state tertiary structure, within the context of a putative ON state secondary structure. We show that these “hybrid” constructs can bind SAM. While ligand affinity is reduced 20- to 50-fold compared with that for the aptamer, binding to the hybrid constructs shows a similar pattern of sensitivity to mutations and ionic conditions as reported for the former. Thus, the mutant binding data are consistent with the conclusion that the hybrid RNAs mimic aspects of aptamer tertiary structure, which mediate SAM binding. Consistent with previous indications that a minimal length P1 helix is necessary for SAM binding, continuous stacking between P1 and AT helical regions is required for SAM binding to hybrids. We discuss the implications for models of conformational switching mechanisms for the SAM-I riboswitch and other riboswitch systems.
RESULTS
Base-pairing probability (BPP) calculations and RNA secondary structure folding data
In a transcriptional riboswitch, the expression platform determines whether transcription is terminated. A competition between the formation of a terminator or antiterminator (AT) hairpin decides the outcome. For the SAM-I riboswitch, the formation of the AT involves a competition with the P1 helix within the aptamer. Since the riboswitch region encompassing the competition between these two helices is transcribed before the terminator hairpin is, the AT/P1 competition is likely to bias the final outcome of riboswitch-regulated transcription (Wickiser et al. 2005a,b).
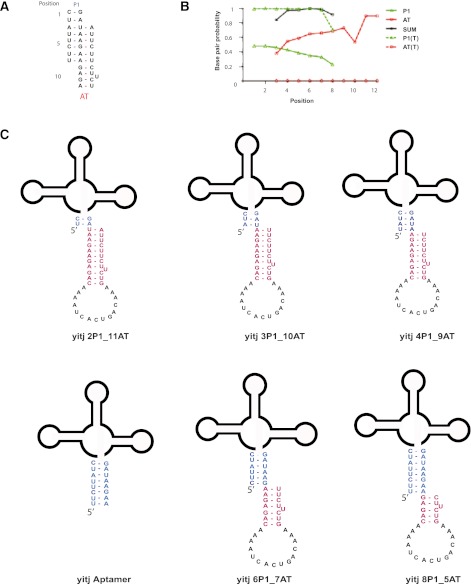
Here we used nucleic acid base-pairing probability calculations, based upon a partition function, to simulate the competition between the P1 and the AT of the B. subtilis yitJ SAM-I riboswitch. Extensive qualitative data regarding the folding of this riboswitch has been reported (Grundy and Henkin 1998; Winkler et al. 2001, 2003; Epshtein et al. 2003; McDaniel et al. 2003, 2005; Heppell and Lafontaine 2008; Lu et al. 2010; Heppell et al. 2011). The base-pairing probability simulations (Fig. 2B) predict that for a transcript length at which the AT helix has been fully transcribed, but the terminator has not, both the P1 (green) and AT (red) helices are populated. The sum of the probability of forming base pairs at positions 4, 5, 6, and 7 (represented by black) is almost one. Moreover, the predicted BPP for P1 helix formation declines, and that for AT helix formation increases for each position moving down the sequence as presented in Figure 2A. This result predicts that the RNA has a propensity to form thermodynamic intermediates that can allow the formation of base pairs in the P1 and also the AT helix simultaneously.
FIGURE 2.
(A) Schematic illustration of base-pair competition between P1 and AT helices. Positions of the base pairs are numbered from 1 to 12. Note that positions 1–2 can only form P1 helix pairs, while positions 9–12 can only form AT pairs. Positions 3–8 can form either. (B) Base-pairing probability (BPP) calculation for B. subtilis yitJ SAM-I riboswitch. For each position, the BPP is plotted for P1 helix base pairing (green), AT pairing (red), the sum of the two (black) for transcripts truncated at the 3′ end of the AT helix (solid) or at the 3′ end of the terminator helix (broken lines) (C) B. subtilis yitJ SAM-I riboswitch AT and aptamer hybrid constructs used for SAM binding studies.
This prediction raises the question as to whether SAM would bind to these “intermediate” conformers. Addressing this question can help delineate the determinants of SAM binding specificity for the aptamer structure, since discrimination between folds should be crucial for the riboswitch mechanism.
A SAM-I riboswitch hybrid binds to SAM with affinity in the micromolar range
The sulfur cationic moiety of SAM interacts electrostatically with the minor groove of the first two base pairs of the P1 helix (Montange and Batey 2006). We constructed a model RNA molecule that is constrained to form a hybrid structure containing the first 3 bp of the P1 helix, and 10 bp of an AT helix (3P1_10AT). This construct resembles structures predicted as minor conformers by the calculations in Figure 2. Residues in the T are truncated to constrain the formation of the partial AT (Fig. 2C).
The dissociation constant for the SAM-I riboswitch aptamer and the hybrid (3P1_10AT) of SAM were determined using scatchard analysis from equilibrium dialysis (Winkler et al. 2003; Supplemental Fig. 1). The dissociation constant for the aptamer measured in this way is 32 nM. This value is within the range of what has been previously reported (Supplemental Table 2). Scatchard analysis yielded Kd for SAM binding to the hybrid 3P1_10AT of ∼790 nM. As a negative control, SAM binding was tested by equilibrium dialysis for ribosomal A-site RNA and a putative (thiamine pyrophosphate [TPP]) Mycobacterium tuberculosis thiC riboswitch aptamer RNA control (data not shown). No potential SAM binding is detected with the A-site RNA or the thiC riboswitch.
The Kd estimates for the aptamer and 3P1_10AT calculated using single-point equilibrium dialysis data obtained with 3 μM RNA and 100 nM SAM agree with the value captured by scatchard analysis to ±15%. For subsequent measurements on other RNA constructs we used single-point measurements at ranges of RNA and SAM concentrations that yielded the highest sensitivity data as a means of qualitatively ranking binding affinity of SAM.
Sensitivity of SAM binding to ions and mutations for hybrid constructs parallels that for the aptamer
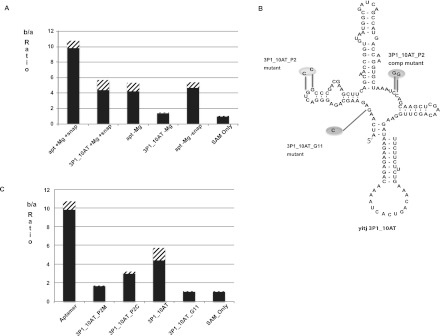
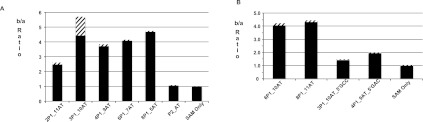
Metal ions play an important role in secondary structure folding of RNA (Woodson 2005; Schroeder and Lilley 2009; Ramesh and Winkler 2010; Royal Society of Chemistry 2011). In particular, Mg2+ has been shown to be required for optimal binding of SAM to the SAM-I riboswitch and for induced folding of the aptamer conformation (Heppell and Lafontaine 2008). X-ray structures of the SAM bound SAM-I riboswitch, and MD simulations derived from the X-ray coordinates indicate the critical role played by Mg2+ ions in facilitating ligand interactions with the P1, P3, and J1/2 region (Montange and Batey 2006; Heppell and Lafontaine 2008; Huang et al. 2009; Lu et al. 2010; Hennelly and Sanbonmatsu 2011). We reasoned that if tertiary folding and the mode of SAM binding in the 3P1_10AT mimics that of the full-length aptamer, then Mg2+ ions would be required to obtain optimal SAM binding to 3P1_10AT. Measurements in the presence and absence of Mg2+ (Fig. 3A) clearly indicate the necessity of Mg2+ for optimal SAM binding. In this figure, SAM binding is indicated by the ratio of counts in chamber b, which contains RNA, to chamber a, which does not. The b/a ratio decreased to barely above 1 when freshly synthesized RNA (without Mg2+) is not heat/snap cooled (as described in the Materials and Methods) before mixing SAM with RNA (Fig. 3A). Freshly synthesized RNA heat/snap cooled in the absence of Mg2+ bound weakly in a detectable range with a b/a ratio above 1.5. RNA samples, which were prehybridized with Mg2+ and stored at −20°C were found to lose SAM-binding activity during storage, but SAM binding was restored by repeating the heat/snap cooling process (data not shown). All single concentration-point binding measurements are presented in tabular form in the Supplemental Material (Supplemental Table 3).
FIGURE 3.
(A) Ratio of number of counts in chamber b to number of counts in chamber a in equilibrium dialysis for B. subtilis yitJ SAM-I riboswitch aptamer and 3P1_10AT hybrid in the presence (+Mg) and absence (−Mg) of Mg2+ and with (+snap) and without (−snap) snap cooling. (B) Schematic representation of B. subtilis yitJ SAM-I riboswitch 3P1_10AT pseudoknot and G11 mutants. (C) Equilibrium dialysis data for Aptamer, Pseudoknot mutant (3P1_10AT_P2M), pseudoknot compensatory mutant (3P1_10AT_P2C), 3P1_10AT, and 3P1_10AT_G11 mutant. Equilibration of SAM in the two chambers is shown with SAM only. The error bars (stripes) represent the standard deviation for at least three replicates for each experiment.
It was reported that mutation of the two guanine residues in the apical loop of the P2 helix in the aptamer (highlighted in Fig. 3B) decreased SAM binding, while compensatory mutation induced by mutating the J3/4 region restored the affinity to SAM (Heppell and Lafontaine 2008). This pattern reflects the formation of a kink-turn stabilized pseudoknot interaction between the P2 loop and J3/4 (Winkler et al. 2001; McDaniel et al. 2005; Montange and Batey 2006; Heppell and Lafontaine 2008; Lu et al. 2010; Montange et al. 2010). We induced the same mutations in the SAM-I riboswitch hybrid 3P1_10AT pseudoknot region (3P1_10AT_P2 mutant). In the equilibrium dialysis assay, the 3P1_10AT_P2 mutant showed decreased b/a ratio (Fig. 3C) as compared with 3P1_10AT. Compensatory mutation in the J3/4 designed to restore pseudoknot base pairing, restores the affinity to SAM. These trends match those reported in SAM binding studies to the wild-type yitJ SAM-I riboswitch aptamer (McDaniel et al. 2005; Heppell and Lafontaine 2008).
The G11 residue of the aptamer in junction J1/2 makes contact with the methionine moiety of SAM through hydrogen bonds (Montange and Batey 2006; Lu et al. 2010; Montange et al. 2010; Schroeder et al. 2011). The carboxylate group of SAM is recognized by the G11 via the N1 and N2 Watson–Crick face (Montange and Batey 2006; Lu et al. 2010). We hypothesized that mutating the G11 will decrease SAM binding to the 3P1_10AT hybrid, as reported within the context of the aptamer alone (Montange and Batey 2006). As expected, mutating the G11 residue decreased the SAM binding drastically in the 3P1_10AT construct to undetectable levels (Fig. 3C).
SAM binding requires stabilization of the P1 helix by coaxial stacking with an AT helix in hybrid constructs
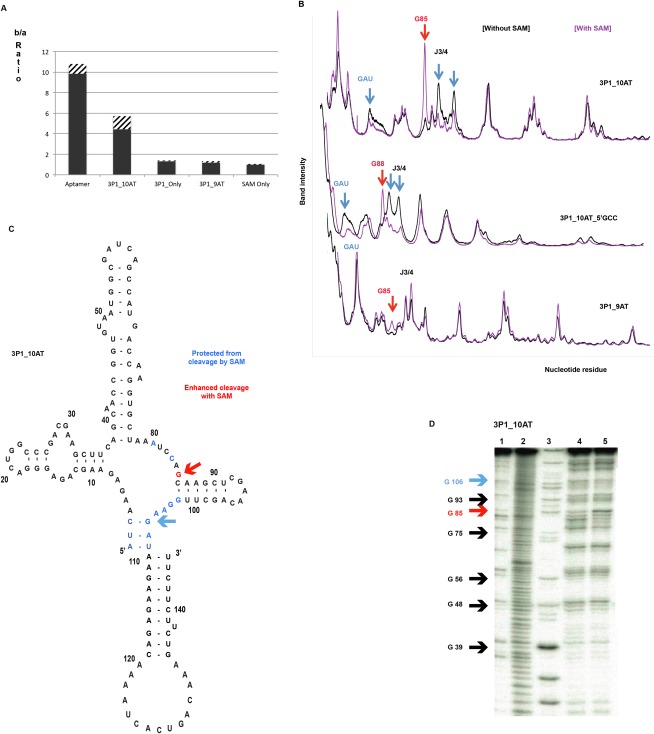
The P1 helix minor groove forms one side of the binding pocket for SAM as shown in the SAM-I riboswitch crystal structures (Montange and Batey 2006; Lu et al. 2010). The sulfur cationic moiety of the ligand interacts electrostatically with the minor groove of the second and third base pairs of the P1 helix (Montange and Batey 2006; Lu et al. 2010). Mutations of these P1 helix residues, (Winkler et al. 2003; Montange et al. 2010; Heppell et al. 2011) disruption of P1 helix base pairs, or shortening of the P1 helix (Heppell et al. 2011) dramatically reduces SAM binding to the aptamer. To test the role of the AT in SAM binding in the hybrid, we tested SAM binding with “3P1_Only” with RNA concentration of 8 μM and ligand concentration 100 nM (Fig. 4A). The 3P1_Only construct that lacks the complete AT and consists of only 3 bp in the P1 helix yielded a b/a ratio of ∼1.2 in the equilibrium dialysis assay, indicative of minimal SAM binding. This minimal SAM binding observed with 3P1_Only indicates that the AT segment contributes to SAM binding in the hybrid construct. We reasoned that the AT helix might be forming a coaxial stack with three P1 base pairs, thus stabilizing the P1 helix formation that is necessary for SAM binding (Winkler et al. 2003; Montange et al. 2010; Heppell et al. 2011). To test this possibility, we deleted a residue at the 3′ end of the AT helix (3P1_9AT), thus preventing the possibility of optimal coaxial stacking. As expected, the 3P1_9AT construct shows minimal binding with SAM with a b/a ratio of 1.16 (Fig. 4A).
FIGURE 4.
(A) The ratio of the number of counts in chamber b to chamber a in equilibrium dialysis assay for the B. subtilis yitJ aptamer, and other hybrid constructs. SAM binding observed for the 3P1_10AT hybrid is reduced dramatically when the possibility for optimal coaxial stacking with the AT helix is disrupted in 3P1_Only and in 3P1_9AT. The error bars represent standard deviation for at least three replicates of each experiment. (B–D) In-line probing data for 3P1_10AT, 3P1_10AT_5′GCC, and 3P1_9AT hybrid constructs: (B) Densitometry traces showing intensity of the base catalyzed cleavage on 8% denaturing acrylamide gel for 3P1_10AT, 3P1_9AT, and 3P1_10AT_5′GCC. The red arrows indicate sites of enhanced cleavage in the presence of SAM and the blue arrows indicate sites of protection in the presence of SAM. (C) Cleavage pattern with and without SAM for 3P1_10AT mapped onto the secondary structure schematic, with color coding as in B. (D) Sample of raw in-line probing data for 3P1_10AT with the lanes representing no reaction (1), alkaline hydrolysis (2), RNase T1 (3), RNA without SAM in in-line probing buffer (4), and RNA with SAM in in-line probing buffer (5).
We utilized in-line probing to obtain a more direct indication of helix formation in the 3P1_10AT and 3P1_9AT constructs (Fig. 4B–D). Unlike chemical modification methods, which rely on primer extension to detect chemical reactivity, this technique enabled us to work with the hybrid constructs with defined 3′ ends. As expected, the addition of SAM leads to changes in the pattern of cleavage for 3P1_10AT (Fig. 4B,C). These include protection of J3/4 residues involved in pseudoknot formation and enhanced cleavage of a G residue closing the P4 helix. The conserved three-nucleotide motif, GAU, which participates in the putative 3P1 helix, is protected in the presence of SAM, while a pattern of reduced and enhanced cleavage is observed in the adjoining J3/4. This pattern parallels that reported for SAM binding to aptamer or full length yitJ riboswitch constructs (Winkler et al. 2003). Far smaller changes in cleavage pattern upon SAM binding are observed for 3P1_9AT, consistent with a lower level of SAM binding as compared with 3P1_10AT (Fig. 4B). Interestingly, the constrained hybrid conformers do not show strand scission in one strand of the P4 helix in the absence of SAM as displayed for the yitJ251 construct (Winkler et al. 2003) that contains the full riboswitch sequence. Raw data for one of the hybrid constructs (3P1_10AT) is also shown in Figure 4D.
SAM binding to the hybrids increases marginally as the length of the P1 helix increases
Previous reports show that a stable P1 helix is crucial for the binding of SAM to the aptamer (Winkler et al. 2003; Stoddard et al. 2010; Hennelly and Sanbonmatsu 2011; Heppell et al. 2011), while our measurements indicate that stacking on an AT helix can, to some degree, substitute for a truncated segment of the P1 helix. We wished to determine how SAM binding would be affected by the relative lengths of P1 and AT helices within the competition region shown in Figure 2. The P2_AT construct, which contains a full-length AT helix but no P1 helix or J1/2, showed minimal binding with a b/a ratio of ∼1. The binding of SAM to a series of hybrids represented by b/a ratio is within 2.5–4.5 in the equilibrium dialysis assay (Fig. 5A) at a RNA concentration of 3 μM and SAM concentration of 100 nM. Even 2P1_11AT, with a P1 helix as short as 2 bp, shows readily detectable binding to SAM. The 2P1_11AT construct nonetheless binds to SAM with weakest affinity amongst all the hybrid constructs. As the number of base pairs in the P1 helix is increased there is a small increase in SAM binding. The differences in Kd are very close to standard deviation for multiple single-point measurements. Nonetheless, the trend of increasing affinity is consistent with a modest “long range” effect of P1 helix length on the stability of the SAM/riboswitch complex (see discussion under heading “accuracy of SAM binding measurements”).
FIGURE 5.
(A) Ratio of number of counts for chamber b to chamber a in equilibrium dialysis for B. subtilis yitJ SAM-I riboswitch hybrid constructs of varying P1 and AT helix length (constructs constrained to form hybrids without any competing nucleotides) and the P2_AT construct in which the J1/2 region and the 5′ of the P1 helix are truncated. (B) Ratio of number of counts for chamber b to chamber a in equilibrium dialysis for B. subtilis yitJ SAM-I riboswitch hybrid constructs with competing nucleotides (6P1_10AT, 8P1_11AT) and non-base-pairing nucleotides (3p_10AT_5′GCC, 4P1_9AT_5′GAC).
SAM binding to the hybrids is sensitive to structural defects near the binding site
The predictions in Figure 2 suggest that under some conditions the riboswitch may form conformations that include the formation of hybrids containing partial P1 and partial AT helices. We were therefore led to speculate as to whether the hybrid constructs shown in Figure 2B could be appropriate model systems for SAM binding to such intermediate conformations. Presumably, the context of a riboswitch containing those 5′ residues that have been truncated in our constructs would present a different binding-site geometry. Addition of three non-base-pairing nucleotides on the 5′ end of the P1 helix represented by the 3P1_10AT_5′GCC construct decreased the SAM binding dramatically to a b/a ratio of 1.4 (Fig. 5B) as opposed to a b/a ratio of 4.4 with 3P1_10AT. The inhibitory effect of adding nonpairing 5′ nucleotides is much less for the 4P1_9AT_5′GAC construct (Fig. 5B). This b/a value is half the value of SAM binding with 4P1_9AT (Fig. 5A).
In spite of the reduced binding observed for 3P1_10AT_5′GCC in Figure 5, addition of SAM induces similar changes to the base-catalyzed cleavage pattern as observed for 3P1_10AT (Fig. 4B). In particular, residues in J3/4 are protected from cleavage when SAM is added, except for the G residue adjoining the P4 helix, which shows enhanced cleavage. Competing nucleotides included from the 3′ end of the AT helix represented by 6P1_10AT and 8P1_11AT constructs do not hinder SAM binding with a b/a ratio of ∼4.0 and 4.3, respectively (Fig. 5B). Thus, addition of 5′ nucleotides inhibits SAM binding if the 5′ overhang has two characteristics: (1) noncomplementarity to residues on the opposing strand, and (2) the noncomplementary residues are near the SAM binding site.
A three-dimensional model for SAM binding to hybrid ON/OFF SAM-I riboswitch conformation
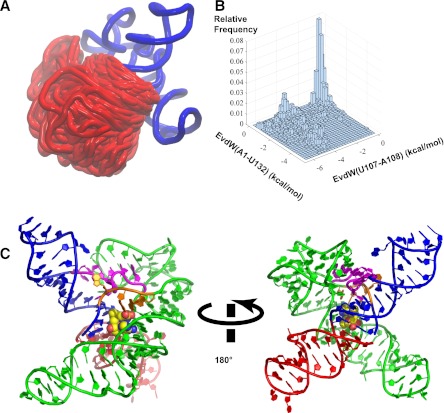
In order to assess the possible steric and structural consequences of incorporating an AT helix within the SAM-I riboswitch aptamer structure, we utilized the three-dimensional modeling package MC-sym (Parisien and Major 2008). As shown in Figure 6A, MC-sym can exhaustively sample the possible integration of the AT helix into the X-ray-derived coordinates for the aptamer core without introducing steric clashes. We used vdW interaction energy to monitor the coaxial stacking between the partial P1 helix and the incomplete AT helix. The stacking for two sets of nucleobases, U107–A108 and A1–U132, are monitored. The two-dimensional histogram of the vdW energies is displayed in Figure 6B. The results show that MC-sym predicts some models with the coaxial stacking feature as demonstrated by the area with favorable vdW energies in both sets of nucleobases. One structural model with the coaxial helical stacking is shown in Figure 6C.
FIGURE 6.
(A) Superposition of models generated from MC-sym sampling for hybrid construct—3P1_10AT. The region shown in blue is from the crystal structure of the yitJ SAM-I riboswitch (PDB:3NPB) (Lu et al. 2010). The part in red is the AT helix as it appears in multiple models generated from MC-sym, with a superposition based upon the fixed region in blue. (B) A two-dimensional histogram of the vdW interaction energies between the nucleobase of U107 and that of A108 and between the nucleobase of A1 and that of U132. A more negative value of vdW energy here indicates better stacking between the nucleobases. (C) An example of models sampling the coaxial helical stacking between the partial P1 and the AT helix with SAM docked into the binding pocket based on the binding mode observed in the aptamer crystal structure. SAM is shown in vdW representation with carbon colored in yellow. The partial P1, J4/1, and P4 helix are shown in blue, J1/2 in orange, J3/4 in magenta, and P2 and P3 in green. The modeling shows that a partial AT helix can stack upon a partial P1 helix without necessarily producing a steric clash with other segments of the structure based upon the aptamer X-ray coordinates.
The hybrid model (Fig. 6) indicates that the partial AT helix can form a continuous stack with the partial P1 helix with little indication of steric clash with other segments of the aptamer structure, when the latter are predicted based upon the X-ray coordinates.
DISCUSSION
The basis for discrimination between riboswitch conformations by SAM is more complex than suggested by secondary structure diagrams
In the case of the SAM-I riboswitch, the aptamer domain has been well characterized (Wang and Breaker 2008; Smith et al. 2010; Batey 2011). It has been more challenging to devise strategies to study the expression platform because of its high-sequence variability (Grundy and Henkin 1998). Secondary structure models for many riboswitch ON states, including SAM-I, differ dramatically from two-dimensional representations of the OFF state (Winkler et al. 2003; Tomsic et al. 2008; Batey 2011; Hennelly and Sanbonmatsu 2011). For the SAM-I (and many other) riboswitches, however, the switching mechanism requires that the base compositions of the P1 and AT helices be similar. In that case, one cannot dismiss the possibility of an ON state secondary structure that mimics some aspects of the OFF state in its tertiary folding, with the AT helix being positioned in a manner to replace the P1 helix.
Moreover, RNA structure, like protein structure, is more accurately described in solution as a Boltzmann distribution of conformer populations, rather than a single structure (Ding 2006; Lu et al. 2009). This may be especially true of unliganded riboswitches, which are the product of evolutionary selection for conformational dynamics. Until now, most experimental studies of the dynamics of riboswitch expression platforms have utilized a two-state model for RNA secondary structure. The chemical and enzymatic probes used in most of these studies, however, do not provide quantitative measures of riboswitch conformer populations (though in favorable circumstances NMR or fluorescence spectroscopy may provide such estimates) (Rieder et al. 2010; Wilson et al. 2011). Predicted secondary structures, base-pairing probability predictions at minimal free energy, and comparative sequence analysis can provide a basis for interpreting riboswitch-folding data (Huynen et al. 1996; Quarta et al. 2009; Halvorsen et al. 2010; Kim et al. 2011). While predictions of a single lowest energy structure (the so-called “MFE”) are often used for this purpose, the secondary structure of any given RNA in solution is determined by the distribution of the RNA's secondary structures at thermodynamic equilibrium (Schmitz and Steger 1992; Dirks and Pierce 2004). Therefore, riboswitches are likely to sample conformers with “hybrid” ON and OFF state character, with the interchange between them facilitated if tertiary folds are similar.
These considerations led us to test the SAM binding properties of a set of SAM-I riboswitch constructs constrained to form likely conformational intermediates. Truncating and mutating riboswitch and other folded RNAs has been a very effective means of constraining conformational dynamics to facilitate biophysical studies (Edwards et al. 2009; Serganov 2009; Batey 2011). This strategy enabled us to detect a reduced level of binding to conformational substrates that may be transiently present in partly or fully transcribed SAM-I riboswitch domains in solution.
SAM recognizes similar tertiary characteristics in hybrid and aptamer SAM-I riboswitch RNA segments
We observed SAM binding to all RNA constructs that contained a partial P1 helix, as long as a potential AT helix extending to the P1 helix boundary was present (Figs. 3–5). The kink turn-mediated pseudoknot interaction acts as an auxiliary for the global folding in the native SAM aptamer structure leading to a tight compaction upon SAM binding (Heppell and Lafontaine 2008; Heppell et al. 2011). According to our measurements for the 3P1_AT_P2 mutant and its compensatory construct (Fig. 3), similar dependence on the pseudoknot for SAM binding is seen in the hybrids. The Mg2+ effect on the aptamer was previously observed from FRET studies, gel-shift assays, and chemical probing (Heppell and Lafontaine 2008; Hennelly and Sanbonmatsu 2011; Heppell et al. 2011). The Mg2+ dependence observed in the hybrids (Fig. 3) is similar to the Mg2+ dependence observed in the isolated aptamer construct. The magnesium-based SAM binding data and the pseudoknot mutational analysis in the hybrids indicate that the junction regions and the helices in the aptamer region of the hybrids retain the global folding and SAM binding mode observed in the isolated aptamer constructs.
We have constructed a model of 3P1_10AT, which shows that an AT helix can be accommodated within a global architecture resembling the aptamer fold observed in X-ray structures (Fig. 6). The model has been constructed using the X-ray coordinates for all elements outside of the AT helix. MD simulations indicate that this configuration with SAM bound is relatively stable, but that the P1 helix base pairs dissociate in the absence of SAM (W Huang, unpubl.).
Minimal P1 helices required for SAM recognition can be stabilized in hybrids by coaxially stacked AT helices
Our hybrid constructs of the B. subtilis yitJ SAM-I riboswitch include the AT helix, which is part of the expression platform. The SAM binding data with the hybrids presented here suggest that the aptamer is not the only domain that can affect SAM binding and recognition. Our data suggest that the nucleotides in the AT helix can, up to a point, substitute for P1 helix base pairs in SAM recognition. Recent studies with the SAM-I riboswitch showed that aptamers having 3 or 4, or even 6 base pairs in the P1 helix in aptamer constructs do not terminate transcription as a result of the addition of 10 μM SAM (Heppell et al. 2011). This finding is in agreement with our SAM binding data with the 3P1 only construct (Fig. 4), since transcription termination is associated with SAM-induced P1 helix formation. The hybrid constructs 2P1_11AT, 3P1_10AT, 4P1_9AT, 5P1_8AT, and 6P1_7AT all bind to SAM, though with reduced affinity compared with the aptamer (Figs. 3–5). Thus, the AT can contribute to SAM recognition along with the remaining crucial components of the aptamer. In particular, in addition to helices P2–P4 and the pseudoknot interaction, a P1 helix nucleation site and constrained J1/2 are required, since binding is not detectable for P2_AT (Fig. 5). As mentioned, the first two base pairs of the P1 helix and residues in J1/2 are involved in critical contacts with SAM according to X-ray data (Montange and Batey 2006; Lu et al. 2010; Montange et al. 2010; Batey 2011) and MD simulations (Huang et al. 2009).
The SAM binding results with the 3P1 only, 3P1_10AT, and 3P1_9AT (Fig. 4) suggest that the 3P1 is stabilized by the AT helix through putative stacking interaction between the two helices (also seen from the MD simulations). Thus, in the absence of a continuously stacked AT helix, the shortened P1 helices do not form, and no SAM binding pocket is present. Our data with the 3P1_10AT_5′GCC (Fig. 5) show that the 5′GCC3′ sequence on the 5′ end of the P1 helix in the 3P1_10AT hybrid construct inhibits SAM binding. This destabilization may be due to disruption of the P1/AT coaxial stack, leading to a dissociation of the three P1 helix base pairs. Alternatively, it may arise from steric interference between the 5′ overhanging nucleotides and the ligand or its binding pocket. When one base-pairing nucleotide is added on the P1 helix represented by the 4P1_9AT_5′GAC, SAM binding is inhibited relative to 4P1_9AT (Fig. 5), but less inhibition is observed than when a similar substitution of unpaired 5′ nucleotides is added to the 3P1_10AT to make 3P1_10AT_5′GCC. As the length of the P1 helix increases, the stability of the P1 helix increases, but steric clash with the binding site may also be reduced.
These results also hint at a dynamic mechanism in which, upon the formation of each base pair in the P1 helix, the equilibrium is forced toward the OFF state. We can hypothesize that this mechanism may be present in the constructs 6P1_10AT and 8P1_11AT, which contain the potential for competing P1 and AT helices. While we cannot say which, if any, conformers predominate in these two constructs in the absence of ligand, we suggest that SAM binding will push the equilibrium toward those states that contain longer P1 helices. This mechanism could be slightly bolstered by the increasing SAM binding trend, however weak, observed with the increasing length of the P1 helix in the hybrids. The apparent interference of 5′ overhangs near the SAM binding pocket with SAM binding will push the equilibrium even more strongly in the direction of an extended P1. Our MD simulations with 6P1_10AT indicate a “branch migration” event in the presence of SAM (W Huang, J Kim, S Jha, and F Aboul-ela, unpubl.). Further research needs to be done in the AT region in conjunction with the P1 helix to pin down the mechanistic aspects of the P1 and AT stacking and branch migration.
The role of conformational dynamics and conformational selection in SAM binding
Earlier studies indicated that an ensemble of conformational states exists in the unliganded SAM-I riboswitch aptamer (Stoddard et al. 2010; Hennelly and Sanbonmatsu 2011). SAM selectively prefers some of these intermediate conformations for binding. The extension of the riboswitch transcript to include 3′ nucleotides with the potential to compete with P1 helix base pairing should increase the conformational heterogeneity of the conformer population. In solution, 2P1_11AT, 3P1_10AT, 4P1_9AT, 5P1_8AT, 6P1_7AT, and 8P1_5AT represent SAM-selected conformational species, along with the aptamer. The hybrids 3P1_Only, P2_AT, and 3P1_9AT represent conformational species of the SAM-I riboswitch that are not preferred by SAM for binding. With stronger affinity for the aptamer (forming the full P1 helix) than for the hybrids, SAM would shift the equilibrium toward the OFF state. The rate of such a shift cannot be predicted based upon the thermodynamic data presented here. 8P1_5AT and 8P1_11AT, which contain the potential to form full P1 helices, bind SAM with weaker affinity than the truncated aptamer (Fig. 5), perhaps because of the presence of alternative conformers, or a long range steric effect from the AT helix and loop. The implication of this finding is that SAM binding can eventually destabilize full AT helix formation, thus facilitating the downstream formation of a rho-independent terminator hairpin. Our rational design of the hybrids reveals the elements of the SAM-I riboswitch that confer the ability to recognize and bind SAM beyond the aptamer domain.
Pathways for the SAM-induced riboswitch conformational folding during transcription
It has been shown that some riboswitches operate on a kinetic and/or thermodynamic control mechanism (Wickiser et al. 2005a,b; Zhang et al. 2010). The SAM-I riboswitch is a transcriptional riboswitch. The timescale at which the decision is to be made between the formation of either dominant conformer is in seconds, considering the processivity of prokaryotic RNAP (Grundy and Henkin 2004). Whether this timescale is sufficient for the growing transcript to fold under conditions of thermodynamic equilibrium depends on rates for folding and ligand binding. This complex interplay between rates for folding, binding, and transcription will also be affected by the presence of proteins and other components of the transcription complex (Mahen et al. 2005, 2010). It has been suggested that in the presence of SAM, the decision to form the aptamer conformation, and therefore to terminate transcription, is fixed by SAM binding before the AT helix is transcribed (Tomsic et al. 2008; Batey 2011). Our findings are consistent with this hypothesis. If SAM is bound to the partial riboswitch transcript containing the aptamer conformation with full P1 helix, the 20- to 50-fold preference of the ligand for the aptamer over AT-helix-containing conformers will prevent AT helix formation.
At the same time, our results also suggest the possibility of a second path for SAM-induced conformational switching. Other studies have suggested that folding of the ON conformer, which consists of proximally formed helical base pairs, may be more rapid than the formation of distal base pairs required for the P1 helix (Huang et al. 2009; Whitford et al. 2009). This may be especially true if the latter is slowed by transient pairing of purine residues from the junction regions with 5′ pyrimidine residues from the P1 helix (Huang et al. 2012). Moreover, literature reports suggest that before SAM binding, P1 helix formation will be somewhat dynamic (Stoddard et al. 2010; Heppell et al. 2011). If AT helix formation can stabilize the nucleation of a nascent P1 helix, then our results show that SAM could bind to the resulting hybrid conformation. SAM binding should result in conversion to a fully formed P1 helix and freeing of potential AT-forming residues to participate in terminator formation. In other words, if hybrid states act as transition states, micromolar SAM levels would accelerate the transition. One attractive aspect of this model is that it predicts that termination will require micromolar concentrations of SAM to stabilize the transition state. In vitro transcription assays with the yitJ SAM-I riboswitch do indicate that much higher levels of SAM are required for transcription termination than would be predicted based upon in vitro equilibrium measurements of SAM binding affinity (Tomsic et al. 2008). A related mechanism has been proposed as an explanation for apparent discrepancies between in vitro and in vivo cotranscriptional folding rates for a ribozyme (Mahen et al. 2010). The investigators pointed out potential analogies to riboswitch folding.
Distinguishing between these two pathways is not possible based upon the thermodynamic measurements presented here alone. It requires consideration of the complex interplay between SAM binding, RNA folding, transcriptional pausing, and the role of the transcription complex itself.
Accuracy of SAM binding measurements
Equilibrium dialysis is a proven method for detecting and quantifying small molecule binding to macro molecules (ligand–protein, ligand–DNA, ligand–RNA, antibody–antigen, etc.) (Winkler et al. 2003; Sudarsan et al. 2005). Artifacts can arise due to minute bubbles near the membranes and adsorption of the ligand to the membranes. We also observed slight variability in measurements obtained with different batches of 3H-SAM. Degradation has been reported for SAM stored above pH 7 (Schlenk and Dainko 1962). For Scatchard analysis an upper limit for the percent of counts associated with degraded SAM has been derived for each batch from the amount of material in chamber a under conditions that most closely approximated 100% binding. Comparing the results determined at different time points for several constructs has shown reproducibility to within ∼98% precision, when the same batch of radiolabeled material is used. Relatively large standard deviations reported in Figures 3–5 and Supplemental Table 3 for the aptamer and 3P1_10AT constructs most likely are a result of the use of different batches of radiolabeled material. A single batch of 3H-SAM was used for measurements involving other constructs. We therefore have some confidence that the trend of increasing affinity for SAM as the P1 helix length is increased for the series 2P1_11AT, 4P1_9AT, 6P1_7AT, and 8P1_5AT (Fig. 5) is significant. Our data leave open the possibility, however, of a deviation from this trend for 3P1_10AT.
Implications for other riboswitch sequences
The motif of competing P1 vs. AT helix formation is present in most, if not all, SAM-I riboswitches. The competition topology varies from one riboswitch sequence to another (Huang et al. 2012). BPP calculations predict differing populations of intermediate conformers for SAM-I riboswitch sequences drawn from 5′ UTR leader regions upstream of different B. subtilis genes (Huang et al. 2012). There is an increasing reason to expect that differing SAM-I riboswitch sequences are “tuned” in a manner that insures an appropriate functional response for the requirements of downstream genes (Tomsic et al. 2008; Heppell et al. 2011; Huang et al. 2012). Varying populations of hybrid conformers may contribute to this tuning effect through a number of mechanisms, but further speculation is premature in the absence of more kinetic and functional data.
CONCLUSIONS
SAM binding studies with the “alternative” structures mentioned in this study indicate three major aspects of the SAM-I riboswitch folding responsible for ligand discrimination between the two functional conformers of the riboswitch: (1) A stable P1 helix is necessary for the SAM binding and recognition. (2) The AT helix can facilitate a transition from the ON state to the OFF state of the riboswitch by stabilizing the P1 helix with stacking interactions. Considering the presence of 5′ residues, as the length of the P1 helix increases, the stability of the P1 helix increases the affinity for SAM. (3) That SAM-binding affinity for the hybrid conformers is weaker by one to two orders of magnitude compared with that for the aptamer implies that SAM binding to hybrids would rapidly convert them to the OFF state. Our study indicates that SAM can bind to a SAM-I riboswitch segment that is constrained to form a full-length antiterminator (2P1_11AT) and also to hybrid constructs which, within the context of a full-length riboswitch, would block terminator formation. This result may appear counterintuitive. Nonetheless, the mechanism for SAM-I riboswitch function is readily compatible with our findings. On the other hand, our results show the minimal secondary structure requirements to form a SAM-binding competent tertiary fold, for SAM-I riboswitches. Other riboswitches, such as the purine (Stoddard and Batey 2009) and lysine (Blouin et al. 2010), as well as simpler systems such as the quenosine (Kang et al. 2009) and SAMIII-SMK (Lu et al. 2011), also contain helix-competing motifs. In some cases, including purine (Lemay and Lafontaine 2007; Greenleaf et al. 2008) and TPP (Anthony et al. 2012) riboswitches, there is evidence that the ligand stabilizes an analog to the SAM-I riboswitch P1 helix. It remains to be seen what role may be played by conformational intermediates, which combine OFF state tertiary structure characteristics with ON state secondary structure elements in the function of SAM-I and other riboswitches.
MATERIALS AND METHODS
DNA oligonucleotides and chemicals
Synthetic DNA oligos were purchased from Integrated DNA technologies, Inc. Equilibrium dialysis (Dispo equilibrium dialyzer 5000 MWCO) chambers were purchased from Harvard apparatus. The S-adenosyl–L-methionine-methyl-3H was purchased from Perkin Elmer. The NTPs for in vitro transcription were purchased from Sigma Aldrich.
DNA templates
Synthetic oligonucleotides were purified on 12% denaturing polyacrylamide gels (PAGE), electro-eluted, and recovered by ethanol precipitation. Oligonucleotide concentration was determined using UV absorbance at 260 nM.
Cassette polymerase chain reaction
The long templates for transcription reactions were synthesized by creating a cassette with two rounds of PCR reactions. The first round of PCR reactions were done with overlapping oligonucleotides (Stokes et al. 2001). Details of the sequences used for each round and the design for the hammerhead ribozyme cleavage are included in Supplemental Material (Supplemental Fig. 2).
In vitro transcriptions
Hammerhead ribozyme integrated RNA (to prevent 5′ heterogeneity) (Price et al. 1995) was included on RNAs transcribed from templates obtained from the PCR cassette using T7 RNA polymerase. The polymerase was prepared as described (Davanloo et al. 1984). Two 5′ methylated nucleotides were introduced to the reverse primer to reduce 3′ heterogeneity (Kao et al. 1999). In vitro transcriptions were carried out for 4 h at 37°C (Milligan and Uhlenbeck 1988). Complete ribozyme cleavage was obtained after 4 h at 37°C without any excess Mg2+. Supplemental Figure 3 shows the cleavage site for the hammerhead ribozyme. The RNA was purified by 12% PAGE, electroeluted, and recovered by ethanol precipitation. Recovered RNA was further purified by high-salt to low-salt dialysis, followed by desalting on PD10 columns (Kieft and Batey 2004). RNA samples were checked for size, concentration, and degradation by 12% analytical PAGE visualized by ethidium bromide staining.
Equilibrium dialysis
Single-point SAM binding assays were done with 100 nM SAM and 3 μM RNA according to established protocols (Winkler et al. 2003). Briefly, 100 nM 3H-SAM (10.0 CI/mmol, 12,000 counts per minute) in a total volume of 30 μL in equilibrium dialysis buffer (500 mM KCl, 50 mM Tris HCL at pH 7.5, 20 mM MgCl2) was added to chamber a of Dispo equilibrium dialyzer. The RNA to be tested for binding was heated to 95°C for 5 min and snap-cooled on ice for 10 min. Three micromolars of RNA to be tested for binding in a total volume of 30 μL in equilibrium dialysis buffer was added to chamber b and equilibrated at room temperature for 12 h with shaking. Aliquots from each chamber were collected and counted separately in the scintillation counter. The ratio of number of counts in chamber b to chamber a is used as a measure of SAM binding. Averaging of at least three replicates with standard deviation is used as the b/a ratio for each sample.
SAM titration (scatchard analysis)
Affinity of SAM to aptamer was calculated from the data obtained from scatchard analysis. Two-hundred nanomolars of yitJ aptamer RNA was added to chamber b of the Dispo equilibrium dialyzer and 3H-SAM was added in chamber a to concentrations ranging from 10 nM to 3 μM, along with equilibrium dialysis buffer (500 mM KCl, 50 mM Tris HCL at pH 7.5, 20 mM MgCl2). The total volume in each chamber was 30 μL. Degradation of SAM was taken into account by eliminating 10% of the counts in each chamber when estimating total SAM in each chamber. Affinity of SAM to the aptamer was calculated from the scatchard plot. SAM bound (Lb), free SAM (Lf), and total SAM (Lt) were all calculated from the number of counts in chamber a and chamber b (Winkler et al. 2003). Data points obtained using dialysis chambers with leaks in the membrane and air bubbles are not included.
BPP calculation
The base-pair probability was calculated at 37°C using the RNAfold program in the Vienna RNA package (Gruber et al. 2008). The probabilities of base pairs in the proposed aptamer and AT models are plotted according to the position as annotated in the Figure 2A. Two versions of sequences are used in the calculation. The one without terminator (T) includes the sequence to form either aptamer or AT without the 3′ strand of the terminator. The other one is the full-length sequence of the riboswitch, in which the terminator can completely form.
In-line probing assay
In-line probing reactions with the 3P1_10AT, 3P1_9AT, and 3P1_10AT_5′GCC hybrid constructs were done in the presence and absence of SAM. The RNA required was generated as mentioned in the in vitro transcriptions above. The dephosphorylation and 5′ end 32P labeling, and in-line probing reactions in the presence of 100 μM SAM are done according to the methods mentioned in Winkler et al. (2003). Denaturing 10% PAGE and its analysis using a molecular dynamics PhosphorImager was done to analyze the RNA cleavage patterns.
Tertiary structural models for SAM binding to hybrid ON/OFF SAM-I riboswitch conformation
Three-dimensional coordinates were constructed using the modeling package, MC-SYM, as described (Parisien and Major 2008). The model was constructed assuming that (1) the P1 and AT helices are continuously stacked, (2) the portion of the RNA that is identical in sequence to the aptamer (P2, P3, and P4 helices, the junction regions, and the truncated P1 helix) forms a similar three-dimensional fold, (3) SAM binds in the pocket in a mode similar to that observed in the aptamer X-ray structure. These assumptions were based upon the findings described in Figures 3 and 4, which indicate that the hybrid binding mode and three-dimensional folding mimic, to some degree, those of the aptamer.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We acknowledge Dr. Anne Grove (Department of Biological Sciences, Louisiana State University) and Dr. Robert Batey (Department of Chemistry and Biochemistry, University of Colorado Boulder) for valuable discussions. This work was supported by NSF (2010)-Pfund-164 and Award Number P20RR020159 from the National Center For Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation, National Center for Research Resources, or the National Institute of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032177.111.
REFERENCES
- Anthony PC, Perez CF, García-García C, Block SM 2012. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci 109: 1485–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger S, Yuen WH, Danchin A, Martin-Verstraete I 2002. The metIC operon involved in methionine biosynthesis in Bacillus subtilis is controlled by transcription antitermination. Microbiology-Sgm 148: 507–518 [DOI] [PubMed] [Google Scholar]
- Batey RT 2011. Recognition of S-adenosylmethionine by riboswitches. Wiley Interdisciplinary Reviews: RNA 2: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin S, Chinnappan R, Lafontaine DA 2010. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res 39: 3373–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR 2006. Riboswitches as antibacterial drug targets. Nat Biotechnol 24: 1558–1564 [DOI] [PubMed] [Google Scholar]
- Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR 2007. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol 3: 44–49 [DOI] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR 2007. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447: 497–500 [DOI] [PubMed] [Google Scholar]
- Cressina E, Chen L, Abell C, Leeper FJ, Smith AG 2011. Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chemical Science 2: 157–165 [Google Scholar]
- Daldrop P, Reyes FE, Robinson DA, Hammond CM, Lilley DM, Batey RT, Brenk R 2011. Novel ligands for a purine riboswitch discovered by RNA-Ligand docking. Chem Biol 18: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P, Rosenberg AH, Dunn JJ, Studier FW 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci 81: 2035–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Ferré-D'Amaré AR 2011. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res 44: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YE 2006. Statistical and Bayesian approaches to RNA secondary structure prediction. RNA 12: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RM, Pierce NA 2004. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J Comput Chem 25: 1295–1304 [DOI] [PubMed] [Google Scholar]
- Edwards AL, Garst AD, Batey RT 2009. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol Biol 535: 1–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E 2003. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci 100: 5052–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319: 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL 2008. The Vienna RNA Websuite. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol 30: 737–749 [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM 2004. Regulation of gene expression by effectors that bind to RNA. Curr Opin Microbiol 7: 126–131 [DOI] [PubMed] [Google Scholar]
- Haller A, Souliére MF, Micura R 2011. The Dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res 44: 1339–1348 [DOI] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A 2010. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 6: e1001074 doi: 10.1371/journal.pgen.1001074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennelly SP, Sanbonmatsu KY 2011. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res 39: 2416–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppell B, Lafontaine DA 2008. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry 47: 1490–1499 [DOI] [PubMed] [Google Scholar]
- Heppell B, Blouin S, Dussault A-M, Mulhbacher J, Ennifar E, Penedo JC, Lafontaine DA 2011. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol 7: 384–392 [DOI] [PubMed] [Google Scholar]
- Huang W, Kim J, Jha S, Aboul-ela F 2009. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: A small molecule with a strong arm. Nucleic Acids Res 37: 6528–6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Kim J, Jha S, Aboul-ela F 2012. Conformational heterogeneity of the SAM-I riboswitch transcriptional ON state: a chaperone-like role for S-adenosylmethionine. J Mol Biol doi: 10.1016/j.jmb.2112.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MA, Perelson A, Vieira WA, Stadler PF 1996. Base pairing probabilities in a complete HIV-1 RNA. J Comput Biol 3: 253–274 [DOI] [PubMed] [Google Scholar]
- Kang M, Peterson R, Feigon J 2009. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell 33: 784–790 [DOI] [PubMed] [Google Scholar]
- Kao C, Zheng M, Rüdisser S 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Batey RT 2004. A general method for rapid and nondenaturing purification of RNAs. RNA 10: 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR 2009. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem Biol 4: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W, Maddineni S, Aboul-ela F, Jha S 2011. Energy landscape analysis for regulatory RNA finding using scalable distributed cyberinfrastructure. Concurr Comput 23: 2292–2304 [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390: 249–256 [DOI] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR 2009. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol 6: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J-Fá, Lafontaine DA 2007. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA 13: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link KH, Breaker RR 2009. Engineering ligand-responsive gene-control elements: lessons learned from natural riboswitches. Gene Ther 16: 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, Gloor JW, Mathews DH 2009. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA 15: 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A 2010. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S Box/SAM-I Riboswitch. J Mol Biol 404: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Smith AM, Ding F, Chowdhury A, Henkin TM, Ke A 2011. Variable sequences outside the Sam-binding core critically influence the conformational dynamics of the SAM-III/SMK box riboswitch. J Mol Biol 409: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciagiewicz I, Zhou S, Bergmeier SC, Hines JV 2011. Structure-activity studies of RNA-binding oxazolidinone derivatives. Bioorg Med Chem Lett 21: 4524–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahen EM, Harger JW, Calderon EM, Fedor MJ 2005. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol Cell 19: 27–37 [DOI] [PubMed] [Google Scholar]
- Mahen EM, Watson PY, Cottrell JW, Fedor MJ 2010. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol 8: e1000307 doi: 10.1371/journal.pbio.1000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM 2003. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci 100: 3083–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Henkin TM 2005. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol Microbiol 57: 1008–1021 [DOI] [PubMed] [Google Scholar]
- Médigue C, Moszer I, Viari A, Danchin A 1995. Analysis of a Bacillus subtilis genome fragment using a co-operative computer system prototype. Gene 165: GC37–GC51 [DOI] [PubMed] [Google Scholar]
- Milligan JF, Uhlenbeck OC 1988. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51–62 [DOI] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756 [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Montange RK, Mondragón E, van Tyne D, Garst AD, Ceres P, Batey RT 2010. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol 396: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR 2002. Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Parisien M, Major F 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452: 51–55 [DOI] [PubMed] [Google Scholar]
- Price SR, Ito N, Oubridge C, Avis JM, Nagai K 1995. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol 249: 398–408 [DOI] [PubMed] [Google Scholar]
- Quarta G, Kim N, Izzo JA, Schlick T 2009. Analysis of riboswitch structure and function by an energy landscape framework. J Mol Biol 393: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A, Winkler WC 2010. Magnesium-sensing riboswitches in bacteria. RNA Biol 7: 77–83 [DOI] [PubMed] [Google Scholar]
- Rieder U, Kreutz C, Micura R 2010. Folding of a transcriptionally acting PreQ1 riboswitch. Proc Natl Acad Sci 107: 10804–10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal Society of Chemistry. 2011 Structural and catalytic roles of metal ions in RNA. Metal ions in life sciences (ed. A Sigel et al.). RSC Publishing, Cambridge, UK. [Google Scholar]
- Schlenk F, Dainko JL 1962. The alkaline hydrolysis of S-adenosylmethionine in tritiated water. Biochem Biophys Res Commun 8: 24–27 [DOI] [PubMed] [Google Scholar]
- Schmitz M, Steger G 1992. Base-pair probability profiles of RNA secondary structures. Comput Appl Biosci 8: 389–399 [DOI] [PubMed] [Google Scholar]
- Schroeder KT, Lilley DM 2009. Ion-induced folding of a kink turn that departs from the conventional sequence. Nucleic Acids Res 37: 7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, Daldrop P, Lilley DM 2011. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure 19: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A 2009. The long and the short of riboswitches. Curr Opin Struct Biol 19: 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Fuchs RT, Grundy FJ, Henkin T 2010. Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal. RNA Biol 7: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Batey RT 2009. Beyond crystallography: Investigating the conformational dynamics of the purine riboswitch. In Non-protein coding RNAs (ed. NG Walter et al.), pp. 215–228. Springer Berlin, Heidelberg, Germany [Google Scholar]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT 2010. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 18: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes HW, Holmes AJ, Nield BS, Holley MP, Nevalainen KM, Mabbutt BC, Gillings MR 2001. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl Environ Microbiol 67: 5240–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR 2003. Metabolite binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR 2005. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol 12: 1325–1335 [DOI] [PubMed] [Google Scholar]
- Tomsic J, McDaniel BA, Grundy FJ, Henkin TM 2008. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-Box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol 190: 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp S, Gallivan JP 2010. Emerging applications of riboswitches in chemical biology. ACS Chem Biol 5: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Breaker RR 2008. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol 86: 157–168 [DOI] [PubMed] [Google Scholar]
- Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY 2009. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys J 96: L7–L9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM 2005a. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44: 13404–13414 [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM 2005b. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18: 49–60 [DOI] [PubMed] [Google Scholar]
- Wieland M, Hartig JS 2008. Artificial riboswitches: Synthetic mRNA-based regulators of gene expression. ChemBioChem 9: 1873–1878 [DOI] [PubMed] [Google Scholar]
- Wilson RC, Smith AM, Fuchs RT, Kleckner IR, Henkin TM, Foster MP 2011. Tuning riboswitch regulation through conformational selection. J Mol Biol 405: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Grundy FJ, Murphy BA, Henkin TM 2001. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707 [DOI] [PubMed] [Google Scholar]
- Woodson SA 2005. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lau MW, Ferré-D'Amaré AR 2010. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry 49: 9123–9131 [DOI] [PMC free article] [PubMed] [Google Scholar]