Introduction of a rare variant into the SAM-I riboswitch results in normal binding of SAM ligand, indicating that tertiary RNA contacts lead to a k-turn folding with a standard k-turn geometry.
Keywords: RNA structure, k-turn, SAM-I riboswitch, X-ray crystallography, Thelohania solenopsae
Abstract
The kink turn (k-turn) is a frequently occurring motif, comprising a bulge followed by G•A and A•G pairs that introduces a sharp axial bend in duplex RNA. Natural k-turn sequences exhibit significant departures from the consensus, including the A•G pairs that form critical interactions stabilizing the core of the structure. Kt-23 found in the small ribosomal subunit differs from the consensus in many organisms, particularly in the second A•G pair distal to the bulge (2b•2n). Analysis of many Kt-23 sequences shows that the frequency of occurrence at the 2n position (i.e., on the nonbulged strand, normally G in standard k-turns) is U>C>G>A. Less than 1% of sequences have A at the 2n position, but one such example occurs in Thelohania solenopsae Kt-23. This sequence folds only weakly in the presence of Mg2+ ions but is induced to fold normally by the binding of L7Ae protein. Introduction of this sequence into the SAM-I riboswitch resulted in normal binding of SAM ligand, indicating that tertiary RNA contacts have resulted in k-turn folding. X-ray crystallography shows that the T. solenopsae Kt-23 adopts a standard k-turn geometry, making the key, conserved hydrogen bonds in the core and orienting the 1n (of the bulge-proximal A•G pair) and 2b adenine nucleobases in position facing the opposing minor groove. The 2b and 2n adenine nucleobases are not directly hydrogen bonded, but each makes hydrogen bonds to their opposing strands.
INTRODUCTION
The kink turn (k-turn) is a widespread and important structural motif in duplex RNA that typically comprises a 3-nt bulge followed on its 3′ side by consecutive G•A and A•G pairs (Fig. 1A). k-turns generate a sharp kink into the axis of the helix and are very important in the architecture of large RNA species, where they can exert an important influence on the long-range structure and the formation of RNA-protein complexes. k-turns were first identified as a structural motif in both subunits of the ribosome (Klein et al. 2001), although they were independently observed as a repeated sequence in regulatory elements in RNA (Winkler et al. 2001). They are also present in the nucleolar RNA species that guide RNA modification (Hamma and Ferré-D'Amaré 2004; Moore et al. 2004; Szewczak et al. 2005; Youssef et al. 2007), U4 snRNA (Vidovic et al. 2000; Wozniak et al. 2005), and in untranslated regions of mRNA (Mao et al. 1999; White et al. 2004), including within riboswitches (Montange and Batey 2006; Blouin and Lafontaine 2007; Heppell and Lafontaine 2008; Smith et al. 2009). k-turns are, consequently, involved in virtually every aspect of RNA function, including the translation and modification of RNA, spliceosome assembly, and the control of gene expression.
FIGURE 1.
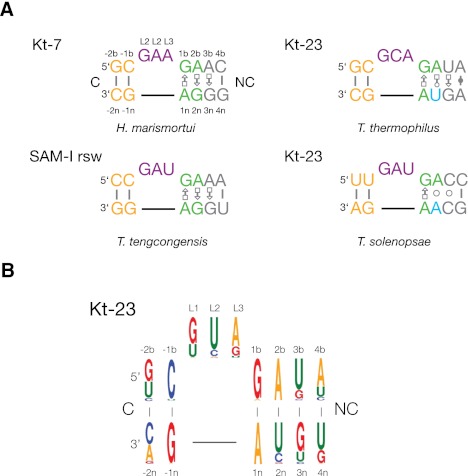
Sequences of k-turns and consensus sequence of Kt23. (A) Sequences of Kt-7, SAM-I, and Kt-23 k-turns. The k-turn sequence nomenclature (Liu and Lilley 2007) is indicated on the Kt-7 sequence; this scheme is used throughout this paper. (B) WebLogo (Crooks et al. 2004) analysis of Kt-23 sequences.
The tightly kinked geometry of the k-turn motif requires stabilization, in the absence of which the structure adopts a more gently bent structure, as expected for a normal 3-nt bulge. The folded k-turn can be stabilized in three different ways: first, by the addition of metal ions (Matsumura et al. 2003; Goody et al. 2004); second, protein binding can induce the formation of the kinked conformation (Turner et al. 2005), as exemplified by (though not limited to) the L7Ae family of proteins; and last, we have recently shown that tertiary interactions within a larger RNA species can stabilize the folded k-turn structure (Schroeder et al. 2011). It is likely that all these effects play a role in the ordered assembly of large RNA-protein complexes such as the ribosome, where the folding of different k-turns mediates long-range structural interactions and the structure becomes fixed by the binding of different proteins. This would probably require that k-turns not be equally intrinsically stable and that proteins would bind with a range of affinities. We have shown that the near-consensus k-turn Kt-7 is induced to fold by the binding of Archeoglobus fulgidus L7Ae with an apparent affinity of Kdapp = 10 pM (Turner and Lilley 2008). However, other k-turns exhibit a significant departure from the consensus sequence and are likely to require greater stabilization of the kinked conformation. Examination of many k-turn sequences (Schroeder et al. 2010) reveals two kinds of departure from the standard sequence. We term simple k-turns as those where the ordering of the nucleotides within the sequence matches those of the standard k-turn such as Kt-7. However, within that framework, simple k-turns can exhibit sequence substitutions that differ from the consensus. In complex k-turns, the position of the nucleotides of the two critical G•A pairs in the structure does not map linearly onto the sequence. For example, the adenine in the 2b position of Haloarcula marismortui Kt-15 actually arises from the nonbulged strand.
We have shown previously that the metal ion-induced folding of Kt-7 is completely intolerant to sequence changes in the two G•A pairs at the 1b•1n and 2n•2b positions (Goody et al. 2004). Yet, some natural k-turn sequences do depart from the consensus, including in these G•A pairs. Kt-23 found in the small ribosomal subunit is commonly observed to have a nucleotide other than guanine at the 2n position (Fig. 1A). For example, in Thermus thermophilus, the 2n nucleotide is uridine, creating a non-Watson-Crick A•U pair at the 2b•2n position (Wimberly et al. 2000). While making this change in Kt-7 generates a sequence that cannot be folded into k-turn geometry by addition of metal ions (Goody et al. 2004), the T. thermophilus Kt-23 sequence folds very well in the presence of Mg2+ or Na+ ions (Schroeder and Lilley 2009). The sequence context was shown to be critical to the ion-induced stabilization of this Kt-23 sequence, with the nature of the 3b•3n position especially important.
In the present study, we have used bioinformatics to investigate the range of Kt-23 sequences more broadly, finding that the majority of sequences lack guanine at the 2n position. A very minor subset of the sequences has adenine in the 2n position, creating a potential A•A pair at the 2b•2n position. The potential equivalence of the structures of trans sugar-Hoogsteen G•A and A•A pairs is shown by the isostericity matrices of Leontis et al. (2002). While we have recently solved the crystal structure of a variant k-turn with such an A•A pair at that position stabilized by tertiary interactions in the SAM-I riboswitch, there are no structural data available for any natural, simple k-turn with this sequence. Using bioinformatics, we discovered that Thelohania solenopsae Kt-23 has just such a possible A•A pair at the 2b•2n position. We have, therefore, studied this sequence, showing that it can adopt the k-turn conformation when stabilized either by L7Ae binding or by tertiary interactions in the SAM-I riboswitch. Using the latter, we have solved the crystal structure of this novel k-turn.
RESULTS
Sequence variation in Kt-23
Kt-23 of the small ribosomal subunit rRNA exhibits a significant degree of sequence variation, particularly at the 2b•2n position that is normally an A•G pair in standard k-turns such as Kt-7. Study of an alignment of 6325 sequences of 16S and 18S rRNA (Cannone et al. 2002) revealed that the presence of an adenine at position 2b is almost invariant (99.9% of all sequences), but the 2n position is more variable (for examples, see Supplemental Fig. S1). The 2n nucleotide is uridine in 96% of bacteria and 97% of archaeal sequences but in only 20% of eukaryotic sequences.
We have shown previously that the Kt-23 of T. thermophilus folds into a normal k-turn conformation in the presence of metal ions, despite the A•U pair at the 2b•2n position (Schroeder and Lilley 2009). We have generated a consensus sequence for Kt-23 from the alignment of 16S and 18S rRNA, represented using WebLogo 3 (Fig. 1B; Crooks et al. 2004). While this underlines that adenine at the 2b position is strongly conserved, the frequency of occurrence of nucleotides at the 2n position is U>C>G>A. The occurrence of an A at this position is ∼1% in all domains of life. This raises the question of whether a potential k-turn having an A•A pair at the 2b•2n position can fold into the kinked geometry, but there are no structural data available for any natural, simple k-turn with this sequence. We, therefore, sought an example of a Kt-23 sequence with an A at the 2n position to investigate structurally.
T. solenopsae is a microsporidial parasite of the fire ant, Solenopsis invicta (Knell et al. 1977). Its Kt-23 sequence is shown in Figure 1A. The sequence can be readily drawn as a simple k-turn, with a conventional G•A pair at the 1b•1n position and an A•A pair at the 2b•2n position. We have, therefore, explored the ability of this sequence to adopt the k-turn conformation.
T. solenopsae Kt-23 is weakly folded by Mg2+ ions
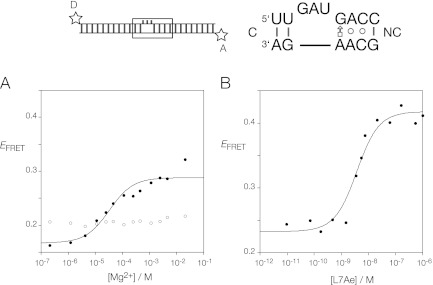
Standard k-turns such as Kt-7 are induced to fold into their characteristic tightly kinked conformation as isolated duplexes by the addition of metal ions. This can be conveniently studied by fluorescence resonance energy transfer (FRET), using a short duplex RNA with the putative k-turn-forming sequence at the center and fluorescein (donor) and Cy3 (acceptor) fluorophores attached at the two 5′ termini. When the RNA adopts the kinked structure, the inter-fluorophore distance is reduced, with a corresponding increase in FRET efficiency (EFRET). This was measured as a function of added Mg2+ ion concentration and plotted in Figure 2A. Addition of Mg2+ ions results in an increase in EFRET, and this is not observed in a variant where the 2′O atom has been deleted from the L1 position, thereby removing the possibility of forming the critical hydrogen bond to N1 of the conserved A1n nucleobase observed in the majority of k-turns of known structures (Liu and Lilley 2007). However, the extent of the change in FRET (ΔEFRET) is 0.12, compared to ≥0.25 for the majority of k-turn structures whose sequences approximate the consensus (Liu and Lilley 2007; Turner and Lilley 2008). The transition is relatively broad; fitting to a two-state model with a Hill coefficient constrained to n = 1 gave a value of [Mg2+]1/2 = 36 μM, but this is not well defined by the data.
FIGURE 2.
FRET analysis of the folding of T. solenopsae Kt-23 induced by addition of Mg2+ ions and by L7Ae binding. The k-turn sequence was centrally located within a 25-bp RNA duplex terminally labeled with fluorescein (donor, D) and Cy3 (acceptor, A) fluorophores. FRET efficiency was measured in the steady-state. (A) Mg2+ ion-induced folding of the natural T. solenopsae Kt-23 (closed circles) and the L1 2′H variant (open circles). The change in EFRET was fitted using Equation (1). (B) L7Ae-induced folding of the natural T. solenopsae Kt-23. The change in EFRET was fitted using Equation (2).
Folding of T. solenopsae Kt-23 induced by the binding of L7Ae protein
Protein binding provides a second way to induce the folding of some k-turn structures (Turner et al. 2005; Turner and Lilley 2008). In view of the rather weak folding of T. solenopsae Kt-23 by the addition of Mg2+ ions, we examined the structure of the same fluorescein, Cy3-labeled Kt-23 RNA upon addition of A. fulgidus L7Ae protein, which induces the folding of standard k-turns such as Kt-7 and that of the box C/D RNA (Turner et al. 2005). We measured EFRET as a function of protein concentration, plotted in Figure 2B. A clear transition was observed, with an increase of FRET efficiency of ΔEFRET = 0.23, comparable to the range expected for full k-turn folding. Using a simple binding model, the apparent affinity of binding was Kdapp = 2.6 nM; this is several orders of magnitude lower than that measured for the same protein binding to Kt-7, where Kdapp = 10 pM (Turner and Lilley 2008). Evidently, L7Ae induces the folding of T. solenopsae Kt-23 RNA but with a markedly reduced affinity. This is likely to result from the lower stability of the folded k-turn due to the A•A pair at the 2b•2n position. If we assume that the lowering of affinity is solely due to the A•A pair, this indicates a destabilization of 13.9 kJ mol−1.
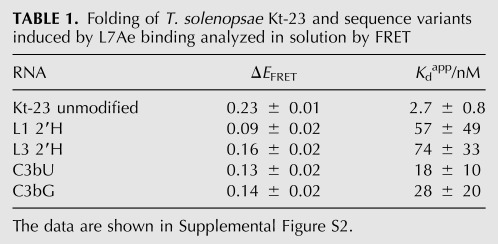
We have examined the effect of some atomic and sequence substitutions on the L7Ae-induced folding of T. solenopsae Kt-23 RNA. FRET efficiencies were measured as a function of L7Ae concentration and the data fitted as above (Supplemental Fig. S2). Values of ΔEFRET and Kdapp are presented in Table 1. The largest effect arises from the L1 2′H substitution, where the increase in FRET efficiency is only ΔEFRET = 0.09, and the apparent affinity is lowered by two orders of magnitude. These data have significant scatter, and the calculated affinity has a large error; nevertheless, a small degree of folding appears to have been induced by L7Ae protein, compared to none for the addition of Mg2+ ions. A second well-conserved hydrogen bond is donated by the L3 2′OH to the proS nonbridging O of the phosphate linking L1 and L2 in all k-turns (Liu and Lilley 2007). However, once again, deletion of this O atom does not completely prevent L7Ae-induced folding. The L3 2′H substitution results in a reduced extent of folding (ΔEFRET = 0.16) and an apparent affinity that is lowered by two orders of magnitude.
TABLE 1.
Folding of T. solenopsae Kt-23 and sequence variants induced by L7Ae binding analyzed in solution by FRET
In our earlier study of the T. thermophilus Kt-23 k-turn, where there is an A•U pair at the 2b•2n position, we found that the identity of the adjacent 3b•3n pair was critical to the stability of the kinked conformation (Schroeder and Lilley 2009). In the T. solenopsae Kt-23, the 3b•3n pair is C•C. Replacement of this by either the potential Watson-Crick pair G•C (C3bG) or a U•C pair (C3bU) did not prevent L7Ae-induced folding but significantly impaired the extent of folding and lowered the affinity of binding by an order of magnitude.
The T. solenopsae Kt-23 is functional in the SAM riboswitch
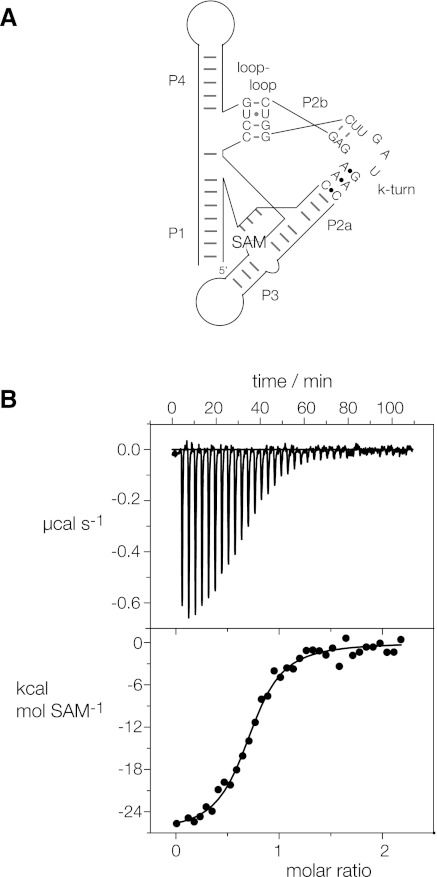
We have recently demonstrated a third process that can lead to stabilization of k-turns in the kinked conformation—the manipulation of the RNA by tertiary interactions within a larger RNA species (Schroeder et al. 2011). The Thermoanaerobacter tengcongensis SAM-I riboswitch contains a standard k-turn that kinks a long helix so that its terminus can make a loop-receptor interaction that is important for functional folding. Disruption of the k-turn leads to an RNA structure that no longer binds the S-adenosyl methionine (SAM) ligand that is straightforwardly analyzed by isothermal titration calorimetry (ITC). We showed that if we changed the standard A•G pair at the 2b•2n position of the riboswitch k-turn to A•A, this prevented folding of the RNA as an isolated duplex, but that the same change within the riboswitch allowed SAM binding to occur unaffected, unlike changes at the 1b•1n position. Clearly the tertiary interactions within the overall riboswitch structure stabilize the structure of the substituted k-turn, overcoming the effect that prevented folding of the isolated k-turn. We, therefore, engineered the T. solenopsae Kt-23 sequence into the corresponding position in the SAM-I riboswitch (Fig. 3A), and performed ITC using SAM as the ligand.
FIGURE 3.
Isothermal titration calorimetric analysis of SAM binding to the SAM-I riboswitch in which the k-turn has been replaced by that of T. solenopsae Kt-23. A solution of SAM was titrated into a SAM-I riboswitch RNA solution, and the heat evolved was measured by ITC as the power required to maintain zero temperature difference with a reference cell. Integration over time gives the heat required to maintain thermal equilibrium between cells. (A) Scheme of SAM-I riboswitch with T. solenopsae Kt-23. (B) Calorimetric data. The upper panel shows the raw data for sequential injections of 8-μL volumes (following an initial injection of 1 μL) of a 100 μM solution of SAM into a 1.4 mL 10 μM RNA solution in 50 mM HEPES (pH 7.5), 100 mM KCl, 10 mM MgCl2. This represents the differential of the total heat (i.e., enthalpy ΔH° under conditions of constant pressure) for each SAM concentration. The lower panel presents the integrated heat data fitted to a single-site binding model (Equation [3]). The thermodynamic parameters calculated are summarized in Table 2.
The ITC results are fully consistent with normal SAM binding to the hybrid riboswitch (Fig. 3B). SAM binds to the riboswitch in an exothermic reaction, and the data may be fitted to a binding model giving an affinity of Kd = 0.49 μM (Table 2), closely similar to 0.54 μM measured for the riboswitch with the natural k-turn sequence (Montange et al. 2010; Schroeder et al. 2011). This is consistent with folding of the T. solenopsae Kt-23 sequence into normal k-turn geometry in the context of the riboswitch, indicating that the tertiary interactions facilitate folding of this k-turn.
TABLE 2.
ITC-derived thermodynamic data for SAM binding to the SAM-I riboswitch
Crystal structure of the T. solenopsae Kt-23 in the SAM riboswitch
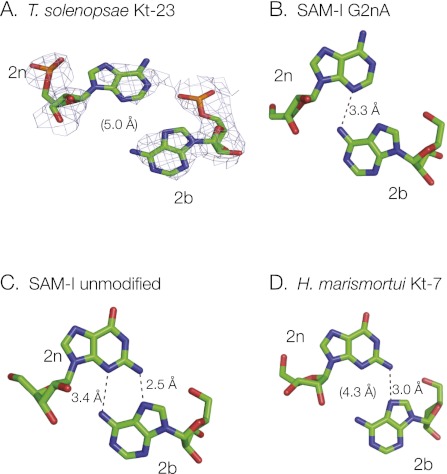
The demonstration that the T. solenopsae Kt-23 sequence is functional in the SAM-I riboswitch and, therefore, probably folded into the k-turn conformation provided an opportunity to determine its structure by X-ray crystallography. We have shown previously that we can crystallize variants of the SAM-I riboswitch containing altered k-turn sequences and solve their structure straightforwardly by molecular replacement (Schroeder et al. 2011). Using the riboswitch as a kind of structural “cassette” in this way, we found that the Kt-23-containing RNA crystallized under closely similar conditions to those used for the natural riboswitch, with retention of the P43212 space group and unit cell parameters (Montange et al. 2010). Diffraction data were collected and processed to a resolution of 2.95 Å (Table 3), and initial phases were obtained by molecular replacement using the previously reported structure of the natural riboswitch, PDB code 2GIS (Montange and Batey 2006).
TABLE 3.
Details of data collection and refinement statistics for the structure of the SAM-I riboswitch with the inserted Kt-23 from T. solenopsae
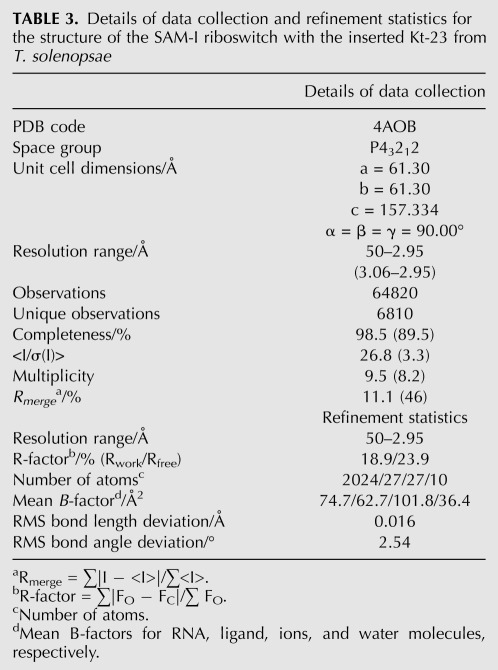
The overall structure of the modified SAM-I riboswitch was retained from the natural species (Supplemental Fig. S3), with an RMSD calculated over all P atoms of 0.63 Å. Both the riboswitch and its bound SAM ligand were well defined by the electron density. The k-turn is clearly visible within the structure (Fig. 4A), kinking the P2 helix so that it can dock its terminal loop into the receptor, a key interaction in the folding of the riboswitch into its functional form that creates the SAM binding site. The k-turn can be superimposed with the Kt-7 k-turn with an RMSD of 1.6 Å (Fig. 4B). Although the superimposition was carried out using the phosphorus atoms, the A1n and A2b nucleobases are almost perfectly superimposed, showing that these key adenines are placed normally to make their interactions with the minor groove of the C helix. A composite omit map was calculated (Fig. 4C; Supplemental Figs. S4, S5) to exclude the possibility of model bias. This omit map clearly supports the obtained model with the great majority of the nucleotides very well resolved. Significantly for this study, the A2n and A2b nucleotides were well resolved (Fig. 4C).
FIGURE 4.
The crystal structure of the SAM-I riboswitch in which the k-turn has been replaced by that of T. solenopsae Kt-23. Parallel-eye stereo views are shown. (A) The structure of the k-turn within the modified riboswitch. The 2Fobs-Fc electron density map is shown contoured at 1σ. The nucleotides are colored to match the schemes shown in Figure 1. (B) Superposition of Kt-23 (magenta) with the standard k-turn Kt-7 (green). The two k-turns were superimposed using their phosphorus atoms only. Note the close superimposition of A1n and A2b nucleobases (highlighted in stick form). (C) View from the nonbulged strand side of the core of the k-turn. The electron density shown on the A2b and A2n nucleotides is taken from the composite omit map. (D) View down onto the loop to show the conserved hydrogen bonds.
The T. solenopsae Kt-23 structure has the near-universal standard hydrogen bonds involving 2′-hydroxyl groups (Fig. 4D; Liu and Lilley 2007). Both the GL1 O2′ to A1n N1 and the UL3 O2′ to PL1/PL2 proS O bonds are 2.7 Å in length, both having good geometry. As shown above, disruption of either leads to impairment of L7Ae-induced folding of the k-turn. k-turns in which the second nucleotide of the bulge (L2) is adenine frequently make an additional hydrogen bond between AL2 N6 and A2n O2′. This is also present in the T. solenopsae Kt-23 structure, with a length of 2.8 Å and good geometry.
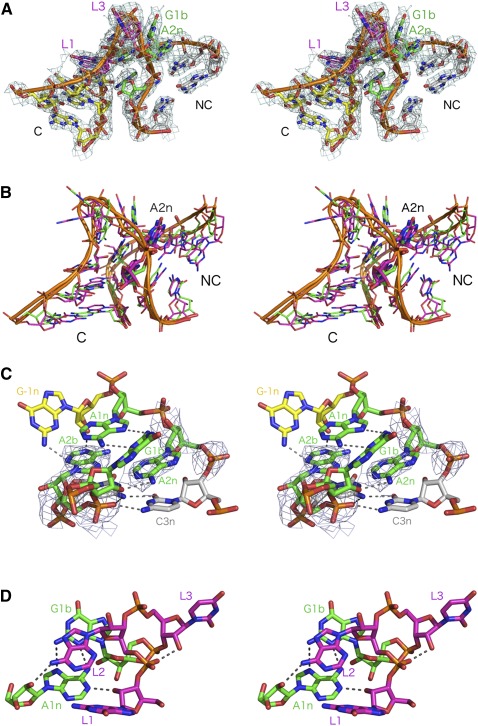
The core of the k-turn is shown in Figure 4C. As expected, the 1b•1n base pair is a standard trans sugar-Hoogsteen pair, with two hydrogen bonds, from G1b N2 to A1n N7 and from A1n N6 to G1b N3, both 3.2 Å in length. In the 2b•2n position, we see that the A•A pair is oriented similarly to the A•A pair in the SAM-I riboswitch G2nA k-turn structure (see Fig. 5A,B), with the nucleobase of the A2n stacked between those of G1b and C3n. However, in contrast to the SAM-I G2nA riboswitch A•A pair, there is no direct hydrogen bonding between the two adenine bases; the position and orientation of the nucleobases are defined by the electron density unambiguously. The A•A pair of the SAM-I G2nA riboswitch is connected by a hydrogen bond from A2b N6 to A2n N3 (Fig. 5B), but the corresponding distance in the T. solenopsae Kt-23 structure is 5.0 Å. However, the two adenine bases are individually bonded into the k-turn structure (Fig. 4C). N6 of the A2n nucleobase hydrogen bonds with the O2′ of G1b (length 3.1 Å). In addition, adenine 2b makes two hydrogen bonds across the interface with the C helix; N3 accepts a bond from G-1n N2 (length 2.8 Å), and N1 accepts a bond from G-1n O2′ (length 2.5 Å).
FIGURE 5.
The structure of the A2b and A2n nucleotides in the T. solenopsae Kt-23 and comparison with 2b•2n pairs observed in other k-turns. (A) T. solenopsae Kt-23. The electron density is taken from the composite omit map. (B) The SAM-I riboswitch G2nA k-turn (2YGH). (C) The SAM-I riboswitch k-turn (3GX5). (D) H. marismortui Kt-7 (1FFK).
Moving to the 3b•3n position, we have seen that the C3bU and C3bG substitutions resulted in significant impairment of L7Ae-induced folding. We have previously observed that the nature of the 3b•3n pair in T. thermophilus Kt-23 (with U at the 2n position) has a strong influence on the folding of that k-turn. A C•C pair occupies the 3b•3n position in the T. solenopsae Kt-23 structure. The cytosine nucleobases are coplanar, and well stacked into the helix. The C•C pair is linked by a single hydrogen bond between C3b N4 and C3n O2 (length 2.9 Å). It would be disrupted by the C3bG substitution, but not by C3bU since this preserves the C3n O2, yet both changes destabilize k-turn folding in the presence of Mg2+ ions. However, C3b makes an additional hydrogen bond from N4 to the nonbridging proR O of the phosphate linking 1n and 2n that requires that 3b be C and not U. The C1′-C1′ distance of the C•C pair (length 10.4 Å) is a close match to that of the A•A pair at the 2b•2n position (length 10.6 Å), which may provide some stabilization. The 4b•4n position is a standard Watson-Crick C-G base pair.
DISCUSSION
Kt-23 is a simple k-turn that frequently departs from the consensus sequence found for the motif. In particular, the majority of Kt-23 sequences do not have a guanine at the 2n position. Comparison of many Kt-23 sequences shows that the presence of adenine at the 2n position is extremely rare, and yet natural examples do exist. One such is the Kt-23 of T. solenopsae. We find that this sequence is only weakly folded by metal ions as an isolated duplex but that it is induced to fold either by the binding of L7Ae protein or by tertiary interactions within the SAM-I riboswitch. The latter allowed us to solve the X-ray crystal structure of this novel k-turn.
In most respects, the T. solenopsae Kt-23 structure is completely standard, both in global conformation and the formation of the normal hydrogen bonding patterns. The nucleobases of A1n and A2b have their usual position and orientation seen in k-turns (Fig. 4B), enabling them to make A-minor interactions with the minor groove of the C helix. The only significant difference is the presence of the adenine at the 2n position (Fig. 5A). The orientation of the two purine nucleobases is closely similar to that in a normal k-turn (cf. that in the natural SAM-I riboswitch seen in Fig. 5C, for example), but there are no direct hydrogen bonds formed. By comparison, when we made the G2nA substitution in the SAM-I riboswitch, the resulting A•A pair was linked by a single hydrogen bond donated from A2b N6 to A2n N3 of length 3.3 Å; the corresponding distance in the T. solenopsae Kt-23 structure is 5.0 Å. However, if A2n were replaced by G in the same position, the resulting exocyclic amine N2 would be able to hydrogen bond to A2n N7 with minimal movement. Interestingly, it seems that the hydrogen bond between A2b N6 to G2n N3 is not formed in many k-turns. For example, the corresponding N6-N3 distance in Kt-7 is 4.3 Å (Fig. 5D), and the average distance observed in the simple k-turns Kt-7, box C/D, U4snRNA, Kt-38, Kt-58, and SAM-I riboswitch is 3.7 ± 0.6 Å. The shortest of these, found in the U4 snRNA k-turn, is 3.1 Å, which is still a little longer than the optimal distance for hydrogen bonding. Perhaps the most extreme case of disruption of a 2b•2n pair was found recently in a complex between a k-turn and the bacterial L7Ae homolog YbxF, where A2b and G2n are neither base-paired nor even coplanar (Baird et al. 2012). In T. solenopsae Kt-23, because of the separation of the nucleobases, the C1′ to C1′ distance at the 2b•2n position is quite long at 10.6 Å, compared to 9.1 Å for the corresponding distance at the G•A pair at the 1b•1n position, and is close to that for a normal Watson-Crick base pair. Although the adenine nucleobases are relatively widely separated in the structure, each is linked to the backbone of the opposite strand by hydrogen bonding. Hydrogen bonds formed between A2b and the ribose and nucleobase of G-1n are a key part of the A-minor interactions with the C helix that probably contribute significantly to the stabilization of the k-turn conformation.
The structure of one other natural k-turn with an A•A pair at the 2b•2n position is known, in the 16S rRNA of T. thermophilus Kt-11. However, this is a complex k-turn with a convoluted connectivity resulting in a different orientation of the nucleobases. The resulting cis Hoogsteen–Watson-Crick base pair is connected by two hydrogen bonds, from A2n N6 to A2b N7 (length 3.1 Å) and from A2b N6 to A2n N1 (length 2.9 Å). This stands in marked structural contrast to the A•A pair of the T. solenopsae Kt-23 k-turn and all the simple k-turns.
The structure of the T. solenopsae Kt-23 k-turn shows how a rare variant of the consensus sequence can be tolerated within the structure, such that the global conformation is preserved with the retention of the majority of the key hydrogen bonding interactions. However, the poor folding induced by Mg2+ ions and the lower apparent affinity for L7Ae indicate a reduced stability of this sequence compared to standard k-turns such as Kt-7. Such differential stability between k-turns could be important in the biogenesis of large RNA-protein assemblies such as the ribosome, and the 2n position may have a major role in this discrimination.
MATERIALS AND METHODS
Bioinformatic methods
Sequences of rRNA from the small ribosomal subunit were obtained from the Comparative RNA Web Site (Cannone et al. 2002). The sequences of 6325 organisms were aligned by the k-turn nucleotides −3b through to 6b, and 6n to −3n. A consensus sequence for Kt-23 was drawn using WebLogo 3 (Crooks et al. 2004).
RNA synthesis and construction of k-turn species
Ribooligonucleotides were synthesized using t-BDMS phosphoramidite chemistry (Beaucage and Caruthers 1981), as described in Wilson et al. (2001). Fluorescein- and Cy3-conjugated oligonucleotides were attached at 5′ termini as phosphoramidites during synthesis as required. Oligoribonucleotides were deprotected in 25% ethanol/ammonia solution at 55°C for 2 h and evaporated to dryness. Oligoribonucleotides were redissolved in 300 μL 1 M tetrabutylammonium fluoride (Aldrich) in tetrahydrofuran to remove t-BDMS protecting groups and agitated at 20°C for 16 h prior to desalting by G25 Sephadex (NAP columns, Pharmacia) and ethanol precipitation. All oligonucleotides were purified by gel electrophoresis in polyacrylamide and recovered from gel fragments by electroelution or diffusion in buffer, followed by ethanol precipitation. Fluorescently labeled RNA was subjected to further purification by reversed-phase HPLC on a C18 column (ACE10, Advanced Chromatography Technologies), using an acetonitrile gradient with an aqueous phase of 100 mM triethylammonium acetate, pH 7.0. Duplex species were prepared by mixing equimolar quantities of the appropriate oligoribonucleotides and annealing them in 50 mM Tris-HCl (pH 7.5), 25 mM NaCl by slow cooling from 90°C to 4°C. They were purified by electrophoresis in 12% polyacrylamide under nondenaturing conditions and recovered by either electroelution or diffusion into buffer, followed by ethanol precipitation.
RNA synthesis of the SAM-I riboswitch for ITC and crystallography
The SAM-I riboswitch aptamer domain RNA was synthesized by in vitro transcription using T7 RNA polymerase and subsequently purified by polyacrylamide gel electrophoresis as described previously (Schroeder et al. 2011).
Expression and purification of A. fulgidus L7Ae
A pET28-b+ vector containing the gene for a hexahistidine-L7Ae fusion protein (supplied by Prof. A. Hüttenhoffer, Innsbruck) was transformed into Escherichia coli BL21-Gold (DE3) pLysS cells (Stratagene), and protein expression was induced by the addition of IPTG to 1 mM. The cells were shaken for 4 h at 37°C and harvested by centrifugation at 4000 rpm for 30 min at 4°C. The cells were immediately lysed in 20 mM phosphate buffer (pH 9.0), 0.5 M NaCl, 2.5 mM imidazole (buffer A), 5 mM MgCl2 containing 0.1 mg/mL lysozyme (Sigma) and Complete protease cocktail (Roche). The suspension was heated at 85°C to denature endogenous protein, and this was removed by centrifugation at 10,000 rpm for 30 min at 4°C. L7Ae was purified from the cleared supernatant by application to a Ni2+-chelated HiTrap column (GE Healthcare) installed on an Äkta purifier HPLC (GE Healthcare) using 20 mM phosphate buffer (pH 9.0), 500 mM NaCl (buffer A), and 20 mM phosphate buffer (pH 9.0), 500 mM NaCl, 1 M imidazole (buffer B). After the protein was bound to the column, it was washed with three column volumes of 20 mM phosphate buffer (pH 9.0), 2M NaCl (buffer N). The bound protein was then dissociated from the column by a linear 0–500 mM imidazole gradient, with the protein eluting at ∼200 mM imidazole. The protein was dialyzed into 20 mM phosphate buffer (pH 9.0) and applied to a HiTrap Heparin HP column (GE Healthcare) and eluted using a NaCl gradient; the protein eluted at 600 mM. This step was included to remove any RNA from the protein preparation. Pooled fractions were then dialyzed against 20 mM HEPES-KOH (pH 7.4), 150 mM KCl, 1.5 mM MgCl2 (buffer C) containing 5% v/v glycerol before incubation with bovine thrombin (Sigma) for 16 h at 4°C. After inactivation of the protease by heat treatment, L7Ae was concentrated and stored at −20°C in buffer C containing 40% v/v glycerol. Protein concentration was calculated from its absorbance at 280 nm, using ɛ = 6085 M−1 cm−1, and the protein concentration was verified using the Bradford assay (Bradford 1976), with a kit purchased from BioRad employing bovine serum albumin as a standard. The purified protein was analyzed by electrophoresis in polyacrylamide in the presence of SDS, alongside a mixture of proteins (2–212 kDa, New England BioLabs) to act as size standards.
Study of Mg2+- and L7Ae-induced folding by fluorescence resonance energy transfer
Absorption spectra were measured in 90 mM Tris-borate (pH 8.3) in 2-μL volumes using a Thermo Scientific NanoDrop 2000c spectrophotometer. Spectra were deconvoluted using a corresponding RNA species labeled only with Cy3 and fluorophore absorption ratios calculated using a MATLAB program. Fluorescence spectra were recorded in 90 mM Tris-borate (pH 8.3) at 4°C using an SLM-Aminco 8100 fluorimeter. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of FRET efficiency (EFRET) were measured using the acceptor normalization method (Clegg 1992) implemented in MATLAB. EFRET as a function of metal ion concentration was analyzed on the basis of a model in which the fraction of folded molecules corresponds to a simple two-state model for ion-induced folding, i.e.,
where E0 is the FRET efficiency of the RNA in the absence of added metal ions, ΔEFRET is the increase in FRET efficiency at saturating metal ion concentration, [Mg] is the prevailing Mg2+ ion concentration, KA is the apparent association constant for metal ion binding, and n is a Hill coefficient. Data were fitted to this equation by nonlinear regression. The metal ion concentration at which the transition is half complete is given by [Mg]1/2 = (1/KA)1/n.
The same RNA oligonucleotides as used in the Mg2+-induced folding were used for the L7Ae binding experiments, and FRET was measured and analyzed using the same approach. L7Ae was added from a stock solution to a solution of 2 nM solution of RNA. Apparent association constants (KA) were measured using a model of stoichiometric binding, i.e.,
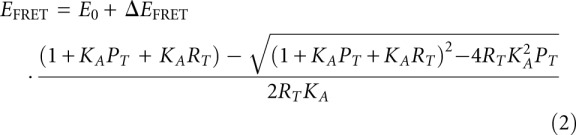 |
where E0 is the initial FRET efficiency in the absence of added protein, ΔEFRET is the full range of the change in FRET efficiency, and PT and RT are the total concentration of L7Ae and RNA, respectively. The dissociation constant Kd = KA−1.
The sequences used in the FRET analyses were as follows (written 5′ to 3′):
Kt-23 upper strand: Fluorescein-CCAGUCAGAUUUGAUGACCCCGGGAGAGG
Kt-23 lower strand: Cy3-CCUCUCCCGGGCAAGAAUCUGACUGG
Single nucleotide substitutions were introduced as required.
Isothermal titration calorimetry
Microcalorimetric measurements of SAM binding to the SAM-I riboswitch and variants were performed by ITC as described by Montange et al. (2010). The sequence of the SAM-I riboswitch was
5′-GGCUUAUCAAGAGAGGGCAAGAGACUGGCUUGAUGACCCCCGGCAACCAAAAAUGGUGCCAAUUCCUGCAGAGGAAACGUUGAAAGAUGAGCCA-3′
together with substitutions noted in the text. Calorimetric data were fitted to a single-site binding model, where possible, using MicroCal ORIGIN software. Individual heat changes ΔQ at constant pressure are given by
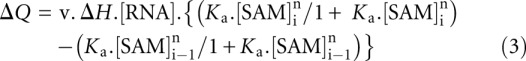 |
where ΔH is the change in enthalpy, v is the reaction volume, Ka is the association constant for SAM binding, and [SAM]i is the SAM concentration at the ith injection.
X-ray crystallography
The SAM-I riboswitch was crystallized using the hanging drop method. The crystal from which the data set used to solve the structure was obtained was grown by mixing 1 μL of 0.5 mM RNA in 40 mM Na-cacodylate (pH 7.0) plus 5 mM SAM with 1 μL of mother liquor. The mother liquor contained 40 mM Na-cacodylate (pH 7.0), 80 mM KCl, 30 mM BaCl2, 12 mM spermine-HCl, 8% (v/v) MPD. The crystal was then cryo-protected with mother liquor containing 30% (v/v) MPD and flash-frozen in liquid nitrogen.
Diffraction data were collected on beamline ID14-1 at the European Synchrotron Radiation Facility in Grenoble, France. The structure was solved by performing molecular replacement using MOLREP with the RNA plus SAM-ligand structure PDB entry 2GIS (Montange and Batey 2006) as a preliminary model. The model was subsequently refined using REFMAC5 (Vagin et al. 2004). Omit maps were calculated using PHENIX (Adams et al. 2010).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Drs. James Procter and Lin Huang for discussion and the Cancer Research UK, the Wellcome Trust, and the Human Frontiers Science Program for financial support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032409.112.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Zhang J, Hamma T, Ferré-D'Amaré AR 2012. YbxF and YlxQ are bacterial homologs of L7Ae, and bind K-turns but not K-loops. RNA 18: 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaucage SL, Caruthers MH 1981. Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett 22: 1859–1862 [Google Scholar]
- Blouin S, Lafontaine DA 2007. A loop–loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13: 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 15 doi: 10.1186/1471-2105-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg RM 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol 211: 353–388 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody TA, Melcher SE, Norman DG, Lilley DMJ 2004. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA 10: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12: 893–903 [DOI] [PubMed] [Google Scholar]
- Heppell B, Lafontaine DA 2008. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry 47: 1490–1499 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knell JD, Allen GE, Hazard EI 1977. Light and electron microscope study of Thelohania solenopsae n. sp. (Microsporida: Protozoa) in the red imported fire ant, Solenopsis invicta. J Invertebr Pathol 29: 192–200 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lilley DMJ 2007. The role of specific 2′-hydroxyl groups in the stabilization of the folded conformation of kink-turn RNA. RNA 13: 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, White SA, Williamson JR 1999. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat Struct Biol 6: 1139–1147 [DOI] [PubMed] [Google Scholar]
- Matsumura S, Ikawa Y, Inoue T 2003. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res 31: 5544–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Montange RK, Mondragon E, van Tyne D, Garst AD, Ceres P, Batey RT 2010. Discrimination between closely related cellular metabolites by the SAM-I riboswitch. J Mol Biol 396: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Zhang Y, Fenley MO, Li H 2004. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12: 807–818 [DOI] [PubMed] [Google Scholar]
- Schroeder KT, Lilley DMJ 2009. Ion-induced folding of a kink turn that departs from the conventional sequence. Nucleic Acids Res 37: 7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DM 2010. A structural database for k-turn motifs in RNA. RNA 16: 1463–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, Daldrop P, Lilley DMJ 2011. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure 19: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16: 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczak LB, Gabrielsen JS, Degregorio SJ, Strobel SA, Steitz JA 2005. Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein. RNA 11: 1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B, Lilley DM 2008. The importance of G · A hydrogen bonding in the metal ion- and protein-induced folding of a kink turn RNA. J Mol Biol 381: 431–442 [DOI] [PubMed] [Google Scholar]
- Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DMJ 2005. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA 11: 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN 2004. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60: 2184–2195 [DOI] [PubMed] [Google Scholar]
- Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 6: 1331–1342 [DOI] [PubMed] [Google Scholar]
- White SA, Hoeger M, Schweppe JJ, Shillingford A, Shipilov V, Zarutskie J 2004. Internal loop mutations in the ribosomal protein L30 binding site of the yeast L30 RNA transcript. RNA 10: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Zhao Z-Y, Maxwell K, Kontogiannis L, Lilley DMJ 2001. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry 40: 2291–2302 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Grundy FJ, Murphy BA, Henkin TM 2001. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak AK, Nottrott S, Kuhn-Holsken E, Schroder GF, Grubmuller H, Luhrmann R, Seidel CA, Oesterhelt F 2005. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA 11: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef OA, Terns RM, Terns MP 2007. Dynamic interactions within sub-complexes of the H/ACA pseudouridylation guide RNP. Nucleic Acids Res 35: 6196–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]