It has been over half a century since Salvador Luria experienced an epiphany while watching a (no doubt illegal) slot machine in a country club in Bloomington, IN (1). The experiment that resulted, the Luria–Delbrück fluctuation test, resides in the molecular biology wing of the Museum of Elegant Science along with the Hershey–Chase, the PaJaMo, the Crick et al. triplet code, and the Meselson–Stahl experiments. What endears the Luria–Delbrück experiment to scientists, particularly geneticists, is that it proved a hypothesis—that mutations arise at random during nonselective growth of cells—without any physical evidence whatsoever. Luria and Delbrück deduced that if a mutant happened to arise early during the growth of a culture, it would produce a large clone of identical descendants. Because such early mutants would be rare, the final numbers of mutant bacteria among a sufficiently large number of parallel cultures would have “a distribution with an abnormally high variance” (2). By showing that the variance was, indeed, far greater than the mean, Luria and Delbrück proved their case. But, what has continued to fascinate for over 50 years is the Luria–Delbrück distribution itself. For those of us who study spontaneous mutation, understanding how to derive mutation rates from fluctuation tests is something of an initiation rite (see ref. 3 for examples).
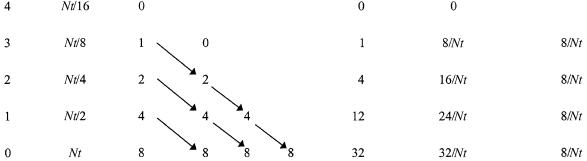
So, it comes as a shock to be told that the fluctuations intrinsic to the Luria–Delbrück experiment constitute not a brilliant way to determine mutation rates but a problem to be overcome (4). The theory underlying the solution to the problem is as follows (Table 1). After an exponentially growing population reaches a sufficient size (≫1/mutation rate per cell), the combination of new mutants plus the growth of preexisting ones results in a constant increase in the mutant fraction. The mutation rate is equal to this increase divided by some measure of time (conventionally cell generations) (2). Therefore, the simplest of all ways to determine a mutation rate is to measure the change in the mutant fraction in a growing population. However, by the time the population reaches the required size, mutations have already occurred and polluted the culture (the fluctuation effect), so that the signal-to-noise ratio is impossibly low. In the June 8, 1999 issue of the Proceedings, Bachl et al. (4) have solved the problem by eliminating preexisting (fluorescent) mutants with a cell sorter. The resulting cultures were nearly mutant free and then, during subsequent growth, accumulated new mutants at the predicted constant rate.
Table 1.
Bachl et al. (4) are interested in a fascinating mutational phenomenon, hypermutation of the Ig locus in activated B-lymphocytes. During B-cell maturation, the rearranged V(D)J region is subjected to a mutation rate some 105- to 106-fold higher than the rest of the genome. This hypermutation produces base-substitution mutations and requires both cis- and trans-acting elements. Bachl et al. placed a mutant nonfluorescent version of the green fluorescent protein (GFP) downstream of the thymidine kinase promoter on a plasmid bearing the large intron enhancer of the Ig heavy chain. When stably transformed into a pre-B cell line, the mutant GFP gene reverted at a high rate, yielding fluorescent cells. At the beginning of their mutation rate experiment, Bachl et al. eliminated preexisting fluorescent cells by fluorescence-activated cell sorting; thereafter, they detected new fluorescent mutants by flow cytometry.
Until now, the ability to eliminate preexisting mutants before measuring mutation rates has been restricted to a few special cases in which target genes can be selected both for and against (in genetic jargon, counterselectable markers). For example, mutations in the hypoxanthine-guanine phosphoribosyl transferase (hprt) locus on the single-copy X chromosome make cells sensitive to HAT medium (which contains hypoxantine, aminopterin, and thymidine) but resistant to 6-thioguanine. In experiments very similar to the ones described by Bachl et al. (4), Glaab and Tindall (5) determined the mutation rates of cancer-cell lines by measuring the fraction of cells resistant to 6-thioguanine in HAT-purged cultures. Reddy and Gowrishankar (6) performed a similar trick for Escherichia coli by engineering strains with conditional lethal mutants. For example, Lac+ cells carrying a temperature-sensitive galE mutation lysed at the nonpermissive temperature (because one product of β-galactosidase is galactose, and accumulation of UDP-galactose is toxic in the absence of the epimerase encoded by galE), but new Lac+ mutants could be selected on lactose medium at the permissive temperature. Obviously, the beauty of the technique pioneered by Bachl et al. is that it can be applied to any cell that can be subjected to (and survive) fluorescence-activated cell sorting.
The study by Bachl et al. (4) appears to be only the second time that GFP has been used as a mutational target. The previous study was of induced mutations in E. coli, and the target was a frameshifted GFP gene under control of the arabinose promoter carried on a ColE1 plasmid (7). Colonies of fluorescent revertants were detected with a hand-held UV lamp. These types of experiments are limited by the intrinsic “brightness” of the GFP protein and the level of its expression. Cariello et al. (7) used an enhanced GFP protein (GFPuv) but a very insensitive detection method, and only a small fraction of the plasmids in fluorescent colonies were, in fact, GFP+. So, it is not out of the question that this level of expression would allow single mutant bacteria to be detected, a necessity if spontaneous mutation rates in normal bacterial cells are to be measured. Bachl et al. give other possible uses of their system in cells and in whole animals (also see ref. 8). Thus, there appears to be a bright future for GFP in mutation research.
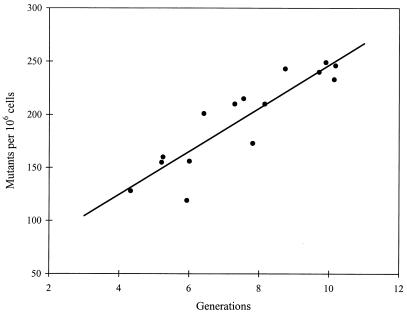
Finally, we can ask how well the experiment actually turned out. Very well, in fact. After an initial period to allow expression of nascent GFP+ mutants, the fluorescent mutant fraction increased linearly for 2 days. Overall, the variance in mutant fraction among five parallel cultures was not greatly different than the mean (a variance-to-mean ratio of one is indicative of the Poisson distribution). Bachl et al. (4) point out that the variance among the cell numbers in the five cultures was rather higher and suggest that the mutation rate may be time dependent instead of replication dependent. But this is unconvincing. The variances will depend on the sampling errors, and these were likely to have been very different for the total cell number, determined microscopically, and the mutant fraction, determined by flow cytometry. And, indeed, the mutant fraction is as strongly correlated with generations as with time (r2 = 0.8 vs. 0.9) (Fig. 1). From the regression in Fig. 1, the mutation rate is 2 per 105 cell generations, roughly equivalent to mutation rates at the hprt locus in some mismatch repair-deficient cancer cell lines (5). However, this is merely a coincidence, as mismatch repair appears not to play a direct role in somatic hypermutation, but only to modify the spectrum of mutations observed (9). The underlying mechanism by which the mutations are produced in the Ig variable region remains a mystery.
Figure 1.
The mutant fraction vs. the number of cell generations from the data of Bachl et al. (4). The line is the least-squares regression: y = 20x + 43; r2 = 0.81; P ≪ 0.001.
ABBREVIATION
- GFP
green fluorescent protein
Footnotes
The companion to this Commentary begins on page 6847 in issue 12 of volume 96.
References
- 1.Luria S E. In: Phage and the Origins of Molecular Biology. Cairns J, Stent G S, Watson J D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1966. pp. 173–179. [Google Scholar]
- 2.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosche, W. A. & Foster, P. L. (1999) Methods, in press.
- 4.Bachl J, Dessing M, Olsson C, von Borstel R C, Steinberg C. Proc Natl Acad Sci USA. 1999;96:6847–6849. doi: 10.1073/pnas.96.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaab W E, Tindall K R. Carcinogenesis. 1997;18:1–8. doi: 10.1093/carcin/18.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Reddy M, Gowrishankar J. Genetics. 1997;147:991–1001. doi: 10.1093/genetics/147.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cariello N F, Narayanan S, Kwanyuen P, Muth H, Casey W M. Mutat Res. 1998;414:95–105. doi: 10.1016/s1383-5718(98)00043-6. [DOI] [PubMed] [Google Scholar]
- 8.Bachl J, Olsson C. Eur J Immunol. 1999;29:1383–1389. doi: 10.1002/(SICI)1521-4141(199904)29:04<1383::AID-IMMU1383>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 9.Wiesendanger M, Scharff M D, Edelmann W. Cell. 1998;94:415–416. doi: 10.1016/s0092-8674(00)81581-0. [DOI] [PubMed] [Google Scholar]
- 10.Luria S E. Cold Spring Harbor Symp Quant Biol. 1951;16:463–470. doi: 10.1101/sqb.1951.016.01.033. [DOI] [PubMed] [Google Scholar]