Summary
Sophisticated mechanisms are employed to facilitate information exchange between interfacing bacteria. A type VI secretion system (T6SS) of Pseudomonas aeruginosa was shown to deliver cell wall-targeting effectors to neighboring cells. However, the generality of bacteriolytic effectors, and moreover, of antibacterial T6S, remained unknown. Using parameters derived from experimentally validated bacterial T6SS effectors and informatics, we identified a phylogenetically disperse superfamily of T6SS-associated peptidoglycan-degrading effectors. The effectors separate into four families composed of peptidoglycan amidase enzymes of differing specificities. Effectors strictly co-occur with cognate immunity proteins, indicating that self-intoxication is a general property of antibacterial T6SSs and effector delivery by the system exerts a strong selective pressure in nature. The presence of antibacterial effectors in a plethora of organisms, including many that inhabit or infect polymicrobial niches in the human body, suggests that the system could mediate interbacterial interactions of both environmental and clinical significance.
Introduction
Bacteria are under immense competitive pressure for limited resources, which has shaped the evolution of a number of antagonistic antibacterial pathways (Hayes et al., 2010; Konovalova and Sogaard-Andersen, 2011; Rendueles and Ghigo, 2012). These pathways often target conserved processes that are not easily modified. One such target is bacterial cell wall peptidoglycan, a structure that is essential for maintaining osmotic stability and cell shape (Vollmer et al., 2008). Despite its many variations, the fundamental structure of peptidoglycan is highly conserved throughout the domain Bacteria. The Gram-negative cell wall is sequestered from the extracellular milieu by an outer membrane; nevertheless, some cell wall-targeting molecules can overcome this barrier. For example colicin M, which degrades peptidoglycan precursors, or pesticin, which degrades the mature sacculus (El Ghachi et al., 2006; Vollmer et al., 1997).
The type VI secretion system (T6SS) of Gram-negative bacteria is a contact-dependent protein translocation apparatus that resembles an inverted bacteriophage puncturing device (Jani and Cotter, 2010; Records, 2011; Schwarz et al., 2010a; Veesler and Cambillau, 2011). The system has recently been demonstrated to act as a pathway for the delivery of antagonistic effectors to adjacent bacteria (Hood et al., 2010; Russell et al., 2011). This finding is consistent with the structural homology of the system to bacteriophage, as both effector delivery and phage entry appear to occur in an analogous fashion. Proteins delivered to bacteria by this system include Tse1-3 (type VI secretion exported 1–3), substrates of the hemolysin co-regulated protein secretion island I-encoded T6SS of Pseudomonas aeruginosa (H1-T6SS) (Hood et al., 2010). Tse1 and Tse3 are bacteriolytic enzymes that degrade the peptidoglycan of recipient bacteria, whereas Tse2 is bacteriostatic through an unknown mechanism (Russell et al., 2011).
Unlike other peptidoglycan-targeting molecules that affect Gram-negative bacteria, Tse1 and Tse3 have no intrinsic means of translocating the outer membrane. These effectors instead rely on the T6S apparatus for delivery to the periplasmic compartment of recipient cells. Within the periplasm, Tse1, as a cell wall amidase, cleaves the γ-D-glutamyl-L-meso-diaminopimelic acid bond, while Tse3, as a muramidase, hydrolyzes the β(1,4) linkage between N-acetylmuramic acid and N-acetyglucosamine (Russell et al., 2011). Both Tse1 and Tse3 appear to transit through the T6S apparatus in one step, bypassing the periplasmic space of the donor cell. This prevents the producing cell from intoxicating itself with Tse1 or Tse3 in transit. However, the H1-T6SS of P. aeruginosa can target neighboring, clonal, P. aeruginosa cells (Hood et al., 2010). Due to the ability of the H1-T6SS to engage in self-targeting interactions, P. aeruginosa cells can intoxicate one-another via T6SS-delivered effectors. This intercellular self-intoxication is overcome through the production of specific cognate immunity proteins, Tsi1 and Tsi3, which reside in the periplasmic compartment and protect against the activities of Tse1 and Tse3, respectively (Li et al., 2012; Russell et al., 2011).
Thus far many of our mechanistic insights into bacteria-targeting T6S derive from the H1-T6SS and its substrates. Without further understanding obtained from the identification of other T6SS substrates, it remains unclear whether these insights are generally applicable. For example, our laboratory previously demonstrated that T6SS-1 of the soil saprophyte Burkholderia thailandensis provides cell contact-dependent fitness to the organism during growth competition assays against certain Gram-negative bacteria (Schwarz et al., 2010b). The specific effector proteins utilized by T6SS-1, and for several other demonstrated antibacterial T6SSs, have yet to be defined (MacIntyre et al., 2010; Murdoch et al., 2011). Therefore, a major bottleneck in the study of bacteria-targeting T6S is the identification of the substrates that mediate T6S-dependent effects.
Both bioinformatic and traditional experimental approaches have been fruitful in the identification of proteins exported by alternative bacterial secretion pathways. However, the application of such approaches to the identification of T6S substrates is complicated by the fact that determinants for passage through the T6S apparatus are not currently apparent and the pathway is repressed under standard laboratory conditions in many organisms (Bernard et al., 2010; Silverman et al., 2011). Here we sought to develop a general method for the identification of antibacterial T6S substrates. We initiated our study by defining substrates of B. thailandensis T6SS-1 using mass spectrometry (MS). Analysis of these data revealed characteristics shared between a B. thailandensis T6SS-1 substrate and proteins previously found to transit the H1-T6SS of P. aeruginosa. These commonalities were exploited to develop a heuristic informatic approach for the large-scale identification of T6S substrates. With this approach, we discovered a phylogenetically diverse and broadly distributed superfamily of T6S effectors. Our findings reveal a general paradigm describing the mechanism of interbacterial T6S, and suggest that it plays a broad role in shaping clinically- and environmentally-relevant microbial communities.
Results
Identification and genomic analysis of T6SS-1 substrates
In an effort to deepen our understanding of interbacterial T6S effector function, we sought to define the substrates of B. thailandensis T6SS-1 (Schwarz et al., 2010b). To this end, we first established a reference secretome using wild-type B. thailandensis. Using MS, we identified a total of 232 proteins in the supernatant fraction of log phase cultures. Of these, 114 were present in each of the three replicates conducted (Table S1). This variability stems from low-abundance proteins; the average spectral count of variably present proteins was < 20% (8 spectral counts (SC)) of those detected in all replicates (43 SC). Based on this correlation, we did not include variably present proteins in further analyses.
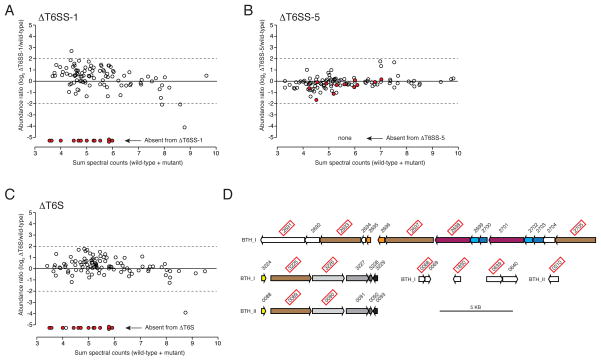
Next we determined the secretome of B. thailandensis ΔT6SS -1 and compared it to the wild-type reference. This analysis identified 13 proteins reproducibly absent from the ΔT6SS-1 secretome (Figure 1A and Table S1). The absence of these proteins appeared specific, as their average abundance was high (34 SC) and the secretion of the remaining reference secretome proteins was largely unaffected in the mutant strain. Our observations also appeared relevant to T6S, as only two of the proteins are predicted substrates of alternative secretory pathways and four are VgrG homologs (Table S1). To determine the specificity of these proteins for export by T6SS-1, we also measured the secretome of a B. thailandensis strain with an inactivating mutation in another of its five T6SSs. We chose T6SS-5 for this experiment, as this system is the only other to have been linked to a phenotype – attenuated virulence in mammalian infection models – in B. thailandensis and closely related organisms (Pilatz et al., 2006; Schell et al., 2007; Schwarz et al., 2010b). Notably, none of the 13 proteins dependent upon T6SS-1 for export were absent or found in significantly lower abundance in the ΔT6SS-5 secretome (Figure 1B). Finally, the secretome of a strain inactivated in all five T6SSs (ΔT6S) lacked only one protein in addition to those that were T6SS-1-dependent (Figure 1C). From these data, we conclude that T6SS-1 is specifically required for the export of at least 13 proteins from B. thailandensis. Furthermore, our analyses demonstrate that the remaining B. thailandensis T6SSs may be quiescent with regard to substrate export under in vitro conditions.
Figure 1. Identification of B. thailandensis T6SS-1 substrates.
(A–C) Comparison of individual protein abundance in wild-type versus ΔT6SS-1 (A), ΔT6SS-5 (B), and ΔT6S (C) B. thailandensis secretomes. Proteins absent from ΔT6SS -1 are indicated in each panel by filled red spheres.
(D) Organization of genes encoding B. thailandensis proteins (boxed red) that specifically require T6SS-1 for export. Color indicates homology.
Substrate export via the T6SS is thought to occur in a Sec-independent fashion. Thus, the lack of two proteins, BTH_II0639 and BTH_I2723, containing predicted N-terminal signal peptides from the ΔT6SS-1 secretome is most likely explained by pleiotropic effects rather than their direct reliance upon T6SS-1 for export. In light of these data, we do not classify these proteins as putative T6SS-1 substrates in this report.
The genes encoding the remaining 11 proteins requiring B. thailandensis T6SS-1 for export clustered non-randomly in the genome. Five are found within a single gene cluster, and four others are distributed among the two copies of a region of the genome that apparently underwent a recent duplication event (Figure 1D). This duplication event gave rise to two vgrG genes encoding identical VgrG proteins, making it impossible to determine the relative contributions of these loci to the secreted protein we observe using MS (Table S1).
Several of the putative T6SS-1 substrates have predicted functions consistent with the known role of T6SS-1 in mediating interbacterial interactions. Structure-based algorithms predict extensive structural similarity between two of the proteins, BTH_II0310 and BTH_I2691, and bacteriocins LlpA and colicin Ia, respectively (Cascales et al., 2007; Parret et al., 2003). Although to our knowledge there is no prior experimental evidence for the secretion of a bacteriocin-like protein by a T6SS, predicted bacteriocins have been found in association with the T6SS (Blondel et al., 2009). Particularly germane to our current study, we found that another putative substrate of T6SS-1, BTH_I0068, has predicted structural homology with peptidoglycan amidase enzymes (Firczuk and Bochtler, 2007). This is of interest, as our laboratory previously showed Tse1, an antibacterial effector of the H1-T6SS of P. aeruginosa, acts as a peptidoglycan-degrading amidase (Russell et al., 2011). Given the genetic linkage and predicted antibacterial properties of many of the putative B. thailandensis substrates, it is likely that these proteins are extracellular components or effectors of T6SS-1. In our effort to better understand the significance of cell wall targeting T6S effectors, we decided to focus further study on BTH_I0068.
BTH_I0068–BTH_I0069 are a T6S amidase effector–immunity pair
BTH_I0068 has a high degree of predicted structural similarity with the CHAP family of peptidoglycan amidases, including catalytic cysteine and histidine residues (Figure 2A) (Bateman and Rawlings, 2003; Rigden et al., 2003). These residues are also conserved in Tse1, which has predicted structural homology with related group of peptidoglycan amidases (NlpC/P60) (Firczuk and Bochtler, 2007). We observed additional similarities between BTH_I0068 and Tse1 beyond predicted catalytic activity. Like Tse1, BTH_I0068 lacks a Sec signal peptide despite its predicted function in the periplasm. Also similar to Tse1, the Burkholderia protein lacks regulatory domains often associated with peptidoglycan amidases. Finally, both proteins are encoded in predicted bicistrons with a gene encoding a periplasmic protein (Figure 1D). In the case of Tse1, this periplasmic protein is Tsi1, an immunity protein that specifically prevents Tse1-dependent intoxication (Russell et al., 2011). Despite their lack of significant primary sequence homology, these observations led us to hypothesize that BTH_I0068–BTH_I0069 constitutes a T6S effector–immunity (EI) pair analogous to Tse1–Tsi1.
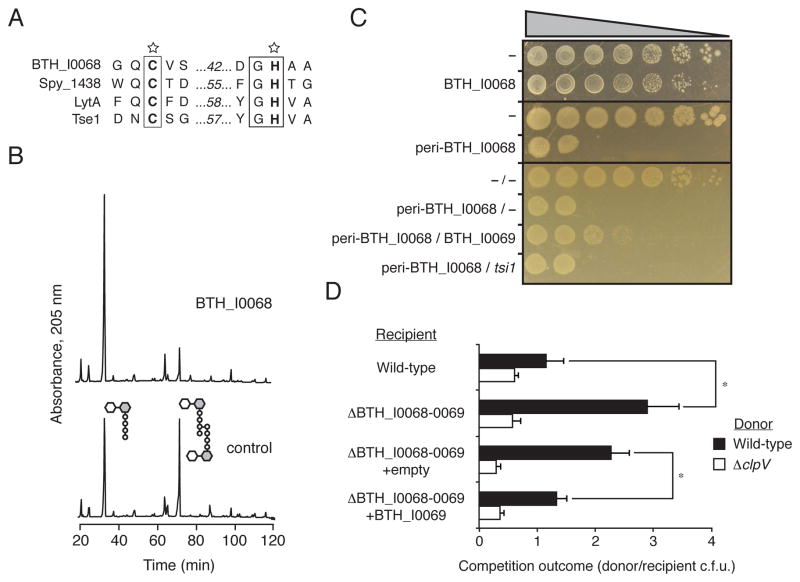
Figure 2. B. thailandensis BTH_I0068 and BTH_I0069 are a T6S amidase effector–immunity pair.
(A) Sequence alignment of conserved catalytic motifs share between BTH_I0068 and characterized cell wall amidase enzymes. SWISS-PROT entry names for the proteins shown are: BTH_I0068 (Q2T2K7_BURTA), Spy_1438 (Q99Z24_STRP1), LytA (LYTA_BACSU), and Tse1 (Q9I2Q1_PSEAE).
(B) BTH_I0068 acts as a peptidoglycan amidase with specificity toward the m-DAP-D-alanine DD-bond. Partial HPLC chromatograms of sodium borohydride-reduced soluble E. coli peptidoglycan products resulting from digestion with BTH_I0068 and subsequent cleavage with cellosyl.
(C) Growth of E. coli harboring one (top panels) or two (bottom panel) vectors expressing the indicated genes. A dash indicates the empty vector. From left to right are increasing serial ten-fold dilutions. Expression data for this experiment are shown in Figure S2.
(D) BTH_I0068 and BTH_I0069 act between cells as a T6S-dependent toxin–immunity pair between B. thailandensis cells. Growth competition assays between the indicated B. thailandensis donor and recipient strains under T6S-conducive conditions. The ΔclpV1 strain is a T6S-deficient control. Asterisks mark significantly different competition outcomes (p < 0.01). Error bars represent ± s.d. n=6.
To determine the extent of functional similarity between BTH_I0068 and Tse1, we first sought to establish whether BTH_I0068 displays amidase activity. To test this, we incubated purified BTH_I0068 with peptidoglycan sacculi prepared from E. coli. Subsequent muramidase digestion followed by HPLC and MS analysis of soluble products indicated that BTH_I0068 cleaves peptidoglycan tetrapeptide-tetrapeptide crosslinks at the D,D amide bond between meso-diaminopimelic acid (mDAP) and D-alanine (Figure 2B). As certain species have higher degrees of the nascent crosslink form (tetrapeptide–pentapeptide), we also determined whether BTH_I0068 is able to process peptidoglycan crosslinks in this configuration. The activity of BTH_I0068 was not affected by the presence of the additional D-alanine residue, suggesting that BTH_I0068 could act broadly as an amidase to cleave peptidoglycan crosslinks (Figure S1).
Overexpression of typical cell wall-degrading amidases is deleterious to E. coli. However, BTH_I0068 lacks a signal peptide and thus outside of the T6SS should be toxic only when directed to the periplasm through the addition of an N-terminal signal peptide. As predicted for a bacteriolytic T6S substrate analogous to Tse1, E. coli viability was reduced only when BTH_I0068 was artificially targeted to the periplasm (Figures 2C and Figure S2).
Next we asked whether the predicted periplasmic protein encoded adjacent to BTH_I0068, BTH_I0069, functions as a cognate immunity protein – analogous to Tsi1. Co-expression experiments in E. coli showed that BTH_I0069 provides significant rescue to cells expressing BTH_I0068, whereas Tsi1 failed to provide such rescue (Figure 2C). Taken together, we conclude that BTH_I0068–BTH_I0069 define an amidase–immunity pair analogous – and not homologous – to P. aeruginosa Tse1–Tsi1.
If BTH_I0068, like Tse1, is an effector, we reasoned that B. thailandensis should be able to intoxicate neighboring bacteria with the protein in a T6SS-dependent manner. We tested this by performing growth competition assays under cell contact-promoting conditions. These assays revealed that B. thailandensis recipient cells bearing a deletion of the BTH_I0068–BTH_I0069 bicistron display a fitness defect relative to wild-type donor strains (Figure 2D). Recipient fitness was rescued either through inactivation of T6SS-1 in the donor strain or by expression of BTH_I0069 in the recipient cell. Together with our in vitro studies, these data demonstrate that BTH_I0068–BTH_I0069 constitute a T6S EI pair. Furthermore, these data suggest the model for T6S interbacterial effector targeting derived from studies of P. aeruginosaH1 -T6SS is applicable to B. thailandensis T6SS-1. We propose the name Tae2 (type VI amidase effector) for BTH_I0068 and Tai2 (type VI amidase immunity) for BTH_I0069.
Identification of a T6S EI pair superfamily
All currently defined T6SS EI pairs lack identifiable primary sequence homology. In spite of an inability to link EI pairs phylogenetically, certain physical and contextual properties appear intrinsic to their function. We hypothesized that parameters describing these properties could be used in a heuristic informatic method for the de novo prediction of EI pairs from genomic sequences. Such an approach was previously successful in the identification of type III secretion effectors from the Pseudomonas syringae DC3000 genome (Collmer et al., 2002; Petnicki-Ocwieja et al., 2002).
We initiated our search by developing a comprehensive set of parameters describing characterized T6S EI pairs. These include parameters common to all pairs (Figure 3A, orange), those indicative of periplasmically-targeted pairs (Figure 3A, blue), and those specific to amidase pairs (Figure 3A, brown). Individually, the parameters describing T6S EI pairs are weak in their discriminatory capacity. However, we reasoned that by restricting our search to amidase EI pairs, thus allowing the application of all parameters serially, we could achieve enrichment sufficient for computationally intensive secondary analyses.
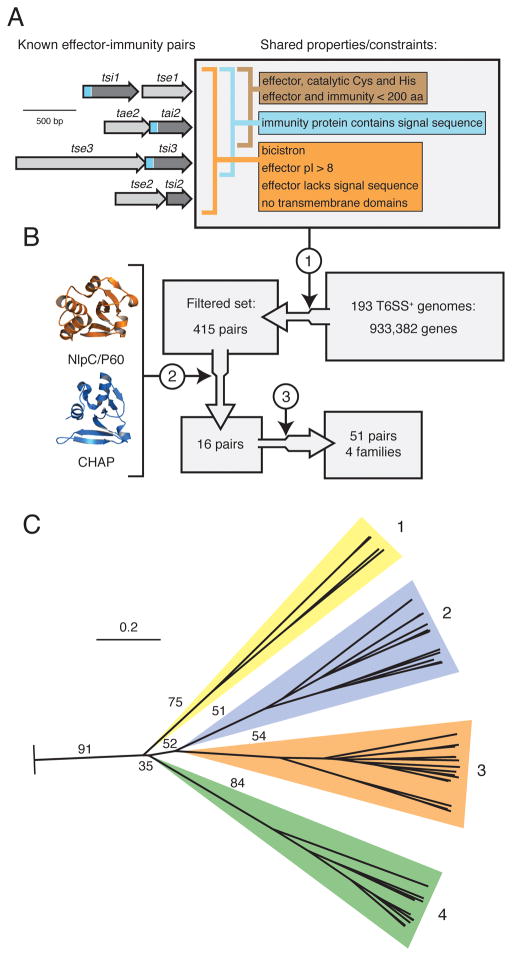
Figure 3. Identification of a T6S effector superfamily.
(A) Overview of shared T6S EI pair properties. Depicted at left are the four bicistrons encoding all characterized EI pairs (this study and (Hood et al., 2010; Russell et al., 2011)). Properties are divided among those shared by all EI pairs (orange), periplasmically-targeted pairs (blue), and amidase pairs (brown). Sequences encoding signal peptides within immunity proteins are represented in blue.
(B) Schematic of informatic effector identification workflow. Key steps in the workflow are indicated: 1) filter by constraints depicted in (A), 2) application of structure prediction criteria, 3) expansion by homology searching of the non-redundant nucleotide database.
(C) Phylogenetic tree of T6S effectors identified by the methods depicted in A and B. The tree was based on alignment of catalytic motifs (Figure S3). Effectors distribute into four branches, referred to as Families 1–4. The background colors assigned to the families are used henceforth. Critical boostrap values are indicated (n = 100). The tree was rooted using equivalent catalytic motifs of papain-like fold enzymes (Pfam clan, CL0125). Scale bar indicates evolutionary distance in amino acid substitutions per site.
To identify T6S amidase EI pairs, we applied our parameters as constraints to a set of 193 phylogenetically diverse organisms encoding T6SSs (Figure 3B). This procedure provided a group of 419 candidate EI pairs, a 99.9% reduction in the sample set. The extent of this reduction enabled detailed examination of each EI pair, including structure prediction analysis – already established as a valuable means for identifying sequence divergent amidase effectors (Russell et al., 2011). This second step further reduced our set of candidate EI pairs to 16 non-orthologous pairs (Table S2). Based on genomic analyses, and biochemical and genetic experiments discussed in subsequent sections, we considered the pairs in this group as our set of high-confidence peptidoglycan amidase effectors. In the last step of our pipeline, we expanded the number of EI amidase pairs to 51 by searching for homologous sequences in the entire non-redundant sequence database (Figure 3B).
Although our search for EI pair homologs was conducted in an unbiased dataset, with only one exception, all homologous pairs we identified reside in Gram-negative organisms encoding T6SSs. This finding is unlikely to occur through chance, as even when we consider the phylogenetic group most enriched in T6S, the Proteobacteria, only a minority possess the system (17.6%, p < 0.0001) (Boyer et al., 2009). Vertical inheritance is one factor that clearly contributes to the non-random association between EI pairs and the T6SS. However, we note that many homologous EI pairs are found both in distantly related organisms and in species with close-relatives that lack T6S. The only organism lacking a T6SS that we found to contain an EI pair is Salmonella enterica subspecies enterica serovar Paratyphi B (S. Paratyphi B); however, Blondel and colleagues noted the loss of T6S in this organism was a recent event (Blondel et al., 2009). Importantly, all effector homologs in our expanded set contain sequence motifs characteristic of CHAP and NlpC/P60 amidase enzymes (Anantharaman and Aravind, 2003; Bateman and Rawlings, 2003).
EI pairs segregate into four families
The 51 putative effectors discovered in our screen segregate into four families based on overall primary sequence homology (Figure S3). These families are disparate, confounding attempts to place them in a phylogenetic context using full-length alignments. However, we did identify two relatively conserved motifs surrounding predicted catalytic residues (Figure S3). Phylogenetic analyses based on these regions, which owing to their involvement in catalysis are subject to less genetic drift, yielded a tree with family assignments matching those derived from full-length sequence homology (Figure 3C).
The majority of the immunity proteins identified in our search mapped to EI bicistrons. Among these, homologous immunity proteins distribute with homologous effectors; therefore, we assigned immunity proteins into families reflecting their cognate effectors. Divergence within immunity protein families was considerably higher than that found within the effector families (Figure S4). This is not surprising, as functional constraints on immunity proteins (effector binding) are likely less restrictive than those on the effectors (immunity binding and catalysis). The remaining 23 immunity proteins group into two categories: those associated with EI pairs (3) and those coded for by orphan immunity genes (20). Immunity genes associated with EI pairs appear to have arisen through gene duplication events, as in each instance they belong to the same immunity family as the neighboring immunity gene. In summary, we have found that the putative T6S effectors identified in our screen distribute coincident with cognate immunity proteins into four phylogenetically discernable families. As expected based on results garnered from detailed studies in P. aeruginosa and B. thailandensis, this observation strongly implies coevolution of effector and immunity proteins (Russell et al., 2011).
Characterization of EI families
An important feature of the informatic approach we utilized for defining T6S effectors is its lack of reliance on primary sequence homology. While this non-sequence-biased approach allowed the discovery of four highly divergent families, it also increases the probability that one or more of these might include either peptidoglycan amidase effectors with differing catalytic specificity, effectors with unexpected activity, or false-positives not representing effector proteins.
To test whether putative effector families 3 and 4 include cell wall amidase enzymes, we purified one member from each family and ascertained its activity towards E. coli peptidoglycan sacculi. Similar to Tae2, the family 3 enzyme from S. Typhi (Tae3TY) hydrolyses DD-crosslinks between D-mDAP and D-alanine (Figure 4A). In contrast, the family 4 enzyme from S. Typhimurium (Tae4TM) hydrolyzes peptide crosslinks at the γ-D-glutamyl-mDAP LD-bond, like Tse1 (Figure 4A). However, unlike Tse1, Tae4TM cleaves acceptor and non-crosslinked tetrapeptide stems, and does not cleave the donor peptide stem (Figure 4B). While pentapeptide-enriched peptidoglycan is readily degraded by Tae3TY, it is a poor substrate for Tae4TM.
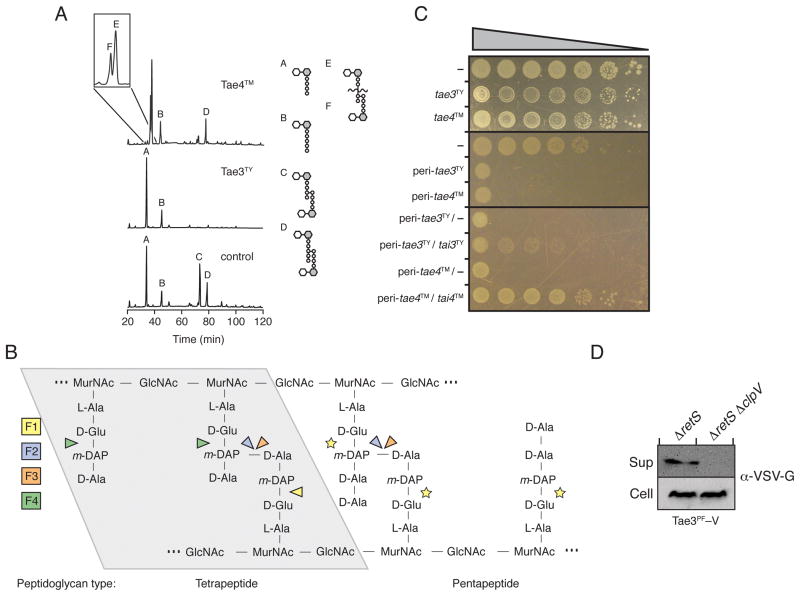
Figure 4. Representatives of Families 3 and 4 are T6S amidase EI pairs.
(A) Tae3TY and Tae4TM are peptidoglycan amidases with specificity for the m-DAP-D-alanine DD-bond and the γ-D-glutamyl-L-m-DAP bond, respectively. Partial HPLC chromatograms of sodium borohydride-reduced soluble E. coli peptidoglycan products resulting from digestion with Tae3TY or Tae4TM and subsequent cleavage with cellosyl. The control sample was digested with cellosyl alone.
(B) Simplified representation of Gram-negative peptidoglycan showing cleavage sites of effector families 1–4 (F1-4) based on HPLC data. Cleavage specificity on peptidoglycan with tetrapeptide (left) and pentapeptide (right) stems are depicted. Tse1 activity against pentapeptide-rich peptidoglycan has not been tested, as indicated by yellow stars. Abbreviations: GlcNAc, N-acetyl-glucosamine, MurNAc, N-acetyl-muramic acid.
(C) Tae3TY and Tae4TM are toxic in the periplasm and this toxicity is rescued specifically by cognate immunity proteins, Tai3TY and Tai4TM, respectively. Growth of E. coli harboring one (top panels) or two (bottom panel) vectors expressing the indicated genes. A dash indicates the empty vector. From left to right are increasing serial ten-fold dilutions. Expression data for this experiment are shown in Figure S2.
(D) Tae3PF is secreted in a T6SS-dependent manner. Western blot analysis of supernatant (Sup) and cell-associated (Cell) fractions of the indicated P. fluorescens strains expressing vesicular stomatitis virus glycoprotein (VSV-G) tagged Tae3PF (Tae3PF –V).
Having demonstrated that Tae3TY and Tae4TM are amidases, we next investigated whether these enzymes and their corresponding immunity proteins fit additional characteristics of T6S EI proteins that have emerged from studies of Tse1, Tse3, Tae2, and cognate immunity proteins. Consistent with previously validated effectors, we found that Tae3TY and Tae4TM are highly toxic when artificially directed to the periplasm of E. coli, but not when expressed in their native form (Figure 4C and Figure S2). Also, we observed that the periplasmic proteins, Tai3TY and Tai4TM, encoded adjacent to tae3TY and tae4TM, respectively, could rescue this toxicity.
As a final means of functionally validating the EI pairs identified by our heuristic approach, we tested the capacity of one informatically-identified effector to serve as a T6S substrate. Since most T6SSs are repressed under in vitro cultivation condition and there is as yet no general means of T6S activation, we restricted our efforts to systems with known regulation. In Pseudomonads, certain T6SSs are posttranscriptionally regulated by the Gac/Rsm pathway (Brencic and Lory, 2009; Hassan et al., 2010; Lapouge et al., 2008; Workentine et al., 2009). This pathway is modulated by two hybrid sensor kinases, LadS and RetS, which activate and repress T6S, respectively. In P. aeruginosa, deletion of retS leads to constitutive effector export by the H1-T6SS (Hood et al., 2010).
P. fluorescensPf -5 is closely related to P. aeruginosa and encodes a single T6SS that is regulated by the Gac/Rsm pathway (Hassan et al., 2010). Despite these similarities, P. fluorescens lacks homologs of H1-T6SS substrates, Tse1-3. Interestingly, our screen identified a family 3 EI pair in this organism (PFL_5498-PFL_5499, Tae3PF-Tai3PF), a family not represented in P. aeruginosa (Figures S3 and S4). While wild-type P. fluorescens did not secrete detectable amounts of Tae3PF, anin -frame deletion of retS in this organism resulted in constitutive secretion of the putative effector (Figure 4D, data not shown). In order to determine if Tae3PF export occurs in a T6S-dependent manner, we introduced an in-frame deletion of clpV into the P. fluorescens ΔretS background. This deletion abrogated secretion, demonstrating that Tae3PF is a substrate of the P. fluorescens T6SS. In total, these data strongly suggest that the proteins identified in our screen include four evolutionarily distinct amidase families that act, along with their cognate immunity proteins, as T6S EI pairs.
EI diversity reflects function
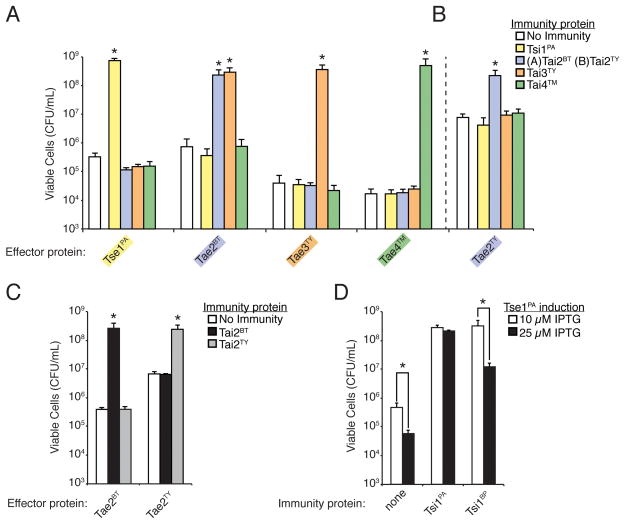
Immunity proteins from bacteria-targeting pathways such as bacteriocins and CDI systems tend to provide specific protection against only their cognate toxins (Aoki et al., 2010; Papadakos et al., 2011; Riley and Wertz, 2002). If immunity to effector Families 1–4 behaves in an analogous fashion we would expect that there would not be cross-complementation of immunity proteins between families, even for those effector families with overlapping enzymatic activity. When we test all combinations of confirmed EI pairs we observe that contrary to this model, one of the immunity proteins can provide significant protection against two effector families (Figure 5A and Figure S2). Specifically, the Tai3TY immunity protein protects against the B. thailandensis Tae2 (Tae2BT) effector in addition to its cognate Tae3TY effector. The Tai2BT immunity protein does not protect against the Tae3TY effector, indicating that immunity does not hold for the inverse configuration.
Figure 5. Immunity proteins display varying non-cognate effector neutralization.
(A) Tai3TY is an exception to a simple cognate effector–immunity protection model. All panels in this figure show the growth of E. coli harboring vectors co-expressing the indicated effector and immunity proteins. Immunity proteins were induced identically in all panels. Error bars represent ± s.d. (n=3). Expression data for all experiments are provided in Figure S2. Asterisks in (A), (B), and (C) indicate immunity proteins that provided significant protection above the empty vector control (p < 0.05).
(B) The immunity provided by Tai3TY against Tae2BT does not extend to all family 2 effectors. Data demonstrating the catalytic activity of Tae2TY on peptidoglycan are shown in Figure S5.
(C, D) Effector proteins of the same family are not always recognized by all immunity proteins of that family. (C) Co-expression of either Tae2BT or Tae2TY with Tai2BT or Tai2TY. (D) Co-expression of Tse1PA with Tsi1PA or Tsi1BP with either lower (10μM IPTG) or higher (25μM IPTG) induction of Tse1PA. Asterisks denote instances in which immunity proteins provided significantly lower protection against Tse1PA at higher induction levels of the effector (p < 0.05).
As both Tae2BT and Tae3TY have identical activity, Tai3TY could be capable of broadly neutralizing all DD-endopeptidases. To examine this possibility we used another Tae2 homolog present in S. Typhi (Tae2TY), which we confirmed to display D,D-endopeptidase activity (Figure S5). Tai3TY is unable to rescue E. coli expressing periplasmic Tae2TY, demonstrating that cross-family immunity between T6SS effector families can be specific (Figure 5B and Figure S2). The non-overlapping immunity spectra provided by Tai2TY and Tai3TY is consistent with the presence of both genes in S. Typhi; if Tai3TY provided immunity to both Tae2TYand Tae3TY there would be no pressure to retain Tai2TY. Furthermore, the finding that immunity to effectors is specific demonstrates that even effector families that are identical in catalytic activity are not redundant in function.
As the differences between effector families are adaptive, we sought to determine whether the variation within effector families also represents functional diversity. Given the observation that Tai3TY provides immunity differentially to Tae2BT and Tae2TY we began by testing whether the cognate immunity proteins of these two effector homologs have the capacity – unlike Tai3TY – to neutralize the toxic activity of both effectors. Our data show that Tai2TY does not provide immunity to Tae2BT, and that Tai2BT also does not protect against Tae2TY (Figure 5C and Figure S2). This demonstrates that even though Tae2TY and Tae2BT are homologs, their neutralization pattern with respect to varying immunity proteins differs. However, we did observe instances of cross-reactivity of cognate immunity proteins between effectors within the same family. Both Tsi1 from P. aeruginosa (Tsi1PA) and Burkholdeira phytofirmans Tsi1 (Tsi1BP) can rescue cells from the toxicity of Tse1PA, albeit rescue by the latter is less efficient (Figure 5D and Figure S2).
We have found that the diversity in effector sequence is adaptive, not only with regard to catalytic activity, but also in terms of immunity protein recognition. This observation extends from inter-family to intra-family diversity, indicating that there is not a stringent selection to maintain identical EI interactions. In contrast, the ability of one immunity protein to neutralize non-homologous effectors suggests that there has been selection for cross-immunity.
Distribution of effector and immunity proteins
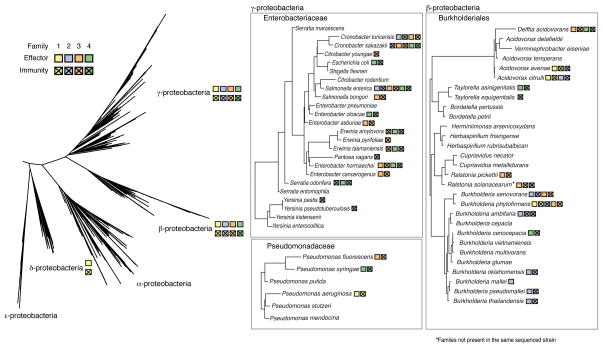
The T6S amidase EI pairs are found among β-, δ-, and γ-proteobacteria. As depicted in Figure 6, they are particularly prevalent in the Enterobacteriaceae, Pseudomonadaceae, and the Burkholderiales. EI pairs appear to be inherited both vertically and horizontally within phylogenetic groups, resulting in a discontinuous distribution of the families. Although the pairs occupy a variety of genomic contexts, at least one member of Families 2–4 is encoded within a T6S gene cluster (Figure S6). Within T6S gene clusters, EI pairs are often encoded adjacent to hcp genes. For one family 4 pair, this association also extends to members encoded by genes outside of T6S gene clusters. Close association of hcp with EI pair loci is consistent with the previous finding that a T6S-exported protein of Edwardsiella tarda, EvpP, directly interacts with an Hcp protein (Zheng and Leung, 2007).
Figure 6.
T6S cell wall amidase effector and immunity proteins are broadly distributed. Phylogenetic tree depicting the distribution of select effector and immunity proteins in Families 1–4. Trees are based on the 16s rRNA tree of life from Silva’s Living Tree project (http://www.arb-silva.de/projects/living-tree/). For each species all effector and immunity proteins present in any genome are noted, however due to variability at the species level, not all members organisms in the group may have all effector and immunity proteins shown. Additional data concerning the genomic context of effectors and immunity proteins are found in Figure S6.
Interestingly, certain homologous EI pairs appear to be recognized by disparate T6SSs. For example, Tae2BT has been experimentally demonstrated to require T6SS-1 for export (Figures 1 and 2), whereas tae2TYis located within Salmonella pathogenicity island 6 (SPI-6) – the only T6SS present in this organism (Figure S6). While the effectors belong to the same family, their associated T6SSs are distantly related. The secretion of multiple effector families by a single secretion system also appears to occur. The genomes of several S. enterica serovars containing only the SPI-6 T6SS encode EI pair Families 2–4 (Blondel et al., 2009). In total, these data strongly suggests that a single effector family can be secreted by divergent T6SSs and that a single T6SS can utilize a diversity of amidase effectors.
A critical observation made in our study is that effector proteins strictly co-occur with cognate immunity proteins (Figure 6). As peptidoglycan amidase effectors are toxic only in the periplasm, and they access that space exclusively by intercellular transfer through the T6S apparatus, the co-occurrence of effectors with immunity proteins demonstrates a strong selection due to active self-intoxication. The only exception we observed is that in certain B. mallei strains Tai2 is encoded by an apparent pseudogene, while the adjacent tae2 locus remains intact. Importantly, the T6SS responsible for intercellular delivery of Tae2, T6SS-1 (based on B. thailandensis orthologs), is mutationally inactivated in B. mallei (Schwarz et al., 2010a; Schwarz et al., 2010b). This example further underscores the generality of selection for immunity via the process of self-intoxication.
In contrast to our observation that effector genes always co-occur with immunity genes, 27% of the immunity proteins we identified were not encoded adjacent to intact effector genes (Figure S6). This argues that there is a selective pressure to retain immunity even in the absence of cognate effectors, and thus supports a role for T6S in antagonistic interspecies interactions. Notable examples of this phenomenon are found in pathogens that inhabit polymicrobial environments at some stage of their life cycle, such as Yersinia pestis and S. Typhimurium.
Discussion
We have developed and implemented a sequence homology-independent means for confidently identifying T6S effectors from bacterial genomes. Using this approach, we discovered four broadly distributed, phylogenetically distinct families of T6S amidase EI pairs. Given current limitations in identifying T6S effectors through strictly experimental means, this method stands to significantly increase our ability to study the functional significance of the system. Our findings have already led to several key insights into the mechanism, function, and evolutionary significance of T6S. While our current approach enriches for effectors with amidase function, the heuristic nature of our method allows for the addition of parameters as they become known. This could allow the removal of constraints specific to enzymatic activity, thereby facilitating the discovery of effectors with assorted functions.
Structural variability in peptidoglycan can provide protection against lytic proteins (Vollmer, 2008). As Gram-positive organisms lack an outer membrane, this protection is considered critical to their survival in certain environments (Davis and Weiser, 2011). Our work has demonstrated that the peptidoglycan of Gram-negative bacteria may also be subject to frequent attack via T6S. Thus, modifications to its structure, such as D-amino acid substitutions and changes to the crosslink position, might serve a protective role against T6S amidase enzymes (Cava et al., 2011; Lam et al., 2009; Magnet et al., 2008). Interestingly, our analyses of T6S cell wall effectors have revealed distinct cleavage specificities against Gram-negative peptidoglycan. The selection for variable specificity could be indicative of a molecular arms race occurring between donor and recipient bacteria.
Thus far we cannot reliably predict non-cognate effector recognition by immunity proteins. We observe instances that violate the simple model that effector relatedness correlates to immunity recognition. This could be explained by a conserved interaction site on the effector protein that binds highly divergent immunity proteins. Alternatively, effector inactivation might proceed through non-conserved immunity protein interactions, and even through non-conserved mechanisms. In this case, homology may be a poor predictor of binding because residue positions participating in the EI interface could differ. Structural insights into EI interaction will be critical for developing an accurate molecular model of the interplay between effector and immunity sequence variation.
A relatively non-discriminating interbacterial EI pathway such as the T6SS has the potential to assist in our comprehension of bacterial interaction networks and community structure. The implications of orphan immunity proteins might be most easily interpreted, as these proteins are presumably present only for defensive purposes. For example, the presence of an orphan immunity protein within one organism that has evolved to specifically recognize an effector encoded by a second organism, suggests that these two organisms compete and interface in their natural environment. Likewise, dissimilar organisms with a matching repertoire of T6S EI pairs might cooperate, and related organisms with incompatible pairs may not. At a cursory level, the distribution and diversity of T6S EI pairs is consistent with expectations. In general, we observe an enrichment of EI pairs in organisms that occupy habitats with relatively dense and rich populations of bacteria, such as the soil and the gastrointestinal tract (GI tract) (Roesch et al., 2007; Walter and Ley, 2011). This trend also holds for pathogens; EI pairs are overrepresented in pathogens that colonize polymicrobial sites such as the GI tract and chronic wounds in comparison to those that elicit disease from sterile sites. Whether the T6S EI pairs of pathogens are adaptive for life within the host, persistence in the environment, or a combination of these, remains to be determined.
The divergence of effector recognition by immunity proteins both within and between effector families suggests that T6S effector loci might drive speciation and kin-recognition. The evolution or acquisition of EI pairs, incompatible with the ancestral EI pair, would render strains incapable of cooperating in the formation of multicellular aggregates with those carrying the ancestral locus. This would produce a barrier to the flow of genetic information between strains and potentially allow for the divergence of species (Cohan, 2002; Majewski, 2001). EI loci might also function in kin discrimination, allowing bacteria to exclude non-kin organisms from their local environment and thus prevent those organisms from benefitting from the production of common goods (Strassmann et al., 2011). In either case further study will be required to ascertain the role of EI loci in sociomicrobiology and community organization.
Experimental Procedures
Bacterial Strains, Plasmids, and Growth Conditions
B. thailandensis and P. fluorescens strains used in this study were derived from the sequenced strains E264 and Pf-5, respectively (Kim et al., 2005; Paulsen et al., 2005). E. coli strains included in this study included DH5α for plasmid maintenance, BL21 pLysS for expression of effectors for toxicity assays, SM10 for conjugal transfer of plasmids into P. fluorescens, and Shuffle T7 pLysS Express (New England Biolabs) for purification of effectors. Growth conditions for all strains and plasmid and strain construction details are described in Supplemental Experimental Procedures.
E. coli Toxicity Measurements
E. coli toxicity assays were performed as described previously with minor modifications (Russell et al., 2011). Full details in Supplemental Experimental Procedures.
Western Blot Analyses
Western blotting was performed as described previously for α-VSV–G and α-RNA polymerase (Russell et al., 2011). The α-His5 Western blots were performed using the Penta-His HRP Conjugate Kit according to manufacturer’s instructions (Qiagen).
Burkholderia Competition Assays
Cells were grown overnight to stationary phase and diluted to an optical density at 600nm (OD600) of 5 before being mixed 1:1 and spotted on a nitrocellulose membrane on LB-LS 3% agar. Plate counts were taken of the initial inoculum and again after 24 hours of competition. Donor cells for all experiments were labeled with a GFP-expression construct integrated into the attTn7 site as previously described, allowing donor and recipient colonies to be disambiguated through fluorescence imaging (Schwarz et al., 2010b). Statistical analyses performed using a two-tailed Student’s t-test.
Purification of Effector Proteins
For purification effector proteins were expressed in pET29b+ vectors in Shuffle Express T7 lysY cells (New England Biolabs). The proteins were purified to homogeneity using previously reported methods, except that in all steps no reducing agents or lysozyme were used (Mougous et al., 2004).
Bioinformatics Screen
A bioinformatics search of type VI secretion positive genomes for bicistronic operons that may encode EI pairs was performed with the constraints described in Figure 3A. Homologs of candidate EI pairs from this search were identified using a tblastn search of the NCBI non-redundant nucleotide database (ftp://ftp.ncbi.nih.gov/blast/db/). Additional details and methods to calculate both prevalence of T6SS in sequenced Proteobacteria and the probability of association of putative EI pairs with T6SSs are described in Supplemental Experimental Procedures.
Alignments and Phylogenetic Reconstruction
All sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). Full-length alignments created in Geneious software using the MUSCLE algorithm (Edgar, 2004). Catalytic alignments created manually based on conserved regions surrounding the predicted catalytic cysteine and histidine residues. For outgroup analysis, the homologous catalytic regions were obtained from the PFAM seed alignment for family Peptidase C1 ((PF00112), http://pfam.sanger.ac.uk/). Phylogenetic tree constructed using Geneious software using the PhyML algorithm with 100 bootstraps for statistical analysis (Edgar, 2004).
Secretome Preparation
B. thailandensis secretome preparations were prepared similarly to those used to identify substrates of the H1-T6SS of P. aeruginosa (Hood et al., 2010). Cells were grown in Vogel-Bonner minimal medium containing 19 mM amino acids as defined in synthetic CF sputum medium (Palmer et al. 2007), 1% Tween 80, and 0.5% v/v glucose. Log-phase cells were harvested at a final OD600 of 1.0. Proteins were prepared as described previously (Wehmhoner et al., 2003).
Pseudomonas fluorescens secretion assays
Overnight cultures of P. fluorescens strains harboring a Tae3-V-expressing plasmid were subinoculated 1:1000 into LB supplemented with 100 μM IPTG and grown at 30 °C to mid-log phase. Cultures were then harvested and cell and supernatant fractions prepared as previously described (Mougous et al., 2006).
MS sample preparation and analysis
Three biological replicates of wild-type samples were used to establish the reference secretome of B. thailandensis. All T6S mutants were analyzed as biological duplicates. Each biological sample was analyzed in triplicate as described elsewhere (Hood et al., 2010). Full details in Supplemental Experimental Procedures.
Enzymatic Assay
The activities of amidase effector proteins on E. coli peptidoglycan sacculi were performed as described previously with minor modifications, as detailed in Supplemental Experimental Procedures (Russell et al., 2011).
Supplementary Material
Highlights.
Mass spectrometry defines several antibacterial T6SS substrates
T6SS substrates from disparate species reveal an effector–immunity paradigm
Informatic analysis based on shared properties defines a large T6S effector family
Effector–immunity distribution indicates a broad role for T6SS in bacterial interactions
Acknowledgments
The authors wish to thank Colin Manoil for sharing B. thailandensis transposon mutants, Joe Gray of the Pinnacle Laboratory of Newcastle University for MS analysis, Nels Elde for advice on sequence analysis, Ferric Fang and Larissa Singletary for insights pertaining to Salmonella, Sonya Heltshe for assistance with statistical analyses, Josie Chandler for critical review of the manuscript, and members of the Mougous laboratory for helpful discussions. Research was supported by grants to J.D.M from the National Institutes of Health (AI080609 and AI057141) and W.V. from the European Commission within the DIVINOCELL programme. A.B.R. was supported by a grant from the National Science Foundation (DGE-0718124). J.D.M. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome biology. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Rawlings ND. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends in biochemical sciences. 2003;28:234–237. doi: 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- Bernard CS, Brunet YR, Gueguen E, Cascales E. Nooks and Crannies in type VI secretion regulation. J Bacteriol. 2010;192:3850–3860. doi: 10.1128/JB.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC genomics. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. The EMBO journal. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. Sexual isolation and speciation in bacteria. Genetica. 2002;116:359–370. [PubMed] [Google Scholar]
- Collmer A, Lindeberg M, Petnicki-Ocwieja T, Schneider DJ, Alfano JR. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 2002;10:462–469. doi: 10.1016/s0966-842x(02)02451-4. [DOI] [PubMed] [Google Scholar]
- Davis KM, Weiser JN. Modifications to the peptidoglycan backbone help bacteria to establish infection. Infection and immunity. 2011;79:562–570. doi: 10.1128/IAI.00651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghachi M, Bouhss A, Barreteau H, Touze T, Auger G, Blanot D, Mengin-Lecreulx D. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. The Journal of biological chemistry. 2006;281:22761–22772. doi: 10.1074/jbc.M602834200. [DOI] [PubMed] [Google Scholar]
- Firczuk M, Bochtler M. Folds and activities of peptidoglycan amidases. FEMS Microbiol Rev. 2007;31:676–691. doi: 10.1111/j.1574-6976.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LD, Hartney S, Duboy R, Goebel NC, Zabriskie TM, et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environmental microbiology. 2010;12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annual review of genetics. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell host & microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell host & microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Schell MA, Yu Y, Ulrich RL, Sarria SH, Nierman WC, DeShazer D. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC genomics. 2005;6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Sogaard-Andersen L. Close encounters: contact-dependent interactions in bacteria. Mol Microbiol. 2011;81:297–301. doi: 10.1111/j.1365-2958.2011.07711.x. [DOI] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Li M, Le Trong I, Carl MA, Larson ET, Chou S, De Leon JA, Dove SL, Stenkamp RE, Mougous JD. Structural basis for type VI secretion effector recognition by a cognate immunity protein. PLoS Pathog. 2012;8:e1002613. doi: 10.1371/journal.ppat.1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol. 2008;190:4782–4785. doi: 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J. Sexual isolation in bacteria. FEMS Microbiol Lett. 2001;199:161–169. doi: 10.1111/j.1574-6968.2001.tb10668.x. [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Petzold CJ, Senaratne RH, Lee DH, Akey DL, Lin FL, Munchel SE, Pratt MR, Riley LW, Leary JA, et al. Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nature structural & molecular biology. 2004;11:721–729. doi: 10.1038/nsmb802. [DOI] [PubMed] [Google Scholar]
- Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilises Type VI Secretion to target bacterial competitors. J Bacteriol. 2011 doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakos G, Wojdyla JA, Kleanthous C. Nuclease colicins and their immunity proteins. Q Rev Biophys. 2011:1–47. doi: 10.1017/S0033583511000114. [DOI] [PubMed] [Google Scholar]
- Parret AH, Schoofs G, Proost P, De Mot R. Plant lectin-like bacteriocin from a rhizosphere-colonizing Pseudomonas isolate. J Bacteriol. 2003;185:897–908. doi: 10.1128/JB.185.3.897-908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Schneider DJ, Tam VC, Chancey ST, Shan L, Jamir Y, Schechter LM, Janes MD, Buell CR, Tang X, et al. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A. 2002;99:7652–7657. doi: 10.1073/pnas.112183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilatz S, Breitbach K, Hein N, Fehlhaber B, Schulze J, Brenneke B, Eberl L, Steinmetz I. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infection and immunity. 2006;74:3576–3586. doi: 10.1128/IAI.01262-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Records AR. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant Microbe Interact. 2011;24:751–757. doi: 10.1094/MPMI-11-10-0262. [DOI] [PubMed] [Google Scholar]
- Rendueles O, Ghigo JM. Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol Rev. 2012 doi: 10.1111/j.1574-6976.2012.00328.x. [DOI] [PubMed] [Google Scholar]
- Rigden DJ, Jedrzejas MJ, Galperin MY. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends in biochemical sciences. 2003;28:230–234. doi: 10.1016/s0968-0004(03)00062-8. [DOI] [PubMed] [Google Scholar]
- Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annual review of microbiology. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrazek J, Nierman WC, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010a;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010b:6. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annual review of microbiology. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev. 2011;75:423–433. doi: 10.1128/MMBR.00014-11. first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Pilsl H, Hantke K, Holtje JV, Braun V. Pesticin displays muramidase activity. J Bacteriol. 1997;179:1580–1583. doi: 10.1128/jb.179.5.1580-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annual review of microbiology. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wehmhoner D, Haussler S, Tummler B, Jansch L, Bredenbruch F, Wehland J, Steinmetz I. Inter- and intraclonal diversity of the Pseudomonas aeruginosa proteome manifests within the secretome. J Bacteriol. 2003;185:5807–5814. doi: 10.1128/JB.185.19.5807-5814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workentine ML, Chang L, Ceri H, Turner RJ. The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: a bacterial two-hybrid analysis. FEMS Microbiol Lett. 2009;292:50–56. doi: 10.1111/j.1574-6968.2008.01445.x. [DOI] [PubMed] [Google Scholar]
- Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.