Abstract
The influenza virus hemagglutinin polypeptides, HA1 and HA2, have been purified by gel filtration in the presence of sodium dodecyl sulfate from a vaccine preparation of the recombinant strain Heq1N2. Use of this technique for purification of the hemagglutinin polypeptides eliminated the need for proteolytic agents for removal of the hemagglutinin from the virus particles and 100-300 mg of virus yielded 10-30 mg of viral protein per chromatographic cycle. Because proteolysis is not required to remove the spikes from the viral envelope, the envelope-embedded HA2 polypeptide was purified in its entirety for structural analysis. Amino-terminal sequence analysis of the smaller polypeptide, HA2, revealed a cyclic repetition of glycyl residues through the first 24 residues at every third to fourth position. The sequence through the first 10 residues was identical to that presented by Skehel and Waterfield for other type A influenza viruses [(1975) Proc. Nat. Acad. Sci. USA 72, 93-97]. The HA1 (Heq/) polypeptide, on the other hand, had different amino acids at three or four out of the first 10 residues of the amino-terminal sequence when compared to HA1 from H0, H1, or H2 subtypes (Skehel and Waterfield). The present study has demonstrated the feasibility of the use of vaccine virus as a source of large quantities of viral protein for determination of primary structure.
Full text
PDF
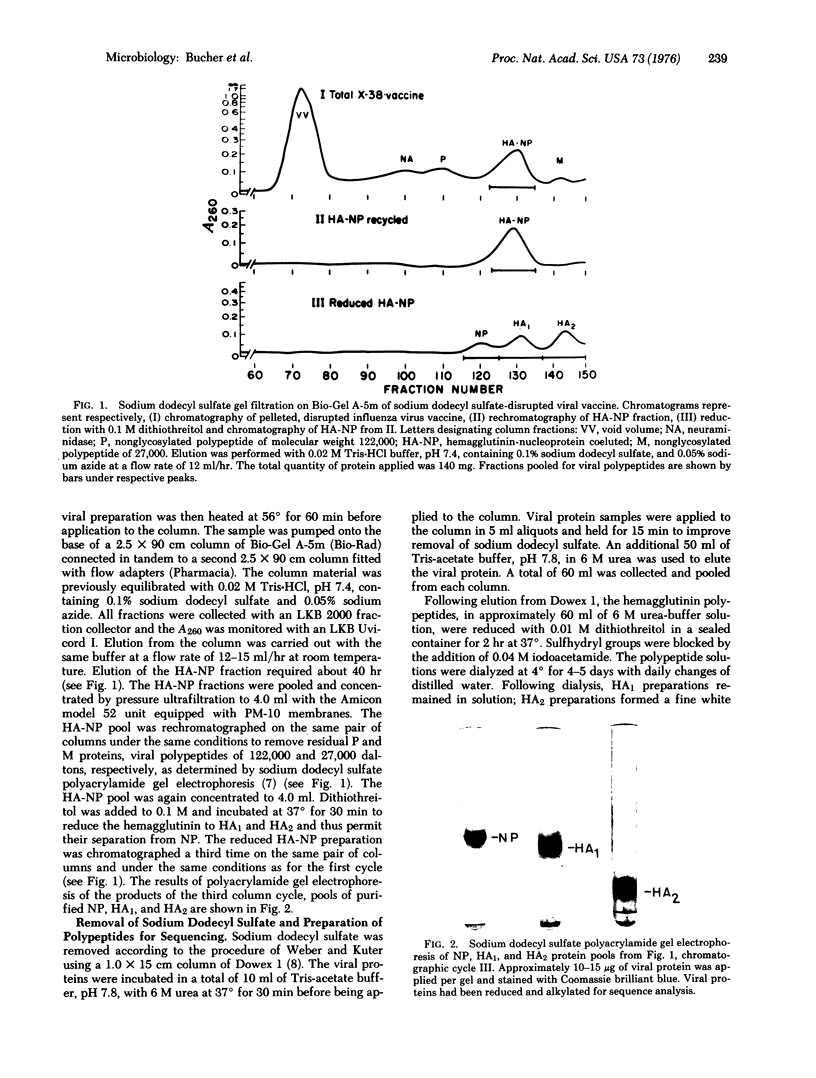
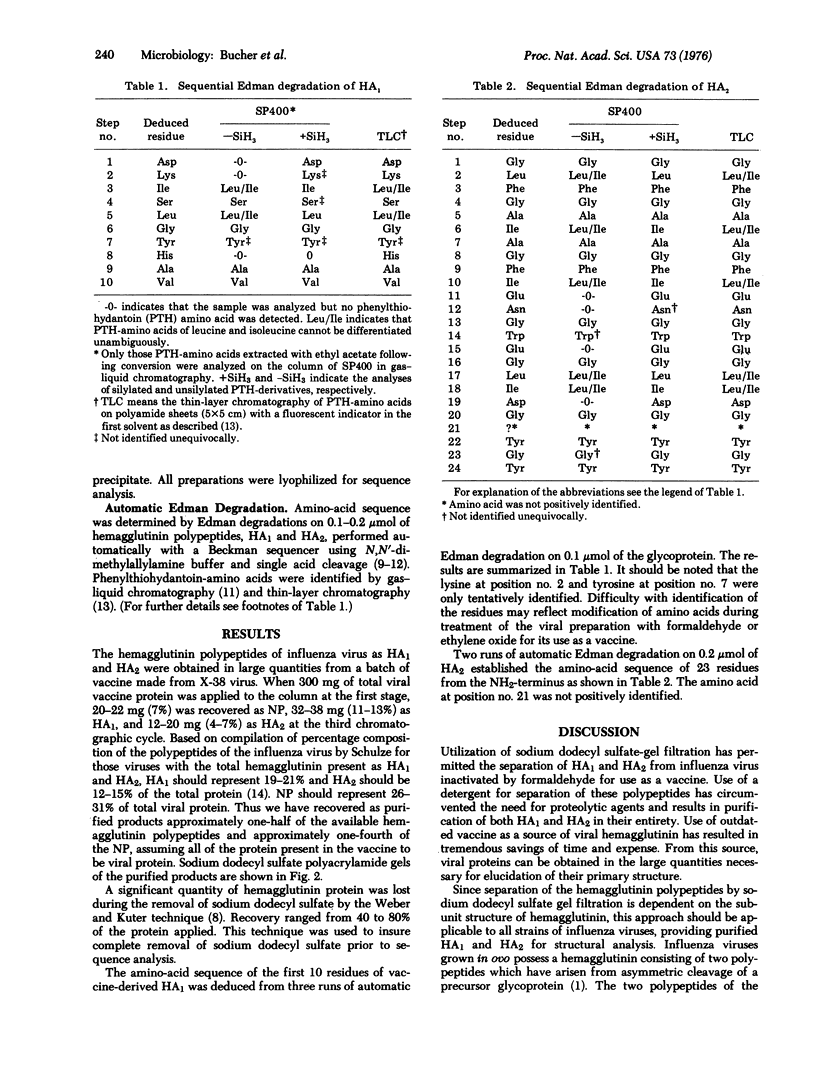


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brand C. M., Skehel J. J. Crystalline antigen from the influenza virus envelope. Nat New Biol. 1972 Aug 2;238(83):145–147. doi: 10.1038/newbio238145a0. [DOI] [PubMed] [Google Scholar]
- Bucher D. J., Kilbourne E. D. A 2 (N2) neuraminidase of the X-7 influenza virus recombinant: determination of molecular size and subunit composition of the active unit. J Virol. 1972 Jul;10(1):60–66. doi: 10.1128/jvi.10.1.60-66.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert E. A. Properties of an antigenic glycoprotein isolated from influenza virus hemagglutinin. J Virol. 1973 Feb;11(2):183–192. doi: 10.1128/jvi.11.2.183-192.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- Kehoe J. M., Capra J. D. Localization of two additional hypervariable regions in immunoglobulin heavy chains. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2019–2021. doi: 10.1073/pnas.68.9.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbourne E. D. Recombination of influenza A viruses of human and animal origin. Science. 1968 Apr 5;160(3823):74–76. doi: 10.1126/science.160.3823.74. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laver W. G., Baker N. Amino acid composition of polypeptides from influenza virus particles. J Gen Virol. 1972 Oct;17(1):61–67. doi: 10.1099/0022-1317-17-1-61. [DOI] [PubMed] [Google Scholar]
- Laver W. G., Webster R. G. Studies on the origin of pandemic influenza. 3. Evidence implicating duck and equine influenza viruses as possible progenitors of the Hong Kong strain of human influenza. Virology. 1973 Feb;51(2):383–391. doi: 10.1016/0042-6822(73)90437-6. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S. G., Compans R. W., Choppin P. W. Influenza virus structural and nonstructural proteins in infected cells and their plasma membranes. Virology. 1971 Dec;46(3):830–843. doi: 10.1016/0042-6822(71)90084-5. [DOI] [PubMed] [Google Scholar]
- Li S. L., Hanlon J., Yanofsky C. Separation of anthranilate synthetase components I and II of Escherichia coli, Salmonella typhimurium, and Serratia marcescens and determination of their amino-terminal sequences by automatic Edman degradation. Biochemistry. 1974 Apr 9;13(8):1736–1744. doi: 10.1021/bi00705a028. [DOI] [PubMed] [Google Scholar]
- Pisano J. J., Bronzert T. J., Brewer H. B., Jr Advances in the gas chromatographic analysis of amino acid phenyl- and methylthiohydantoins. Anal Biochem. 1972 Jan;45(1):43–59. doi: 10.1016/0003-2697(72)90006-1. [DOI] [PubMed] [Google Scholar]
- Skehel J. J., Waterfield M. D. Studies on the primary structure of the influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 1975 Jan;72(1):93–97. doi: 10.1073/pnas.72.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Kuter D. J. Reversible denaturation of enzymes by sodium dodecyl sulfate. J Biol Chem. 1971 Jul 25;246(14):4504–4509. [PubMed] [Google Scholar]