Abstract
Objective
The usefulness of neurosurgical navigation with current visualizations is seriously compromised by brain shift, which inevitably occurs during the course of the operation, significantly degrading the precise alignment between the preoperative MR data and the intra-operative shape of the brain. Our objectives were (i) to evaluate the feasibility of non-rigid registration that compensates for the brain deformations within the time constraints imposed by neurosurgery, and (ii) create augmented reality visualizations of critical structural and functional brain regions during neurosurgery using pre-operatively acquired fMRI and DT-MRI.
Materials and Methods
Eleven consecutive patients with supratentorial gliomas were included in our study. All underwent surgery at our intra-operative MR imaging–guided therapy facility and have tumors in eloquent brain areas (e.g. precentral gyrus and cortico-spinal tract). Functional MRI and DT-MRI, together with MPRAGE and T2w structural MRI were acquired at 3T prior to surgery. SPGR and T2w images were acquired with a 0.5T magnet during each procedure. Quantitative assessment of the alignment accuracy was carried out and compared with current state-of the-art systems based only on rigid-registration.
Results
Alignment between preoperative and intra-operative datasets was successfully carried out during surgery for all patients. Overall, the mean residual displacement remaining after non-rigid registration was 1.82 mm. There is a statistically significant improvement in alignment accuracy utilizing our non-rigid registration in comparison to the currently used technology (p<0.001).
Conclusions
We were able to achieve intra-operative rigid and non-rigid registration of (1) pre-operative structural MRI with intra-operative T1w MRI; (2) pre-operative FMRI with intra-operative T1w MRI, and (3) pre-operative DT-MRI with intra-operative T1w MRI. The registration algorithms as implemented were sufficiently robust and rapid to meet the hard real-time constraints of intra-operative surgical decision making. The validation experiments demonstrate that we can accurately compensate for the deformation of the brain and thus can construct an augmented reality visualization to aid the surgeon.
Keywords: MRI, DT-MRI, FMRI, brain, image-guided neurosurgery, navigation systems, non-rigid registration
Introduction
The American Cancer Society estimates 18,820 new brain tumors will be diagnosed in 2006 in the United States, with an estimated 12,820 deaths. Low-grade gliomas account for 25% of all primary brain tumors.
One of the principle causes of death among patients with low-grade glioma (LGG) is progression of the tumor to a malignant form (ie, anaplastic degeneration). Surgical resection of low-grade gliomas may decrease the rate of recurrence and increase the time to tumor progression (Piepmeier et al. 1996, Berger et al. 1994). Additionally, a maximal resection and a smaller volume of postoperative residual tumor are associated with an improved prognosis for the patient (Philippon et al. 1993, Piepmeier et al. 1996, Janny et al. 1994, Healy et al. 1991). But increased resection margins can increase the risk for postoperative neurologic deficits, due to possible damage of eloquent brain areas, such as the precentral gyrus and cortico-spinal tract, which concern motor function. Therefore, the principle challenge and objective of surgical intervention is to maximize the resection of tumor, while also minimizing the potential for neurological deficit by preserving critical tissue.
One of the challenges for neurosurgeons is to preserve the function during surgery for lesions in the central region. Cortical stimulation is commonly used to localize motor function and has become the “gold standard” when performing surgery in and adjacent to the motor cortex. However, it does not allow for ascertaining the risk of a new postoperative motor deficit before surgery. Moreover, cortical stimulation is a demanding, time consuming, and costly procedure and as such is often not possible during surgery. Therefore, because fMRI of motor and language tasks is feasible in patients with cerebral tumors (Mueller et al. 1996), several groups have proposed the integration of functional data into the neuronavigation system in recent years (Krishnan et al. 2004, Nimsky et al. 1999, Ganslandt et al. 1999, Gralla et al. 2003, Jannin et al. 2002, Roessler et al. 2005, Reithmeier et al. 2003, Talos et al. 2003, O'Shea et al. 2006). And strong evidence that a more radical tumor resection may be achieved by using fMRI information during neurosurgery has been demonstrated by Krishnan et al. 2004, Haberg et al. 2004.
Diffusion tensor imaging (DTI) has recently emerged as a potentially valuable tool for preoperative planning (Tummala et al. 2004, Field et al. 2004, Mori et al. 2002, Clark et al. 2003, Wieshmann et al. 2000, Holodny et al. 2001, Holodny et al. 2001, Witwer et al. 2002, Moller-Hartmann et al. 2002, Coenen et al. 2003.) and postoperative follow-up (Alexander et al. 2003) of surgically treated brain tumors and vascular malformations. DTI provides information about the normal course, displacement, or interruption of white matter tracts in and around a tumor, as well as detecting the widening of fiber bundles due to edema or tumor infiltration (Beppu et al., 2003; Clark et al., 2003; Hendler et al., 2003; Lu et al., 2003; Price et al., 2003; Tummala et al., 2003; Wieshmann et al., 1999; Witwer et al., 2002; Yamada et al., 2003). Consequently, efforts have been made in recent years to integrate DTI data with neurosurgical navigation systems (Nimsky et al. 2005, Coenen et al. 2003, Nimsky et al 2005, Talos et al 2003, Berman et al. 2004, Shinoura et al. 2005, Nimsky et al. 2006). Such a study on the role of diffusion tensor imaging of the corticospinal tract (CST) before and after mass resection, and the correlations with clinical motor findings, was recently published by Laundre et al. 2005.
Interventional MRI (iMRI) has proven to be an effective tool for improving the completeness of low grade glioma resection (Claus et al. 2005, Bradley 2002, Schwartz et al 1999, Schneider et al. 2001, Schneider et al. 2005, Knauth et al. 1999, Wirtz et al. 2000). However, brain deformations typically occur during the neurosurgical procedure, which results in a misalignment between the pre-operatively acquired datasets and the intraoperative brain position. Commonly, commercial systems (such as those developed by Medtronic or BrainLab) only use rigid registration algorithms to project the pre-operatively acquired fMRI and DTI into the navigational system. Intraoperative re-acquisition of fMRI and DTI with iMRI is impractical at present due to long image acquisition and processing times. Non-rigid registration algorithms are therefore necessary to preserve the accuracy of the preoperative fMRI and DTI data.
Intraoperative changes in the shape of the target anatomy impose a stringent requirement upon navigation systems. In order to capture such shape changes it is often necessary to make use of nonrigid registration techniques, which are characterized by a capacity to estimate transformations that model not only affine parameters (global translation, rotation, scale and shear), but also local deformations. This typically requires higher order transformation models, with increased numbers of parameters, and is usually more computationally expensive.
Modeling the behavior of the brain remains a key issue in providing navigation in image-guided neurosurgery. The biomechanical property experiments of Miller (2002) have contributed significantly in the understanding of the physics of brain tissue. He and his colleagues have explored and evaluated several constitutive models (Miller and Chinzei 1997; Miller et al. 2000; Miller and Chinzei 2000; Chinzei and Miller 2001; Miller and Chinzei 2002), which have shown very good concordance of the hyper-viscoelastic constitutive equation with in vivo and in vitro experiments (Miller et al. 2000).
Additionally, Miga, Paulsen and their collaborators (Miga et al. 1999; Miga et al. 1999; Miga et al. 2000, 2000; Miga et al. 2000; Miga et al. 2001; Paulsen et al. 1999; Roberts et al. 1998; Roberts et al. 1999; Roberts et al. 2001) have developed a sophisticated model of brain tissue while undergoing surgery, incorporating simulation of retraction, resection and local stress associated with tumor tissue. Careful validation experiments indicate their model is capable of closely matching observed deformations (Platenik et al. 2002). Their experiments also indicate further improvements in accuracy will be possible by incorporating sparse data from inexpensive intraoperative imaging devices. This work has demonstrated that computer aided updating of preoperative brain images can restore close correspondence between the preoperative data and the intraoperative configuration of the subject. But a practical difficulty of these models is the extensive time necessary to mesh the brain and solve the problem, which is takes too much time for intra-operative purposes. Davatzikos et al. (2001) proposed a statistical framework consisting of pre-computing the main mode of deformation of the brain using a biomechanical model. And recent extensions of this framework show promising results for intra-operative surgical guidance based on manually extracted data (Lunn et al. 2003).
Simple biomechanical models have been used to interpolate the full brain deformation based on sparse measured displacements. Audette (2003) and Miga et al. (2003) measured the visible intra-operative cortex shift using a laser range scanner. The displacement of deep brain structures was then obtained by applying these displacements as boundary conditions to the brain mesh. A similar surface based approach was proposed by Skrinjar et al. (2002) and Roberts et al. (2003), whereby they imaged the brain surface with a stereo vision system.
Previously, we created a full, three-dimensional non-rigid registration implementation using the mean square intensity difference in local regions as the similarity metric, constrained by a linear elastic material (Ferrant et al., 1999). In practice, the method was successful in clinical applications where an assumption of constant image intensities for corresponding structures held true. Our most recent work has built upon our earlier efforts and explorations in non-rigid registration for segmentation, preoperative planning, and enhanced visualization in support of image-guided surgery, and has been described previously (Warfield, Jolesz, and Kikinis 1998; Warfield et al. 2000; Ferrant et al. 2000; Warfield et al. 2000; Ferrant et al. 2000; Warfield et al. 2001; Rexilius 2001; Ferrant et al. 2001; Guimond et al. 2002).
A robust volumetric non-rigid registration scheme for brain deformations has been introduced by our group (Clatz et al. 2005). These studies, using intra-operative brain non-rigid registration, were demonstrated using only retrospective data. To our knowledge, there is no published prospective study on non-rigid registration of pre-operative imaging (T1, fMRI, DTI) with intra-operative images (T1). Five major contributions are presented in our manuscript: (i) our study is prospective, with 11 patients enrolled over one year; (ii) the alignments are achieved in near real-time during the neurosurgical procedure; (iii) we aligned both the fMRI and DTI using non-rigid registration; (iv) we presented quantitative results assessing the accuracy of the rigid and the non-rigid registration accuracy; (v) and we prospectively utilized a new volumetric non-rigid registration scheme, that has previously been assessed only retrospectively (Clatz et al. 2005), offline and outside of the operating room.
Materials and Methods
Patient population
Eleven consecutive patients (6 female, 5 male, age range: 28–62 years; mean: 45.2 years) with supratentorial gliomas (World Health Organization (WHO) grading: II: 5, III: 4, IV: 2) were included in our study. All patients underwent surgery at our institution's intraoperative MR image–guided therapy facility between April 2005-January 2006 for tumors in and adjacent to eloquent brain areas (such as the precentral gyrus and cortico-spinal tract, for motor function; and Broca's and Wernicke's areas, for language function). For these patients, DTI was judged necessary for preoperative surgical planning. The study was carried out with Institutional Review Board approval.
Pre-operative imaging
After providing informed consent, patients underwent the following MR imaging protocol on a General Electric (Milwaukee, WI) 3T Signa scanner several days before their scheduled surgery:
Anatomic Imaging: (1) whole brain sagittal 3D-SPGR (slice thickness 1.3mm, TE/TR=6/35 msec, FA=75°, FOV = 24cm, matrix=256×256); (2) axial T2-weighted fast-spin-echo (slice thickness 5mm, TE/TR 100/3000 msec, FOV=22cm, matrix=512×512).
-
Functional MRI: Functional MRI was performed using a GE (Milwaukee, WI) 3 Tesla Signa scanner. Whole-brain functional images were acquired with a T2*-weighted echo-planar (EPI) sequence sensitive to the blood oxygen-level dependent signal (TR, 2000 ms; TE, 30 ms; matrix, 64 ×64 mm; FOV, 240 mm; imaging 24 contiguous slices of 5 mm thickness). Visual stimuli were presented using a PC laptop (Dell, Inc., Austin,TX) running either E-prime (Psychology Software Tools, Pittsburgh, PA) or the Presentation (Neurobehavioral Systems Inc., Davis, CA) software package, on an MR-compatible goggle system (Resonance Technology, Northridge, CA). Auditory stimuli were presented with MR-compatible headphones (Avotec Inc., Stuart, FL).
Patients performed a clinically-relevant set of tasks from a battery of motor, language, and visual paradigms. All fMRI stimuli were visually-presented (except one language task as noted below). Motor mapping tasks were block paradigms of twelve alternating 20-second blocks of active finger-tapping or hand-clenching and rest. The visual mapping task was a block paradigm of twenty 20-second blocks of pattern-reversal (2-Hz) checkerboards of right, left, and whole visual field stimulation, and rest, whereby patients were instructed to maintain fixation on a point in the center of the screen. And language mapping tasks consisted of: a silent, block paradigm of twelve alternating 20-second blocks of antonym generation and rest; a silent, block paradigm of eighteen alternating 20-second blocks of word generation (i.e., verbal fluency) and rest; a vocalized, event-related antonym generation paradigm (average ISI=8.3 sec., 50 stimuli); a silent, event-related Stroop task, whereby the patient performed congruent and incongruent color naming of color words (2-4 sec. pseudo-random ISI, 90 stimuli); and a vocalized, event-related, aurally-presented, semantic concrete words paradigm (“alive” or “not alive” response; average ISI=8.3 sec., 50 stimuli). Functional images were motion-corrected, smoothed (8 mm Gaussian kernel), and analyzed with SPM2 (Wellcome Department of Cognitive Neurology, London, UK), using the general linear model (Friston et al. 1995). Differences between stimulus and baseline conditions, convolved with the hemodynamic response function, were examined using analysis of co-variance with global signal and low frequency components treated as nuisance covariates. Correction for multiple comparisons was performed using the theory of Gaussian random fields (Friston et al. 1996).
Diffusion Tensor Imaging: axial line scan diffusion images (LSDI) (Slice thickness=5mm, matrix=512×512, FOV=24cm) and EPI DTI (128×128, Phase FOV=1.0, FOV=25.6, Slice thickness=3, Bvalue=800, 31 directions, number of T2=1) were acquired, covering the entire region of interest as well as “landmark” regions, i.e. areas where the relevant fiber tracts show high density (e.g. ventral brain stem for the corticospinal tract, lateral geniculate body for the optic radiation).
MR-Spectroscopy from the tumor was also acquired for three patients.
Intra-operative imaging
After the patients were positioned for craniotomy and their heads were fixated by using an MR-compatible carbon fiber Mayfield clamp (Ohio Medical Instruments, Cincinnati, Ohio), they were examined with the following initial imaging protocol by way of a vertically open 0.5-T iMR imaging unit (SignaSP; GE Medical Systems, Milwaukee, W) with the following parameters: (a) for transverse, sagittal, and coronal T1-weighted fast spin-echo imaging, 700/29 (repetition time msec/echo time msec), a 22-cm field of view, a 256 × 256 matrix, one signal acquired, a 3-mm section thickness, and a 1-mm intersection gap; (b) for transverse T2-weighted fast spin-echo imaging, 5000/99, a 22-cm field of view, a 256 × 256 matrix, two signals acquired, a 3-mm section thickness, and a 1-mm intersection gap; and (c) for transverse three-dimensional spoiled gradient-echo (SPGR) imaging, 15.5/5.2, a 45° flip angle, a 22-cm field of view, a 256 × 256 matrix, one signal acquired, a 2.5-mm section thickness, and a 0-mm intersection gap. During the surgical intervention, T2-weighted fast spin-echo and three-dimensional SPGR MR image updates were obtained after the dural opening, and at any time the neurosurgeon believed that a brain shift had occurred or a substantial amount of tumor tissue had been removed. The final imaging protocol, which was performed after the dura mater was closed, included the same sequences that were performed at initial imaging. The initial and final image datasets were used to determine the initial and residual tumor volumes, respectively.
Rigid registration with the first acquired intra-operative dataset
As surgery began, before opening the dura mater, a first intra-operative T1 scan was always acquired as part of the current protocol. Because there is no brain shift at this stage, a rigid registration between the pre- and first intra-operative T1 is sufficient.
The rigid alignment is performed using software tools developed by our research group, using an approach based on Mattes mutual information metric. This method is automatic and requires no human interaction. Registration between two datasets takes approximately 30 seconds on a conventional workstation, and is quite satisfactory given the time constraints of neurosurgery.
Non-rigid registration
As the tumor resection progressed after opening the dura mater, brain deformation would inevitably occur. Brain shift is influenced by tissue characteristics, intraoperative patient positioning, opening of the ventricular system, craniotomy size, and resected volume.
Intra-operative T1 and T2 scans are acquired to asses the patient's status and the intraoperative shape of the brain. These intraoperative scans now allowed us to update the preoperative high-resolution structural images, by non-rigid registration with the intra-operative T1 dataset.
Non-rigid registration algorithms are time consuming, and therefore, to date several groups have reported these algorithms as being impractical for real-time use in neurosurgical procedures (Nimsky et al., 2006). Therefore, only rigid registration methods (which does not account for brain deformations) are used in commercial navigational systems (such as ones developed by Medtronic and BrainLab). To address this inconvenient issue, our group has developed a software tool that has proven to be sufficiently robust for volumetric, non-rigid registration between two 3D MR images (Clatz et al., 2005), using a high-performance computational infrastructure. Despite being computational expensive, the full biomechanical model has previously shown to be robust and accurate, which is one of the essential requirements in image-guided neurosurgery.
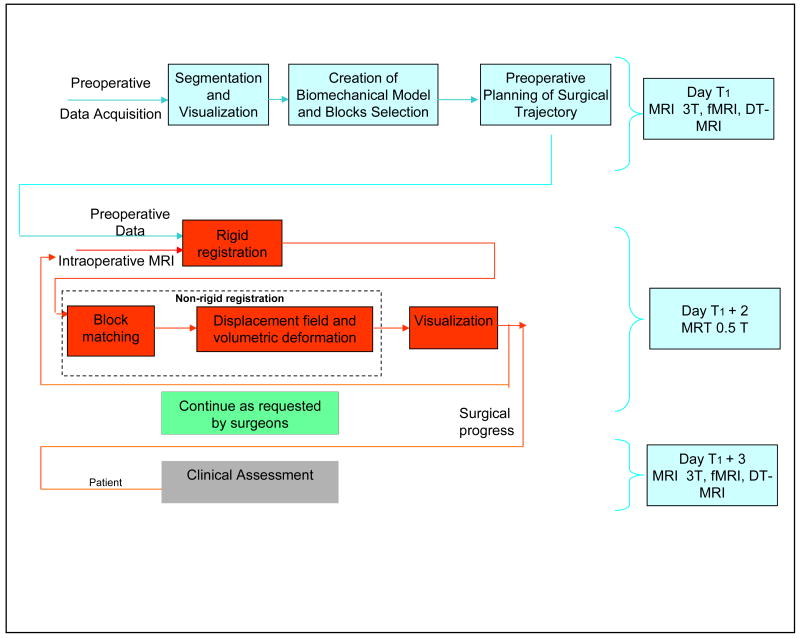
The image-guided neurosurgery architecture is presented in Figure 1. It uses a patient-specific registration algorithm that estimates the brain deformation between two 3D volumetric MRI images (presented in Figure 2). The algorithm can be decomposed into three main parts. The first part consists of building the patient-specific model utilizing intra-operative MRI. The second part is comprised of a block matching computation for selected blocks which estimates a set of displacements across the volume. The third part is an iterative hybrid solver that estimates the 3D volumetric deformation field, utilizing the anisotropic information provided by the structure tensor computed for each block.
Figure 1.
The current system for the image guided neurosurgery that integrates our novel non-rigid registration technology.
Figure 2.
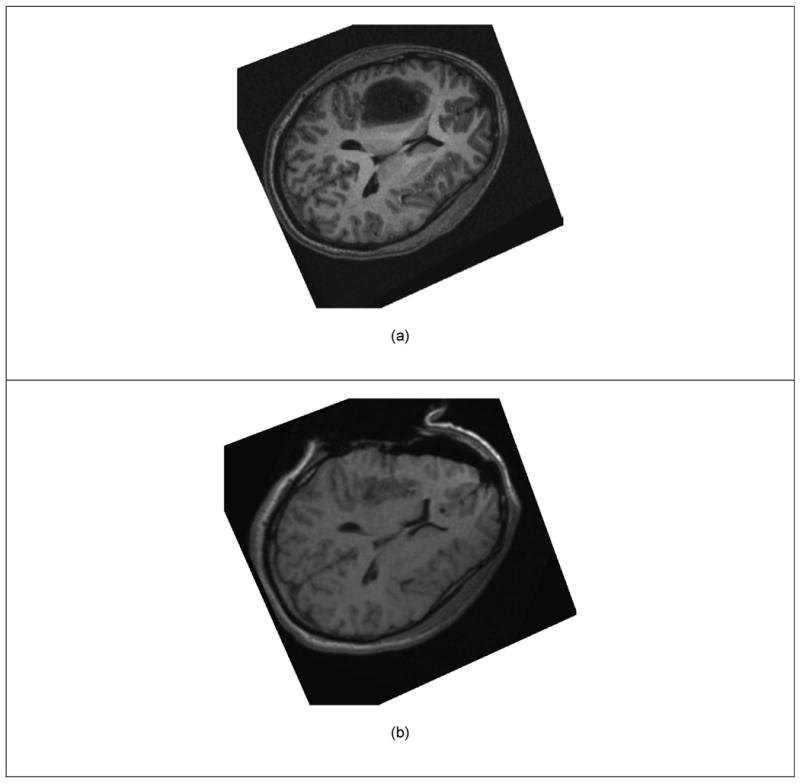
Two Dimensional View of Brain Shift. (a) Pre-operative Image. (b) Intra-operative Image (after the dura has been open and part of the tumor removed).
One advantage of our proposed scheme is that a large part of the computation can be performed before the intra-operative MR images acquisition. In the following section, we describe the sequence of actions of the algorithm and distinguish between the preoperative and intra-operative calculations. Since the pre-operative images are available hours or days before surgery, we can use robust, accurate, time consuming pre-processing algorithms to 1) segment the brain, 2) generate the patient-specific biomechanical model of the brain, 3) select blocks in the pre-operative images with relevant information, and compute the structure tensor in the selected blocks.
1. Pre-operative data processing
Following the pre-operative imaging acquisition, a biomechanical model of the brain is built. The following steps are performed prior the surgery:
mage segmentation: The delineation of brain in the preoperative data is achieved using segmentation strategies optimized for the particular type of acquisition (Warfield et al. 2000). Recently we have also successfully used a method based on a deformable model which evolves to fit the brain's surface by the application of a set of locally adaptive model forces (Smith 2002).
Mesh generation: Tetrahedral discretization (volume mesh) of the segmented intra-cranial cavity serves as the basis for Finite Element Method (FEM) modeling of the physical tissue deformation, and serves as the function of regularization on the estimated displacements obtained as the result of block matching step of non-rigid registration. The technique used for tetrahedral mesh generation is described in (Fedorov et al. 2006). This is a fast and effective mesh generation algorithm that generates elements of high quality.
Biomechanical model: As in our past work (Ferrant et al. 2001; Ferrant et al. 2002; Warfield et al. 2002) we discretized the partial differential equation describing the behavior of the brain under deformation by utilizing the finite element method. We model the brain material using an incompressible linear elastic constitutive equation to characterize the mechanical behavior of the brain parenchyma. Because the ventricles and subarachnoid space are connected, the CSF is free to flow from one to another. We thus assume very soft and compressible tissue for the ventricles. The skull is implicitly modeled by preventing the brain mesh vertices from moving outside the brain segmentation. Biomechanical model is used to bring additional information mostly in the vicinity of the tumor resection. It is also used in areas where outliers are identified and rejected. The outliers typically occur in homogenous regions of the brain structural MRI.
Block Selection: As we described previously (Ruiz-Alzola et al. 2002), we select blocks with high variance of signal intensity to ensure sufficient information is present to compute matches. Additionally, the ill-posed nature of finding correspondences in the resection cavity is anticipated, by performing the block selection in a mask corresponding to the segmentation of the brain without the tumor segmentation.
2. Intra-operative data processing
Extension of Block Matching Algorithm with Outlier Rejection
Also known as template or window matching, the block matching algorithm is a simple method used for decades in computer vision (Dengler and Schmidt 1988; Ruiz-Alzola et al. 2002). It makes the assumption that a global deformation results in translation for small parts of the image. Then the complex global optimization problem can be decomposed into many simple ones: considering a block B(Ok) in the reference image centered in Ok, and a similarity metric between two blocks M(Ba,Bb), the block matching algorithm consists in finding the positions that maximize the similarity:
That is, we seek to identify the smallest block center displacement that maximizes the similarity between the reference image and the target image.
Performing this operation on the selected blocks in the pre-operative image produces a sparse estimate of the displacement between the two images. The choice of the similarity function has largely been debated in the literature, we will refer the reader to the article of Roche (Roche, Malandain, and Ayache 2000) for a detailed comparison of them. In our case, the mono-modal (MR T1 weighted) nature of the registration problem authorizes us to make the strong assumption of an affine relationship between the two image intensity distributions, and so we use the correlation coefficient:
where BF and BT respectively denote the block in the floating and in the reference image, and B̄ is the average intensity in block B. In addition, the value of the correlation coefficient for two matching blocks is normalized between 0 and 1 and reflects the quality of the matching: a value close to 1 indicates two blocks very similar while a value close to 0 for two blocks very different. We use this value as a confidence in the displacement measured by the block matching algorithm.
Estimation of Displacement Field from Block Displacements
The remaining step is to compute a volumetric displacement field from the block displacements. In this section, we summarize o ur concept of approaching the classical energy minimization formulation of this problem by moving from first an approximation problem to interpolating after robust outlier rejection. The reason for this new formulation is that the approximation formulation, which balances match quality against a regularization term, performs well in the presence of noise, but suffers from a systematic error, whereas the exact interpolation problem gives an undesirable solution when it is based on noisy or erroneous data.
We propose an algorithm that takes advantage of both formulations to iteratively estimate the deformation from the approximation to the interpolation formulation, while rejecting outliers. Let K be the stiffness matrix representing the brain mechanical properties, H be a linear interpolation matrix and S the structure tensor. We seek the displacement field represented by U and denote the block displacements by D. The gradual convergence to the interpolation solution is achieved through the use of an external force added to the approximation formulation:
This force Fi is computed at each iteration i to balance the mesh internal mechanical energy UTiKUi. This leads to the iterative scheme:
The transformation is then estimated in a coarse to fine approach, from large deformations to small details up to the interpolation. This scheme is guaranteed to converge toward the interpolation formulation. The above equations are iterated to convergence, incorporating outlier rejection as described below. This new formulation combines the advantages of robustness to noise at the beginning of the algorithm and accuracy when reaching convergence. Because some of the measured displacements are outliers, we propose to introduce a robust block-rejection step based on a least-trimmed squares algorithm (Rousseeuw and Van Driessen 1999). This algorithm rejects a fraction of the total blocks based on an error function measuring for block at Ok the error between the current mesh displacement and the matching target. The error measure is thus simply the estimated displacement error weighted according to the direction of the intensity gradient (structure tensor) of the block.
The mechanical system was solved using the conjugate gradient (Luenberger 1969) method with the GMM++ sparse linear system solver with a maximum error set to 10−4mm. The rejected block fraction for 1 iteration was set to 2.5% and the number of rejection steps to 10.
During the neurosurgical procedures, a three dimensional displacement field between the pre-operative and intra-operative T1 images was estimated using the strategy described above.
High performance computational architecture
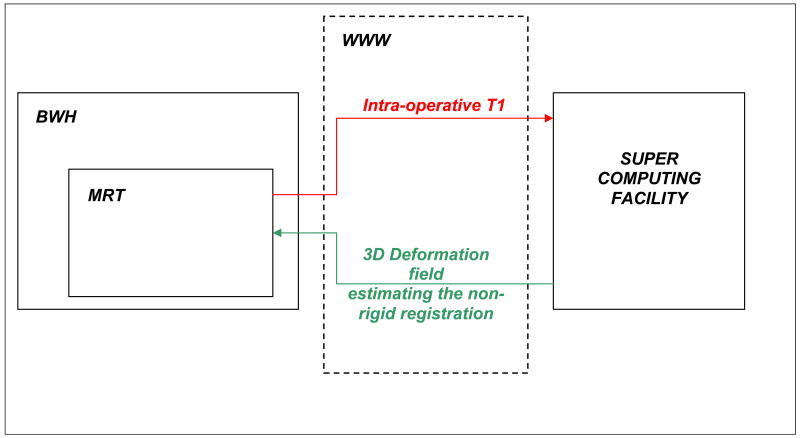
Non-rigid registration algorithms are typically computationally expensive and often proven to be impractical for solving clinical problems. In a previous retrospective study, we employed a cluster of computers to achieve near-real time performance. Our implementation addresses three aspects: (1) load balancing, (2) fault-tolerance and (3) ease-of-use for parallel and distributed registration procedures. With dynamic load balancing we improved by 50% the performance of the most computational intensive part, parallel block matching. Our 2-level fault-tolerance introduced a moderate 6% overhead due to additional communication. With web-services and by hiding pre-processing overheads, we developed faster and easier to use remotely registration procedure. Details of the novel technology can be found in Chrisochoides et al. (2006). The system architecture is presented in Figure 3.
Figure 3.
The architecture used to perform the non-rigid registration computations required by the neurosurgery. The data is transferred from the operating room (MRT) at our institution (BWH). The computations are performed during the case, and then are transferred back to BWH in the OR. The complete process takes approximately 5 minutes.
The datasets are transferred during the surgery from the operating room to the supercomputing facility. The computations are performed and subsequently, the results are transferred back to our institution. And finally, the registration results are displayed in the OR.
Augmented Reality Visualization with FMRI
The fMRI, after processing, is stored as a 3D dataset. The displacement fields estimated with the non-rigid registration algorithm are used to deform the pre-operative image and match the intra-operative anatomy. Regions of activation are rendered with color coding on cross-sectional slices, and as triangle models that illustrate the 3D region of activation.
Augmented Reality Visualization with DT-MRI
The fiber tracts are reconstructed on the pre-operative images, before the surgery starts. For the present study, we have used the 3D tractography method described by Westin et al. (2002 and Park et al. (2004). For initialization of the 3D tractography algorithm, seeding points were placed in the landmark areas. Alternatively, seeding points were placed in the subcortical white matter subjacent to areas of cortical activation seen on fMRI. The motor fibers are identified as described by Mori et al. (2002) and Nimsky et al. (2006).
During surgery, the displacement field between T1w MR images is estimated with the non-rigid registration algorithm. The same displacement field is applied to the pre-operative DTI data, using the method described in Sierra (2001) and Ruiz-Alzola et al. (2002). Then the seeds used in the pre-operative phase for tractography are deformed using the displacement field, so they match the brain anatomy during tumor resection. The novel seeds are used to re-generate the tracts of interests. On a standard workstation it takes less then 30 seconds; therefore, this step is not a concern with regard to the execution time within the context of neurosurgery.
The patient coordinate system is extracted from the DICOM images acquired with the intra-operative MRI. The augmented reality visualization can then be presented in the patient space, and available for navigation with surgical instruments, based on 3D Slicer (www.slicer.org) our open source software.
Estimation of registration accuracy
The non-rigid registration algorithm estimates a displacement field that warps the pre-operative structural MRI to the intra-operative T1w imaging, accounting for brain deformations occurring during tumor resection. The displacement field is subsequently applied to the pre-operative image. This new obtained image must match the intra-operative image of the brain.
We measured the accuracy of alignment between these two images, by extracting the edges from the images using a Canny edge detector, as in Figure 9. The 95% Hausdorff metric (Hausdorff 1962) is used to estimate the registration accuracy. The Hausdorff distance is the maximum distance of a set to the nearest point in the other set. More formally, Hausdorff distance from set A to set B is a maxi-min function, defined as:
Figure 9.
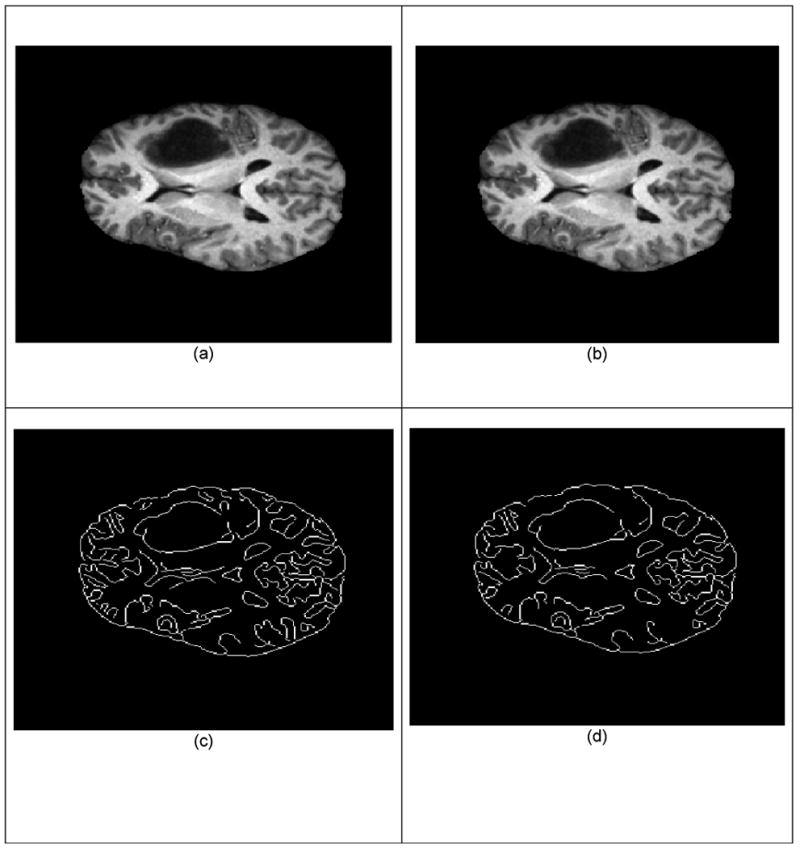
Validation of the non-rigid registration alignment. (a) Non-rigid registered pre-operative T1 3T. (b) Intra-operative image T1 0.5T. (c) Contours extracted from (a) with the Canny edge detector. Contours extracted from (b) with the Canny edge detector. The accuracy of alignment is computed between the points on (c) and (d).
where a and b are points of sets A and B respectively, and d(a, b) is Euclidian metric between these points. The 95% Hausdorff distance is then calculated between the two set of points representing the edges, and it represents the accuracy of the alignment. The ideal case with perfect alignment is when the Hausdorff distance is equal to zero.
The non-rigid registration is compared with the rigid registration only, which represents the current state of the art. Therefore, two Hausdorff distances are computed between (1) the deformed registered image, and the intra-operative image, and (2) the preoperative data are rigidly aligned with the intra-operative image. The ratio between the (1) and (2) is computed and represents the factor of improvement of our novel technology.
Results
The system has been evaluated during 11 consecutive neurosurgery cases. The data has been transferred, processed and displayed in the OR during the neurosurgery. A total of 19 non-rigid registrations have been performed. All the alignments for all the datasets acquired during the tumor resection have been successfully estimated.
Examples of meshes used in the brain biomechanical model are presented in the Figure 4. For all the cases, meshes have been successfully generated. The rigid alignment of the datasets (T1, T2, fMRI, DTI) and the segmentation of anatomical structures of interest are performed. A complex scene is built and used for surgical planning. An example is illustrated in the Figure 5.
Figure 4.
Meshes of the brain used to generate the biomechanical model.
Figure 5.
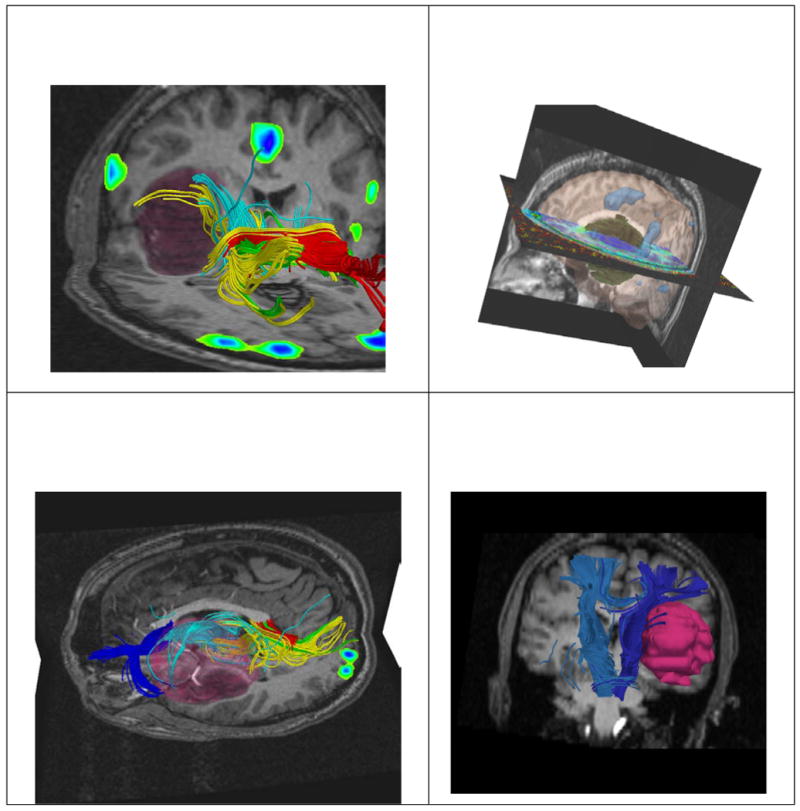
Alignment of preoperative imaging is performed. Tumor segmentation is also carried out, prior the surgery. The images show the T1, DT-MRI tractography, and fMRI alignment together with the 3D reconstruction of the tumor. Neurosurgeons have complex information available to decide the best strategy to adopt for the craniotomy.
For each patient, we measured the alignment accuracy between (1) the deformed registered image, and the intra-operative image, and (2) the preoperative data rigid aligned with the intra-operative image. The ideal case is when the distance in (1) is zero. The (2) is used as a comparison, as only rigid registration is performed in current clinical procedures. Overall, the minimum error for our novel registration technique is 0.85mm, the mean 1.82mm, and the max 3.57mm. The mean ratio between (2) and (1) is 4.58; which reveals that our results can be at least 4.58 times more precise than current technology. There is a statistically significant difference between the accuracy of the alignment of pre-with intra-operative images, with and without non-rigid registration (p<0.001). The complete results are presented in Table 1. The resected tumor can be a maximum 3 voxels off its contour, as seen on the MR imaging. Typically, the voxel size on the axial plane is 0.8 mm. Therefore, the registration accuracy should be < 2.4 mm.
Table 1.
Numerical accuracy results for the non-rigid registration algorithm and its comparison with conventional rigid-registration.
| Tumor position | Sex/Age | Registration between preoperative and intraoperative scans (95% Haussdorf distance) | ||||
|---|---|---|---|---|---|---|
| Max Displacement measured (mm) | Rigid registration accuracy–preop to intraop (mm) | Non-Rigid registration accuracy–preop to intraop (mm) | Ratio Rigid/Non-Rigid | |||
| Case 1 | right posterior frontal | F/29 | 10.68 | 5.95 | 1.90 | 3.13 |
| Case 2 | left posterior temporal | M/54 | 21.03 | 10.7 | 2.90 | 3.69 |
| Case 3 | left medial temporal | F/57 | 15.27 | 1 7.65 | 1.70 | 4.50 |
| Case 4 | left temporal | M/54 | 10.00 | 6.80 | 0.85 | 8.00 |
| Case 5 | right frontal | F/33 | 9.87 | 5.10 | 1.27 | 4.01 |
| Case 6 | left frontal | M/62 | 17.48 | 10.20 | 3.57 | 2.85 |
| Case 7 | right medial temporal | F/50 | 19.96 | 9.35 | 2.55 | 3.66 |
| Case 8 | right frontal | M/40 | 17.44 | 8.33 | 1.19 | 7.00 |
| Case 9 | right frontotemporal | M/28 | 15.08 | 7.14 | 1.87 | 3.81 |
| Case 10 | right occipital | F/56 | 9.48 | 5.95 | 1.44 | 4.13 |
| Case 11 | left frontotemporal | M/34 | 10.74 | 4.76 | 0.85 | 5.60 |
| AVG | 14.27 | 7.44 | 1.82 | 4.58 | ||
Alignment results of the preoperative with intra-operative datasets are presented in Figure 6. Fiber tracts that were deformed due to brain shift were calculated and made available for the neurosurgeon during the tumor resection (Figure 7). The fMRI activation areas were updated, thereby matching the deformed brain, and used to seed the fiber tracts.
Figure 6.
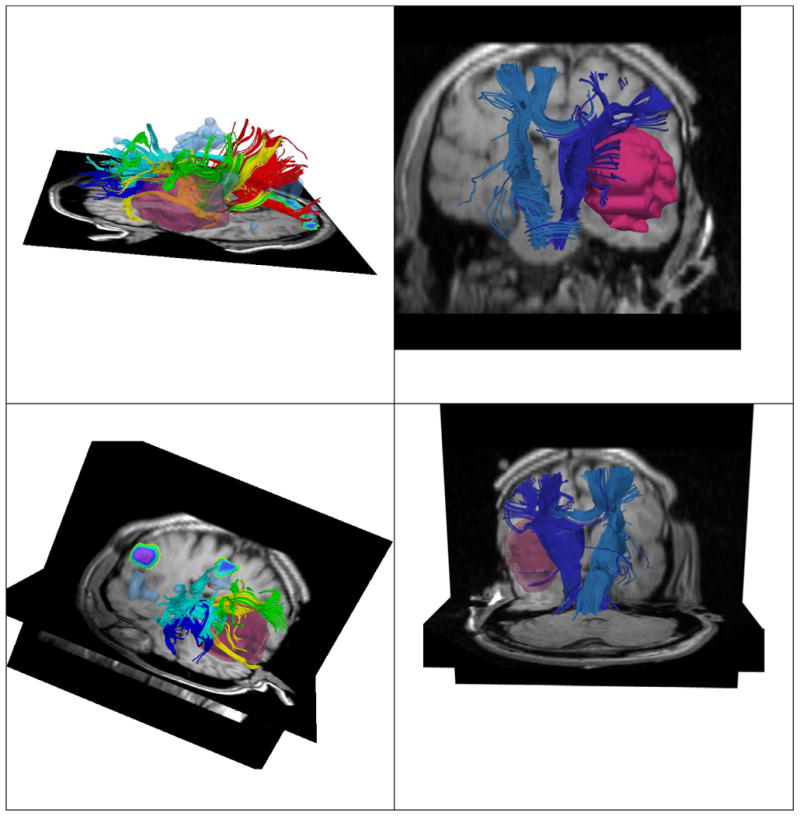
Non-rigid registration of preoperative imaging (T1, fMRI, DTI) with intraoperative imaging. We enhance the current procedure, by aligning preoperative imaging with intraoperative imaging. The DTI, fMRI, T1 images are displayed during tumor resection. Damage of critical structures can be avoided, while achieving gross tumor resection.
Figure 7.
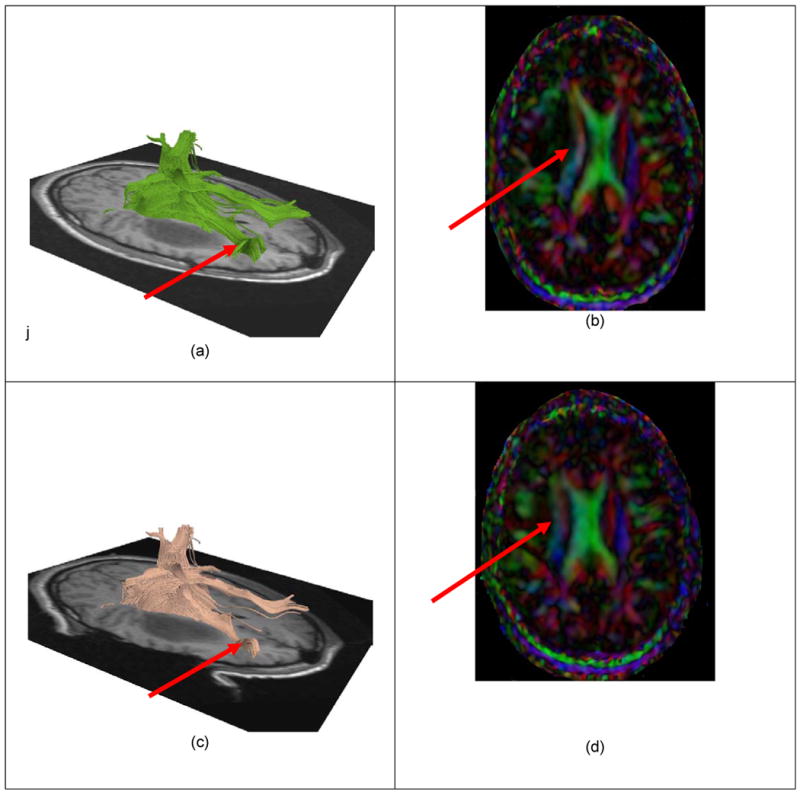
White matter fiber tracts deformation during the neurosurgery. The pre-operative data is shown in the images (a) and (b), while the intra-operative imaging is shown in (c) and (d). A significant displacement of the fiber tracts can be noticed.
One of the hypotheses investigated is that relevant white matter fiber tracts pass through the fMRI activation areas. An example of such a scene created within the time constraints imposed by the neurosurgery procedure is illustrated in the Figure 8.
Figure 8.
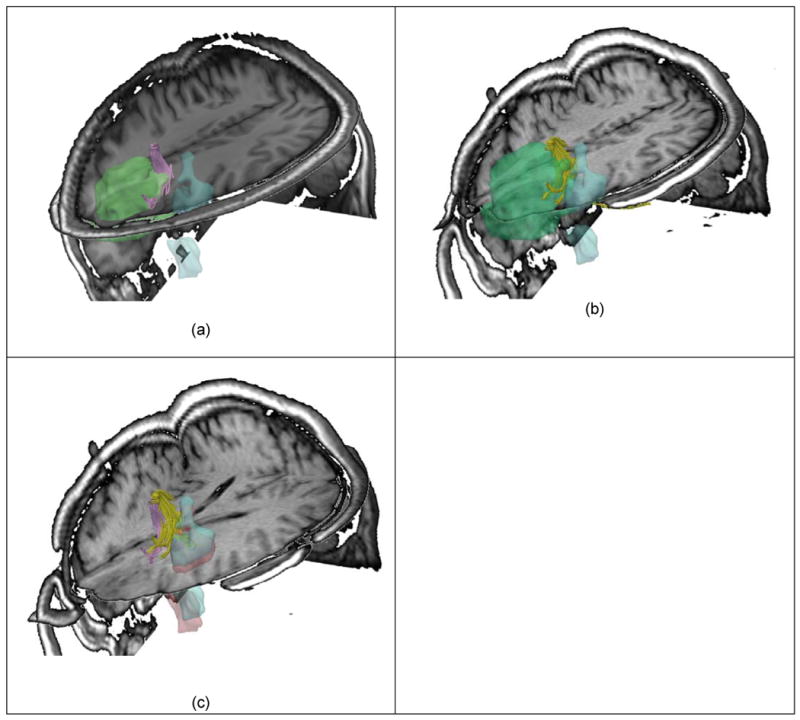
Significant brain deformations require precise re-alignment of pre-operative fMRI and DTI datasets. (a) Illustrates the pre-operative images aligned (T1, fMRI) and several fiber tracts located in the vicinity of the tumor. Some of them are also crossing the tumor. Tumor is represented with green, while the fMRI in blue, and fiber tracts in magenta. (b) The fMRI is realigned with the intra-operative images, while compensating for the brain shift. The preoperative fiber tracts are also realigned and displayed during tumor resection (in yellow). (c) Shows the fMRI and fiber tracts before and after craniotomy. It is important to notice the deformation occurred.
The non-rigid registration algorithm between T1 images (including the loading and saving the output data) is executed in 179 seconds using 240 processors. The execution time for the algorithm itself (once data loaded from the hard-drive) is about 1 minute.
The transfer of images between the two sites takes aproximatelly 1 minute. The orientation of each of the pre-operative dataset is achieved in less than o ne minute. Overall, this is satisfactory given the time constraints imposed by neurosurgeries. Illustrative examples of the validation algorithm are shown in the Figures 9 and 10.
Figure 10.
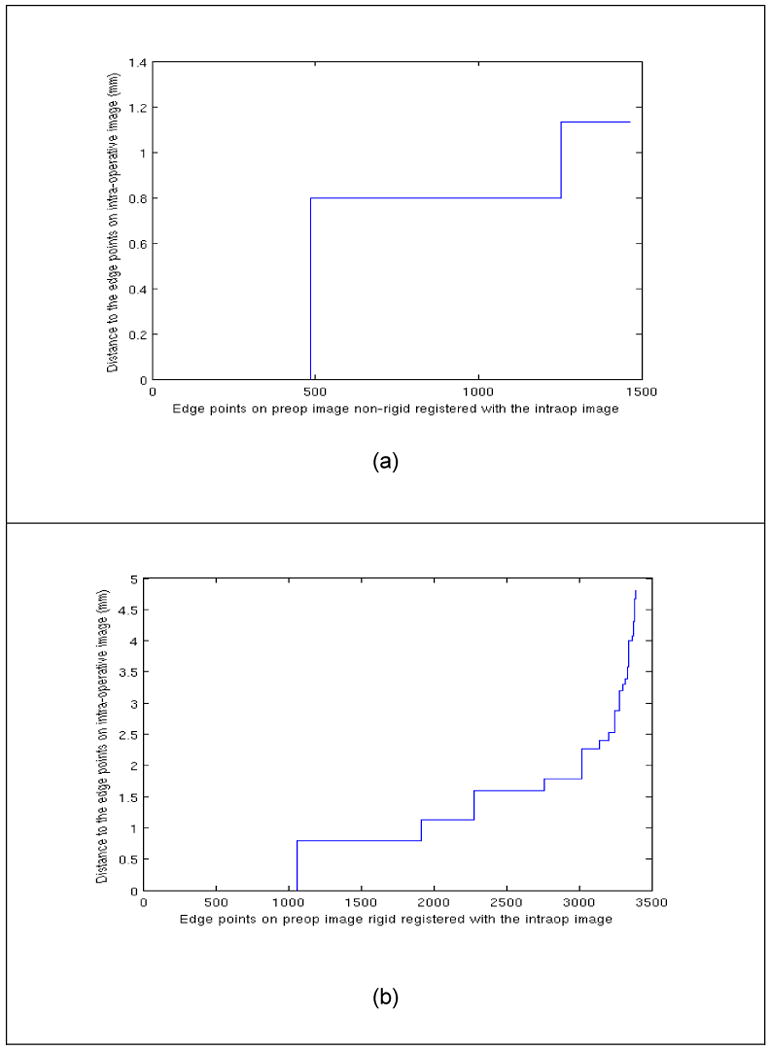
(a) 95% Hausdorff distance between the points on the edges of the registered image and intra-operative image. (b) presents the 95% Hausdorff between points on the edges on the rigid aligned preoperative image and the edges on the intra-operative data. The number of points on the extracted edges is different in the two cases, since typically MR 3T images have higher contrast.
The non-rigid registration algorithm was also tested on a standard workstation (2-way Dell Precision Workstation 650, Intel(R) Xeon(TM) 3.06GHz with 1MB Level 3 cache 512KB L2 cache and 3.5GB RAM). Between 15 and 40 minutes are needed to run the non-rigid registration algorithm on a workstation with 4 processors. Therefore, complex facilities involving high performance computing are currently necessary for use during neurosurgery. However, the performance of the standalone workstations is continuously improving. And, based on the dramatic increase in computer performance over the past decade, we envision in the near future a marked reduction in execution time for this novel technology on standard workstations. The current study establishes its feasibility in a clinical context. The neurosurgeons involved in our study have also reviewed the registration results. Anatomical landmarks (such as ventricles, putamen, gyrus, midline), visible on both pre-operative and intra-operative imaging, were used. For all the patients enrolled in our study, these anatomical landmarks matched o ur non-rigid registration results with intra-operative findings. Therefore, the results were considered clinically useful.
Discussion
Enhanced navigation using pre-operative multi-modal images (fMRI, MRI-DTI) can provide useful information during tumor resection. However, brain shift can produce non-linear changes in the anatomy that will induce inaccuracies of conventional rigid registration techniques (used by current commercial neurosurgery navigation systems).
Modeling the behavior of the brain remains a key issue in providing a priori knowledge for image-guided surgery. Sophisticated models of brain tissue undergoing surgery are presented and validated in Miga et al. (2000) and Platenik et al. (2002). However, a practical difficulty of these models is the extensive time necessary to mesh the brain and solve the problem, which is too long for intra-operative purposes. Similar approaches have investigated the use of brain biomechanical models updated during neurosurgery, using brain surface measurements based on laser range scanner (Miga et al. 2003) or stereo vision system based (Skrinjar, Nabavi, and Duncan 2002).
The use of intraoperative ultrasound in order to provide data that could be used to update pre-operative models to account for brain shift has been also investigated. (Dey et al. 2002; Gobbi and Peters 2003) have utilized a tracked, free-hand ultrasound probe. Pennec, Cachier, and Ayache (2003) demonstrated a system that utilizes full-volume, intensity-based registration and 3D ultrasound, rather than the landmark-based methods discussed above.
These studies show that several intraoperative imaging modalities have the potential to accurately measure brain deformation, but that further study is needed. Methods that require the identification of landmarks in the intraoperative image need to overcome the difficulty of robust landmark detection in those images. Also, further evaluation in the operating room is needed to determine how well these methods can capture, and adjust for, true brain deformation, as opposed to the mechanically-induced simulations that have been investigated thus far.
An interesting aspect is how our mathematical model for DTI reorientation compares with the intra-operative DTI acquired at 0.5T. Our group has demonstrated the feasibility of acquiring intra-operative diffusion weighted imaging in the 0.5 T MRI (Mamata et al. 2001). However, for neurosurgical procedures, this is impractical due to the long acquisition time (scan time per slice 94 seconds for diffusion tensor imaging, and 46 seconds for diffusion trace imaging), and poor spatial resolution of the diffusion weighted images (rectangular FOV = 260×195 mm; effective slice thickness = 7 mm; slice gap = 3mm; matrix size = 128 × 48). Therefore, for our prospective study we did not have diffusion weighted intra-operative images of the patients. Nevertheless, future studies will be performed on animals and phantoms to compare the results of intra-operative DTI with our mathematical estimation.
The similarity between the DTI datasets will be computed based on the metrics recently introduced by Fletcher et al. (2004), Moakher et al. (2005), Arsigny et al. (2006), and Schwartzman (2006).
An important aspect is the way multi-modal imaging information is presented during tumor resection to neurosurgeon. Relevant fiber tracts for each patient are displayed during tumor resection, along with information about the fMRI activation areas. Because the DTI data is also non-rigidly registered with the brain anatomy during the tumor resection, extracted fiber tracts correspond precisely to the brain changes induced by the surgery. And our system works effectively even if some parts of a fiber tract system (such as visual pathway) were removed during the surgical procedure, or in a case where fiber tracts become visible after resection that were not before the resection (due to compression).
The proposed solution fits well within the time constraints imposed by neurosurgery at the MRT. An initial SPGR scan is acquired before the craniotomy (scanning time is approximately 12 minutes). The first rigid registration between the pre-operative T1w MRI and the first intra-operative SPGR is performed, and an affine transformation is estimated. Less than 30 seconds are needed to perform this task on a conventional workstation. The other pre-operative images (T2, fMRI, DTI) are also aligned with the intra-operative SPGR based on the estimated affine transformation (in approximately 30 seconds on a conventional workstation). At this point, neurosurgeons will express an interest in having the fMRI and DTI information during the navigation, to decide about the best strategy for the craniotomy.
As the surgery progresses (depending on the tumor histology and location), brain deformation inevitably occurs, and the initial alignment becomes inaccurate. A new SPGR scan is acquired, typically after 45 minutes. The pre-operative T1w MRI scan is non-rigidly registered with the intra-operative scan, using the grid-computing architecture previously presented, and the results are available in less than 5 minutes. The other pre-operative datasets (T2w, fMRI, DTI) are also aligned with the intra-operative brain anatomy in less than 1 minute on a conventional workstation. The multi-modal images are registered and available for navigation in less than 7 minutes from the scan acquisition time. At this point, the neurosurgeons are interested in the fiber tracts that can be found in the vicinity of the tumor. Towards the end of neurosurgery, additional intra-operative SPGR scans are often acquired, typically every 30 minutes. The non-rigid registration process is performed for each new SPGR scan, and hence, the time constraints are extremely significant.
An interesting question is whether the introduced technology could potentially improve the patients outcome, by reducing the tumor residual, while avoiding clinical deficits. Our group has recently published a study that assesses the main variables that affect the complete magnetic resonance (MR) imaging–guided resection of supratentorial low-grade gliomas (Talos et al. 2006). Of all the variables assessed individually in the univariate analyses, 11 were found to be significantly associated with incomplete tumor resection (Table 1). Among the tumor characteristics, an ill defined tumor margin on T2-weighted MR images, LGO or low-grade mixed oligoastrocytoma histopathologic tumor type (ie, both types appear to be more difficult to resect than LGA), and large tumor volume were found to be associated with incomplete resection. Furthermore, tumor involvement of the following functionally critical structures led to incomplete resection: corpus callosum, CST, insular lobe, middle cerebral artery, primary motor cortex, optic radiation, visual cortex, and basal ganglia (one-sided P < .05 for all correlations).Therefore, in a future prospective study, we will employ our technology on a large number of patients that have predictors of potentially incomplete tumor resection. The intra-operative information from DTI, as provided with our novel system, can contribute to improved patients' outcome.
Conclusions
The utility of enhanced visualization during neurosurgery procedures is clearly demonstrated in the literature. To date, commercial image-guidance neurosurgery systems can only rigidly align the pre-operative, high resolution imaging data with the intra-operative imaging data. However, rigid alignment can induce errors as high as 20 mm, as measured in our study. To date, there is no fully volumetric, non-rigid registration of brain deformations demonstrated in a clinical environment during a neurosurgical procedure.
We demonstrated the feasibility of our system utilizing volumetric non-rigid registration, and quantitatively measured its value with respect to the commercially available state-of-the-art technology, based on rigid registration methods only. We found that a statistically significant increase in alignment accuracy was achieved using non-rigid registration.
Five major contributions are presented in our manuscript: (i) our study is prospective, with patients enrolled over 1 year; (ii) the alignments are achieved during the neurosurgery; (iii) we aligned both fMRI and DTI using non-rigid registration; (iv) we presented quantitative results assessing the accuracy of the rigid and the non-rigid registration; (v) we utilized prospectively a new volumetric non-rigid registration scheme, that has previously been assessed only retrospectively.
Acknowledgments
This investigation was supported in part by NSF ITR 0426558, a research grant from CIMIT, grant RG 3478A2/2 from the NMSS, a grant from the Brain Science Foundation and by NIH grants R21 MH067054, R01 RR021885, P41 RR013218, R03 EB006515, and U41 RR019703.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander AL, Badie B, Field AS. Diffusion tensor MRI depicts white matter reorganization after surgery. Proceedings of the ISMRM 11th Scientific Meeting; Berkeley. International Society of Magnetic Resonance in Medicine; 2003. [Google Scholar]
- Arsigny V, Fillard P, Pennec X, Ayache N. Log-Euclidean Metrics for Fast and Simple Calculus on. Diffusion Tensors Magnetic Resonance in Medicine. 2006 Aug;56(2):411–421. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- Atlas SW, Howard RS, Maldjian J, Alsop D, Detre JA, Listerud J, D'Esposito M, Judy KD, Zager E, Stecker M. Functional magnetic resonance imaging of regional brain activity in patients with intracerebral gliomas: Findings and implications for clinical management. Neurosurgery. 1996;38:329–338. doi: 10.1097/00006123-199602000-00019. [DOI] [PubMed] [Google Scholar]
- Audette MA. PhD. McGill University; 2003. Anatomical Surface Identification, Range-sensing and Registration for Characterizing Intrasurgical Brain Deformation. [Google Scholar]
- Audette MA, Siddiqi K, Ferrie FP, Peters TM. An integrated range-sensing, segmentation and registration framework for the characterization of intra-surgical brain deformations in image-guided surgery. Computer Vision and Image Understanding. 2003;89:226–251. [Google Scholar]
- Beppu T, Inoue T, Shibata Y, Kurose A, Arai H, Ogasawara K, Ogawa A, Nakamura S, Kabasawa H. Measurement of fractional anisotropy using diffusion tensor MRI in supratentorial astrocytic tumors. J Neuro-Oncol. 2003;63:109–116. doi: 10.1023/a:1023977520909. [DOI] [PubMed] [Google Scholar]
- Berman JI, Berger MS, Mukherjee P, Henry RG. Diffusion-tensor imaging-guided tracking of fibers of the pyramidal tract combined with intraoperative cortical stimulation mapping in patients with gliomas. J Neurosurg. 2004 Jul;101(1):66–72. doi: 10.3171/jns.2004.101.1.0066. [DOI] [PubMed] [Google Scholar]
- Black PML, Moriarty T, Alexander E, et al. Development and implementation of intraoperative magnetic resonance imaging and its neurosurgical applications. Neurosurgery. 1997;41:831–845. doi: 10.1097/00006123-199710000-00013. [DOI] [PubMed] [Google Scholar]
- Black PM, Alexander E, 3rd, Martin C, Moriarty T, Nabavi A, Wong TZ, Schwartz RB, Jolesz F. Craniotomy for tumor treatment in an intraoperative magnetic resonance imaging unit. Neurosurgery. 1999 Sep;45(3):423–31. doi: 10.1097/00006123-199909000-00001. [DOI] [PubMed] [Google Scholar]
- Bradley WG. Related Articles, Achieving gross total resection of brain tumors: intraoperative MR imaging can make a big difference. AJNR Am J Neuroradiol. 2002 Mar;23(3):348–9. [PMC free article] [PubMed] [Google Scholar]
- Chinzei K, Miller K. Towards MRI guided surgical manipulator. Med Sci Monit. 2001;7(1):153–63. [PubMed] [Google Scholar]
- Chrisochoides N, Fedorov A, Kot A, Archip N, Black PM, Clatz O, Golby A, Kikinis R, Warfield SK. Toward Real-Time Image Guided Neurosurgery using Distributed and Grid Computing. To be presented at Supercomputing. 2006 [Google Scholar]
- Clark CA, Barrick TR, Murphy MM, Bell BA. White matter fiber tracking in patients with space-occupying lesions of the brain: a new technique for neurosurgical planning? Neuroimage. 2003;20:1601–1608. doi: 10.1016/j.neuroimage.2003.07.022. [DOI] [PubMed] [Google Scholar]
- Clatz O, Delingette H, Talos IG, Golby AJ, Kikinis R, Jolesz FA, Ayache N, Warfield SK. Robust Non-Rigid Registration to Capture Brain Shift from Intra-Operative MRI. IEEE Transactions on Medical Imaging. 2005;24:1417–1427. doi: 10.1109/TMI.2005.856734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus EB, Horlacher A, Hsu L, Schwartz RB, Dello-Iacono D, Talos F, Jolesz FA, Black PM. Survival rates in patients with low-grade glioma after intraoperative magnetic resonance image guidance. Cancer. 2005 Mar 15;103(6):1227–33. doi: 10.1002/cncr.20867. [DOI] [PubMed] [Google Scholar]
- Coenen VA, Krings T, Axer H, et al. Intraoperative three-dimensional visualization of the pyramidal tract in a neuronavigation system (PTV) reliably predicts true position of principal motor pathways. Surg Neurol. 2003;60:381–390. doi: 10.1016/s0090-3019(03)00392-6. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Shen D, Mohamed A, Kyriacou SK. A framework for predictive modeling of anatomical deformations. IEEE Trans Med Imaging. 2001;20(8):836–43. doi: 10.1109/42.938251. [DOI] [PubMed] [Google Scholar]
- Dengler J, Schmidt M. The Dynamic Pyramid - A Model for Motion Analysis with Controlled Continuity. International Journal of Pattern Recognition and Artificial Intelligence. 1988;2(2):275–286. [Google Scholar]
- Dey D, Gobbi DG, Slomka PJ, Surry KJ, Peters TM. Automatic fusion of freehand endoscopic brain images to three-dimensional surfaces: creating stereoscopic panoramas. IEEE Trans Med Imaging. 2002;21(1):23–30. doi: 10.1109/42.981231. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Chrisochoides N, Kikinis R, Warfield SK. An evaluation of three approaches to tetrahedral mesh generation for deformable registration of MR images. Proceedings of IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI 2006) [Google Scholar]
- Ferrant M, Nabavi A, Macq B, Black PM, Jolesz FA, Kikinis R, Warfield SK. Serial registration of intraoperative MR images of the brain. Med Image Anal. 2002;6(4):337–59. doi: 10.1016/s1361-8415(02)00060-9. [DOI] [PubMed] [Google Scholar]
- Ferrant M, Nabavi A, Macq B, Jolesz FA, Kikinis R, Warfield SK. Registration of 3D intraoperative MR images of the brain using a finite-element biomechanical model. IEEE Trans Med Imaging. 2001;20(12):1384–97. doi: 10.1109/42.974933. [DOI] [PubMed] [Google Scholar]
- Ferrant M, Warfield SK, Nabavi A, Macq B, Kikinis R. Registration of 3D Intraoperative MR Images of the Brain Using a Finite Element Biomechanical Model. Paper read at MICCAI 2000: Third International Conference on Medical Robotics, Imaging And Computer Assisted Surgery; Oct 11-14; Pittsburgh, PA, USA. 2000. [Google Scholar]
- Field AS, Alexander AL, Wu YC, Hasan KM, Witwer B, Badie B. Diffusion tensor eigenvector directional color imaging patterns in the evaluation of cerebral white matter tracts altered by tumor. J Magn Reson Imaging. 2004;20:555–562. doi: 10.1002/jmri.20169. [DOI] [PubMed] [Google Scholar]
- Fletcher PT, Joshi SC. Principal geodesic analysisee on symmetric spaces: Statistics of diffusion tensors. Proc. of CVAMIA and MMBIA Workshops; Prague, Czech Republic. May 15, 2004; Springer; 2004. pp. 87–98. LNCS 3117. [Google Scholar]
- Friston K, Holmes A, Worsley K, et al. Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp. 1995;(2):189–210. [Google Scholar]
- Friston K, Holmes A, Poline JB, et al. Detecting activations in PET and fMRI : levels of inference and power. Neuroimage. 1996;(4):223–235. doi: 10.1006/nimg.1996.0074. [DOI] [PubMed] [Google Scholar]
- Ganslandt O, Fahlbusch R, Nimsky C, et al. Functional neuronavigation with magnetoencephalography: outcome in 50 patients with lesions around the motor cortex. J Neurosurg. 1999;91:73–79. doi: 10.3171/jns.1999.91.1.0073. [DOI] [PubMed] [Google Scholar]
- Gobbi DG, Peters TM. Generalized 3D nonlinear transformations for medical imaging: an object-oriented implementation in VTK. Comput Med Imaging Graph. 2003;27(4):255–265. doi: 10.1016/s0895-6111(02)00091-5. [DOI] [PubMed] [Google Scholar]
- Gralla J, Ganslandt O, Kober H, Buchfelder M, Fahlbusch R, Nimsky C. Image-guided removal of supratentorial cavernomas in critical brain areas: application of neuronavigation and intraoperative magnetic resonance imaging. Minim Invasive Neurosurg. 2003 Apr;46(2):72–7. doi: 10.1055/s-2003-39338. [DOI] [PubMed] [Google Scholar]
- Guimond A, Guttmann CRG, Warfield SK, Westin CF. Deformable Registration of DT-MRI Data Based On Transformation Invariant Tensor Characteristics. Paper read at International Symposium on Biomedical Imaging; Washington D.C.. 2002. [Google Scholar]
- Haberg A, Kvistad KA, Unsgard G, Haraldseth O. Preoperative blood oxygen level-dependent functional magnetic resonance imaging in patients with primary brain tumors: clinical application and outcome. Neurosurgery. 2004 Apr;54(4):902–14. doi: 10.1227/01.neu.0000114510.05922.f8. [DOI] [PubMed] [Google Scholar]
- Hall WA, Kowalik K, Liu H, Truwit CL, Kucharezyk J. Costs and benefits of intraoperative MR-guided brain tumor resection. Acta Neurochir Suppl. 2003;85:137–42. doi: 10.1007/978-3-7091-6043-5_19. [DOI] [PubMed] [Google Scholar]
- Hausdorff F. Set Theory. 2nd. Chelsea Pub. co.; 1962. [Google Scholar]
- Hendler T, Pianka P, Sigal M, Kafri M, Ben-Bashat D, Constantini S, Graif M, Fried I, Assaf Y. Delineating gray and white matter involvement in brain lesions: three-dimensional alignment of functional magnetic resonance and diffusion-tensor imaging. J Neurosurg. 2003;99:1018–1027. doi: 10.3171/jns.2003.99.6.1018. [DOI] [PubMed] [Google Scholar]
- Holodny AI, Schwartz TH, Ollenschleger M, Liu WC, Schulder M. Tumor involvement of the corticospinal tract: diffusion magnetic resonance tractography with intraoperative correlation. J Neurosurg. 2001;95:1082. doi: 10.3171/jns.2001.95.6.1082. [DOI] [PubMed] [Google Scholar]
- Holodny AI, Ollenschleger MD, Liu WC, Schulder M, Kalnin AJ. Identification of the corticospinal tracts achieved using bloodoxygen-level-dependent and diffusion functional MR imaging in patients with brain tumors. AJNR Am J Neuroradiol. 2001;22:83–88. [PMC free article] [PubMed] [Google Scholar]
- Jannin P, Morandi X, Fleig OJ, Le Rumeur E, Toulouse P, Gibaud B, Scarabin JM. Integration of sulcal and functional information for multimodal neuronavigation. J Neurosurg. 2002 Apr;96(4):713–23. doi: 10.3171/jns.2002.96.4.0713. [DOI] [PubMed] [Google Scholar]
- Jolesz FA, Nabavi A, Kikinis R. Integration of interventional MRI with computer-assisted surgery. J Magn Reson Imaging. 2001 Jan;13(1):69–77. doi: 10.1002/1522-2586(200101)13:1<69::aid-jmri1011>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kamada K, Houkin K, Takeuchi F, et al. Visualization of the eloquent motor system by integration of MEG, functional, and anisotropic diffusion-weighted MRI in functional neuronavigation. Surg Neurol. 2003;59:352–362. doi: 10.1016/s0090-3019(03)00018-1. [DOI] [PubMed] [Google Scholar]
- Keles GE. Intracranial neuronavigation with intraoperative magnetic resonance imaging. Curr Opin Neurol. 2004 Aug;17(4):497–500. doi: 10.1097/01.wco.0000137543.14610.fd. [DOI] [PubMed] [Google Scholar]
- Knauth M, Wirtz CR, Tronnier VM, Aras N, Kunze S, Sartor K. Intraoperative MR imaging increases the extent of tumor resection in patients with high-grade gliomas. AJNR Am J Neuroradiol. 1999 Oct;20(9):1642–6. [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Raabe A, Hattingen E, Szelenyi A, Yahya H, Hermann E, Zimmermann M, Seifert V. Functional magnetic resonance imaging-integrated neuronavigation: correlation between lesion-to-motor cortex distance and outcome. Neurosurgery. 2004 Oct;55(4):904–14. doi: 10.1227/01.neu.0000137331.35014.5c. [DOI] [PubMed] [Google Scholar]
- Kucharczyk W, Bernstein M. Do the benefits of image guidance in neurosurgery justify the costs? From stereotaxy to intraoperative MR. AJNR Am J Neuroradiol. 1997 Nov-Dec;18(10):1855–9. [PMC free article] [PubMed] [Google Scholar]
- Laundre BJ, Jellison BJ, Badie B, Alexander AL, Field AS. Diffusion tensor imaging of the corticospinal tract before and after mass resection as correlated with clinical motor findings: preliminary data. AJNR Am J Neuroradiol. 2005 Apr;26(4):791–6. [PMC free article] [PubMed] [Google Scholar]
- Laws ER, Taylor WF, Clifton MB, Okazaki H. Neurosurgical management of low-grade astrocytoma of the cerebral hemispheres. J Neurosurg. 1984;61:665–673. doi: 10.3171/jns.1984.61.4.0665. [DOI] [PubMed] [Google Scholar]
- Lehericy S, Duffau H, Cornu P, Capelle L, Pidoux B, Carpentier A, Auliac S, Clemenceau S, Sichez JP, Bitar A, Valery CA, Van Effenterre R, Faillot T, Srour A, Fohanno D, Philippon J, Le Bihan D, Marsault C. Correspondence between functional magnetic resonance imaging somatotopy and individual brain anatomy of the central region: Comparison with intraoperative stimulation in patients with brain tumors. J Neurosurg. 2000;92:589–598. doi: 10.3171/jns.2000.92.4.0589. [DOI] [PubMed] [Google Scholar]
- Lu S, Ahn D, Johnson G, Cha S. Peritumoral diffusion tensor imaging of high-grade gliomas and metastatic brain tumors. Am J Neuroradiol. 2003;24:937–941. [PMC free article] [PubMed] [Google Scholar]
- Lunn K, Paulsen K, Roberts DW, Kennedy F, Hartov A, Platenik LA. Nonrigid brain registration: synthesizing full volume deformation fields from model basis solutions constrained by partial volume intraoperative data. Computer Vision and Image Understanding. 2003;89(2):299–317. [Google Scholar]
- Maldjian JA, Schulder M, Liu WC, Mun IK, Hirschorn D, Murthy R, Carmel P, Kalnin A. Intraoperative functional MRI using a real-time neurosurgical navigation system. J Comput Assist Tomogr. 1997;21:910–912. doi: 10.1097/00004728-199711000-00013. [DOI] [PubMed] [Google Scholar]
- Mamata Y, Mamata H, Nabavi A, Kacher DF, Pergolizzi RS, Jr, Schwartz RB, Kikinis R, Jolesz FA, Maier SE. Intraoperative diffusion imaging on a 0.5 Tesla interventional scanner. J Magn Reson Imaging. 2001 Jan;13(1):115–9. doi: 10.1002/1522-2586(200101)13:1<115::aid-jmri1017>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Miga MI, Paulsen KD, Hoopes PJ, Kennedy FE, Hartov A, Roberts DW. In vivo modeling of interstitial pressure in the brain under surgical load using finite elements. J Biomech Eng. 2000;122(4):354–63. doi: 10.1115/1.1288207. [DOI] [PubMed] [Google Scholar]
- Miga MI, Paulsen KD, Hoopes PJ, Kennedy FE, Jr, Hartov A, Roberts DW. In vivo quantification of a homogeneous brain deformation model for updating preoperative images during surgery. IEEE Trans Biomed Eng. 2000;47(2):266–73. doi: 10.1109/10.821778. [DOI] [PubMed] [Google Scholar]
- Miga MI, Paulsen KD, Kennedy FE, Hoopes PJ, Hartov A, Roberts DW. In Vivo Analysis of Heterogeneous Brain Deformation Computations for Model-Updated Image Guidance. Comput Methods Biomech Biomed Engin. 2000;3(2):129–146. doi: 10.1080/10255840008915260. [DOI] [PubMed] [Google Scholar]
- Miga MI, Paulsen KD, Lemery JM, Eisner SD, Hartov A, Kennedy FE, Roberts DW. Model-updated image guidance: initial clinical experiences with gravity-induced brain deformation. IEEE Trans Med Imaging. 1999;18(10):866–74. doi: 10.1109/42.811265. [DOI] [PubMed] [Google Scholar]
- Miga MI, Roberts DW, Hartov A, Eisner S, Lemery J, Kennedy FE, Paulsen KD. Updated neuroimaging using intraoperative brain modeling and sparse data. Stereotact Funct Neurosurg. 1999;72(2-4):103–6. doi: 10.1159/000029707. [DOI] [PubMed] [Google Scholar]
- Miga MI, Roberts DW, Kennedy FE, Platenik LA, Hartov A, Lunn KE, Paulsen KD. Modeling of retraction and resection for intraoperative updating of images. Neurosurgery. 2001;49(1):75–84. doi: 10.1097/00006123-200107000-00012. discussion 84-5. [DOI] [PubMed] [Google Scholar]
- Miga MI, Sinha TK, Cash DM, Galloway RL, Weil RJ. Cortical surface registration for image-guided neurosurgery using laser-range scanning. IEEE Trans Med Imaging. 2003;22(8):973–85. doi: 10.1109/TMI.2003.815868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. Biomechanics of Brain for Computer Integrated Surgery. Warsaw University of Technology Publishing House; 2002. [Google Scholar]
- Miller K, Chinzei K. Constitutive modelling of brain tissue: experiment and theory. J Biomech. 1997;30(11-12):1115–21. doi: 10.1016/s0021-9290(97)00092-4. [DOI] [PubMed] [Google Scholar]
- Miller K, Chinzei K. New UWA robot--possible application to robotic surgery. Biomed Sci Instrum. 2000;36:135–40. [PubMed] [Google Scholar]
- Miller K, Chinzei K. Mechanical properties of brain tissue in tension. J Biomech. 2002;35(4):483–90. doi: 10.1016/s0021-9290(01)00234-2. [DOI] [PubMed] [Google Scholar]
- Miller K, Chinzei K, Orssengo G, Bednarz P. Mechanical properties of brain tissue in-vivo: experiment and computer simulation. J Biomech. 2000;33(11):1369–76. doi: 10.1016/s0021-9290(00)00120-2. [DOI] [PubMed] [Google Scholar]
- Moakher M. A differential geometry approach to the geometric mean of symmetric positive-definite matrices. SIAM Journal on Matrix Analysis and Applications. 2005;26:735–747. [Google Scholar]
- Moller-Hartmann W, Krings T, Coenen VA, et al. Preoperative assessment of motor cortex and pyramidal tracts in central cavernoma employing functional and diffusion-weighted magnetic resonance imaging. Surg Neurol. 2002;58:302–307. doi: 10.1016/s0090-3019(02)00869-8. [DOI] [PubMed] [Google Scholar]
- Mori S, Frederiksen K, van Zijl PCM, et al. Brain white matter anatomy of tumor patients evaluated with diffusion tensor imaging. Ann Neurol. 2002;51:377–380. doi: 10.1002/ana.10137. [DOI] [PubMed] [Google Scholar]
- Mori S, Van Zijl PC. Fiber tracking: principles and strategies—A technical review. NMR Biomed. 2002;15:468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Mueller WM, Yetkin FZ, Hammeke TA, Morris GL, 3rd, Swanson SJ, Reichert K, Cox R, Haughton VM. Functional magnetic resonance imaging mapping of the motor cortex in patients with cerebral tumors. Neurosurgery. 1996 Sep;39(3):515–20. doi: 10.1097/00006123-199609000-00015. [DOI] [PubMed] [Google Scholar]
- Nabavi A, Black PM, Gering DT, Westin CF, Mehta V, Pergolizzi RS, Jr, Ferrant M, Warfield SK, Hata N, Schwartz RB, Wells WM, 3rd, Kikinis R, Jolesz FA. Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery. 2001 Apr;48(4):787–97. doi: 10.1097/00006123-200104000-00019. [DOI] [PubMed] [Google Scholar]
- Nimsky C, Ganslandt O, Kober H, et al. Integration of functional magnetic resonance imaging supported by magnetoencephalography in functional neuronavigation. Neurosurgery. 1999;44:1249–1256. doi: 10.1097/00006123-199906000-00044. [DOI] [PubMed] [Google Scholar]
- Nimsky C, Ganslandt O, Merhof D, Sorensen AG, Fahlbusch R. Intraoperative visualization of the pyramidal tract by diffusion-tensor-imaging-based fiber tracking. Neuroimage. 2005 Dec 16; doi: 10.1016/j.neuroimage.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Nimsky C, Ganslandt O, Hastreiter P, Wang R, Benner R, Sorensen AG, Fahlbusch R. Intraoperative Diffusion-Tensor MR Imaging: Shifting of White Matter Tracts during Neurosurgical Procedures— Initial Experience. Radiology. 2005;234:218–225. doi: 10.1148/radiol.2341031984. [DOI] [PubMed] [Google Scholar]
- Nimsky C, Ganslandt O, Merhof D, Sorensen AG, Fahlbusch R, Nimsky C, Ganslandt O, Hastreiter P, Wang R, Benner T, Sorensen AG, Fahlbusch R. Preoperative and intraoperative diffusion tensor imaging-based fiber tracking in glioma surgery. Neurosurgery. 2005;56(1):130–7. doi: 10.1227/01.neu.0000144842.18771.30. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci U S A. 1990;87:9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JP, Whalen S, Branco DM, Petrovich NM, Knierim KE, Golby AJ. Integrated image- and function-guided surgery in eloquent cortex: a technique report. Int J Med Robotics Comput Assist Surg. 2006;2:75–83. doi: 10.1002/rcs.82. [DOI] [PubMed] [Google Scholar]
- Park HJ, Kubicki M, Westin CF, Talos IF, Brun A, Peiper S, Kikinis R, Jolesz FA, McCarley RW, Shenton ME. Method for combining information from white matter fiber tracking and gray matter parcellation. AJNR Am J Neuroradiol. 2004 Sep;25(8):1318–24. [PMC free article] [PubMed] [Google Scholar]
- Paulsen KD, Miga MI, Kennedy FE, Hoopes PJ, Hartov A, Roberts DW. A computational model for tracking subsurface tissue deformation during stereotactic neurosurgery. IEEE Trans Biomed Eng. 1999;46(2):213–25. doi: 10.1109/10.740884. [DOI] [PubMed] [Google Scholar]
- Pennec X, Cachier P, Ayache N. Tracking brain deformations in time sequences of 3D US images. Pattern Recognition Letters. 2003;24:810–813. [Google Scholar]
- Philippon JH, Clemenceau SH, Fauchon FH, Foncin JF. Supratentorial low grade astrocytomas in adults. Neurosurgery. 1993;32:554–559. doi: 10.1227/00006123-199304000-00010. [DOI] [PubMed] [Google Scholar]
- Piepmeier J, Christopher S, Spencer D, et al. Variations in the natural history and survival of patients with supratentorial low-grade astrocytomas. Neurosurgery. 1996;38:872–879. doi: 10.1097/00006123-199605000-00002. [DOI] [PubMed] [Google Scholar]
- Platenik LA, Miga MI, Roberts DW, Lunn KE, Kennedy FE, Hartov A, Paulsen KD. In vivo quantification of retraction deformation modeling for updated image-guidance during neurosurgery. IEEE Trans Biomed Eng. 2002;49(8):823–35. doi: 10.1109/TBME.2002.800760. [DOI] [PubMed] [Google Scholar]
- Price S, Burnet N, Donovan T, Green H, Pena A, Antoun N, Pickard J, Carpenter T, Gillard J. Diffusion tensor imaging of brain tumours at 3T: a potential tool for assessing white matter tract invasion? Clin Radiol. 2003;58:455–462. doi: 10.1016/s0009-9260(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Reithmeier T, Krammer M, Gumprecht H, Gerstner W, Lumenta CB. Neuronavigation combined with electrophysiological monitoring for surgery of lesions in eloquent brain areas in 42 cases: a retrospective comparison of the neurological outcome and the quality of resection with a control group with similar lesions. Minim Invasive Neurosurg. 2003 Apr;46(2):65–71. doi: 10.1055/s-2003-39334. [DOI] [PubMed] [Google Scholar]
- Rexilius J. Physics-Based Nonrigid Registration for Medical Image Analysis. Master, Medical University of Luebeck; Luebeck, Germany: 2001. [Google Scholar]
- Rexilius J, Warfield SK, Guttmann CRG, Wei X, Benson R, Wolfson L, Shenton M, Handels H, Kikinis R. A Novel Nonrigid Registration Algorithm and Applications. Paper read at MICCAI '01: Fourth International Conference on Medical Image Computing and Computer-Assisted Intervention; Oct 14-17; Utrecht, the Netherlands. 2001. [Google Scholar]
- Roberts DW, Lunn K, Sun H, Hartov A, Miga M, Kennedy F, Paulsen K. Intra-operative image updating. Stereotact Funct Neurosurg. 2001;76(3-4):148–50. doi: 10.1159/000066712. [DOI] [PubMed] [Google Scholar]
- Roberts DW, Miga MI, Hartov A, Eisner S, Lemery JM, Kennedy FE, Paulsen KD. Intraoperatively updated neuroimaging using brain modeling and sparse data. Neurosurgery. 1999;45(5):1199–206. discussion 1206-7. [PubMed] [Google Scholar]
- Roberts DW, Sun H, Hartov A, Rick K, Paulsen K. Paper read at Medical Imaging 2003: Visualization, Image-Guided Procedures, and Display, at SPIE 2003. 2003. Using cortical vessels for patient registration during image-guided neurosurgery: a phantom study. [Google Scholar]
- Roessler K, Donat M, Lanzenberger R, Novak K, Geissler A, Gartus A, Tahamtan AR, Milakara D, Czech T, Barth M, Knosp E, Beisteiner R. Evaluation of preoperative high magnetic field motor functional MRI (3 Tesla) in glioma patients by navigated electrocortical stimulation and postoperative outcome. J Neurol Neurosurg Psychiatry. 2005 Aug;76(8):1152–7. doi: 10.1136/jnnp.2004.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Alzola J, Westin CF, Warfield SK, Alberola C, Maier S, Kikinis R. Nonrigid registration of 3D tensor medical data. Med Image Anal. 2002;6(2):143–61. doi: 10.1016/s1361-8415(02)00055-5. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ, Van Driessen K. Computing LTS Regression for Large Data Sets: University of Antwerp 1999 [Google Scholar]
- Ruiz-Alzola J, Westin CF, Warfield SK, Nabavi R, Kikinis R. Nonrigid Registration of 3D Scalar, Vector and Tensor Medical Data. Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science. 2000:541–550. [Google Scholar]
- Schmidt F, Dietrich J, Schneider JP, Thiele J, Lieberenz S, Werner A, Grunder W. Technological and logistic problems and first clinical results of an interventional 0.5-T MRI system used by various medical specialties. Radiologe. 1998;38:173–184. doi: 10.1007/s001170050339. [DOI] [PubMed] [Google Scholar]
- Schneider JP, Schulz T, Schmidt F, Dietrich J, Lieberenz S, Trantakis C, Seifert V, Kellermann S, Schober R, Schaffranietz L, Laufer M, Kahn T. Gross-total surgery of supratentorial low-grade gliomas under intraoperative MR guidance. AJNR Am J Neuroradiol. 2001 Jan;22(1):89–98. [PMC free article] [PubMed] [Google Scholar]
- Schneider JP, Trantakis C, Rubach M, Schulz T, Dietrich J, Winkler D, Renner C, Schober R, Geiger K, Brosteanu O, Zimmer C, Kahn T. Intraoperative MRI to guide the resection of primary supratentorial glioblastoma multiforme--a quantitative radiological analysis. Neuroradiology. 2005 Jul;47(7):489–500. doi: 10.1007/s00234-005-1397-1. [DOI] [PubMed] [Google Scholar]
- Schulder M, Carmel PW. Intraoperative magnetic resonance imaging: impact on brain tumor surgery. Cancer Control. 2003 Mar-Apr;10(2):115–24. doi: 10.1177/107327480301000203. [DOI] [PubMed] [Google Scholar]
- Schwartz RB, Hsu L, Wong TZ, et al. Intraoperative MR imaging guidance for intracranial neurosurgery: experience with the first 200 cases. Radiology. 1999;211:477–488. doi: 10.1148/radiology.211.2.r99ma26477. [DOI] [PubMed] [Google Scholar]
- Schwartzman A. Ph D thesis. Stanford University: Jun, 2006. Random ellipsoids and false discovery rates: statistics for diffusion tensor imaging data. [Google Scholar]
- Shinoura N, Yamada R, Kodama T, Suzuki Y, Takahashi M, Yagi K. Preoperative fMRI, tractography and continuous task during awake surgery for maintenance of motor function following surgical resection of metastatic tumor spread to the primary motor area. Minim Invasive Neurosurg. 2005 Apr;48(2):85–90. doi: 10.1055/s-2004-830227. [DOI] [PubMed] [Google Scholar]
- Sierra R. Nonrigid Registration of Diffusion Tensor Images. M.S., Swiss Federal Institute of Technology; Zurich, Switzerland: 2001. [Google Scholar]
- Skrinjar O, Nabavi A, Duncan J. Model-driven brain shift compensation. Med Image Anal. 2002;6(4):361–73. doi: 10.1016/s1361-8415(02)00062-2. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz CR, Knauth M, Staubert A, et al. Clinical evaluation and follow up results for intraoperative magnetic resonance imaging in neurosurgery. Neurosurgery. 2000;46:1120–1122. doi: 10.1097/00006123-200005000-00017. 2000. [DOI] [PubMed] [Google Scholar]
- Talos IF, O'Donnell L, Westin CF, Warfield SK, Wells WM, Yoo SS, Panych L, Golby A, Mamata H, Maier SE, Ratiu P, Guttmann CG, Black PML, Jolesz FA, Kikinis R. Diffusion tensor and functional MRI fusion with anatomical mri for image guided neurosurgery. Sixth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI'03); Montreal, Canada. pp. 407–415. [Google Scholar]
- Talos IF, Zou KH, Ohno-Machado L, Bhagwat JG, Kikinis R, Black PM, Jolesz FA. Supratentorial low-grade glioma resectability: statistical predictive analysis based on anatomic MR features and tumor characteristics. Radiology. 2006;239(2):506–13. doi: 10.1148/radiol.2392050661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala RP, Chu RM, Liu H, Truwit CL, Hall WA. Application of diffusion tensor imaging to magnetic resonance-guided brain tumor resection. Pediatr Neurosurg. 2003;39:39–43. doi: 10.1159/000070879. [DOI] [PubMed] [Google Scholar]
- Warfield SK, Kaus M, Jolesz FA, Kikinis R. Adaptive, template moderated, spatially varying statistical classification. Med Image Anal. 2000 Mar;4(1):43–55. doi: 10.1016/s1361-8415(00)00003-7. [DOI] [PubMed] [Google Scholar]
- Warfield SK, Talos F, Tei A, Bharatha A, Nabavi A, Ferrant M, McL Black P, Jolesz FA, Kikinis R. Real-Time Registration of Volumetric Brain MRI by Biomechanical Simulation of Deformation during Image Guided Neurosurgery. Comput Visual Sci. 2002;5:3–11. [Google Scholar]
- Warfield SK, Jolesz FA, Kikinis R. Real-Time Image Segmentation for Image-Guided Surgery. Paper read at SC 1998: High Performance Networking and Computing Conference; Nov 7-13; Orlando, FL, USA. 1998. [Google Scholar]
- Warfield SK, Ferrant M, Gallez X, Nabavi A, Jolesz FA, Kikinis R. Real-Time Biomechanical Simulation of Volumetric Brain Deformation for Image Guided Neurosurgery. Paper read at SC 2000: High Performance Networking and Computing Conference; 2000 Nov 4--10; Dallas, USA. 2000. [Google Scholar]
- Wells WM, 3rd, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. Med Image Anal. 1996 Mar;1(1):35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- Westin CF, Maier SE, et al. Processing and visualization for diffusion tensor MRI. Med Image Anal. 2002;6(2):93–108. doi: 10.1016/s1361-8415(02)00053-1. [DOI] [PubMed] [Google Scholar]
- Wieshmann UC, Symms MR, Parker GJM, et al. Diffusion tensor imaging demonstrates deviation of fibres in normal appearing white matter adjacent to a brain tumour. J Neurol Neurosurg Psychiatry. 2000;68:501–503. doi: 10.1136/jnnp.68.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieshmann UC, Clark CA, Symms MR, Franconi F, Barker GJ, Shorvon SD. Reduced anisotropy of water diffusion in structural cerebral abnormalities demonstrated with diffusion tensor imaging. Magn Reson Imaging. 1999;17:1269–1274. doi: 10.1016/s0730-725x(99)00082-x. [DOI] [PubMed] [Google Scholar]
- Witwer BP, Moftakhar R, Hasan KM, et al. Diffusion tensor imaging of white matter tracts in patients with cerebral neoplasm. J Neurosurg. 2002;97:568–575. doi: 10.3171/jns.2002.97.3.0568. [DOI] [PubMed] [Google Scholar]
- Witwer BP, Moftakhar R, Hasan KM, Deshmukh P, Haughton V, Field A, Arfanakis K, Noyes J, Moritz CH, Meyerand ME, Rowley HA, Alexander AL, Badie B. Diffusion-tensor imaging of white matter tracts in patients with cerebral neoplasm. J Neurosurg. 2002;97:568–575. doi: 10.3171/jns.2002.97.3.0568. [DOI] [PubMed] [Google Scholar]
- Yamada K, Kizu O, Mori S, Ito H, Nakamura H, Yuen S, Kubota T, Tanaka O, Akada W, Sasajima H, Mineura K, Nishimura T. Brain fiber tracking with clinically feasible diffusiontensor MR imaging: initial experience. Radiology. 2003;227:295–301. doi: 10.1148/radiol.2271020313. [DOI] [PubMed] [Google Scholar]