Abstract
Cellular membranes are heterogeneous in composition, and the prevailing theory holds that the structures responsible for this heterogeneity in vivo are small structures (10–200 nm), sterol- and sphingolipid-enriched, of different sizes, highly dynamic denominated rafts. Rafts are postulated to be platforms, which by sequestering different membrane components can compartmentalize cellular processes and regulate signaling pathways. Despite an enormous effort in this area, the existence of these domains is still under debate due to the characteristics of the structures itself: small in size and highly mobile, which from the technical point of view implies using techniques with high spatial and temporal resolution. In this report we measured rapid fluctuations of the normalized ratio of the emission intensity at two wavelengths of Laurdan, a membrane fluorescent dye sensitive to local membrane packing. We observed generalized polarization fluctuations in the plasma membrane of intact rabbit erythrocytes and Chinese hamster ovary cells that can be explained by the existence of tightly packed micro-domains moving in a more fluid background phase. These structures, which display different lipid packing, have different sizes; they are found in the same cell and in the entire cell population. The small size and characteristic high lipid packing indicate that these micro-domains have properties that have been proposed for lipid rafts.
Keywords: fluctuation spectroscopy, lipid packing defects, membrane microdomains
Membrane microdomains in vivo (rafts) are postulated to be part of a mechanism for fine-regulating a range of biological processes driven by transduction pathways, where the first step involves a protein–membrane interaction. Many authors have discussed the composition, distribution, and significance of the existence of lipid rafts in biological membranes; however, their existence in vivo is still controversial. At the Keystone Symposium on Lipid Rafts and Cell Function in 2006 (1) rafts were defined as small (10–200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains. Because these domains are small and transient, the study of membrane lateral organization requires the use of techniques with simultaneous high spatial and temporal resolution (2). The focus of this article is using a fluorescent probe that is sensitive to lipid packing in combination with scanning fluorescence correlation spectroscopy (FCS) that has very high temporal and spatial resolution.
Several techniques have been used to demonstrate the existence of rafts in vivo with different degrees of success. Methodologies as homo- and hetero-transfer, fluorescence polarization anisotropy, FCS, total internal reflection (TIRF), single-molecule and the super resolution techniques stimulated emission depletion (STED), photo-activated localization microscopy (PALM), and stochastic optical reconstruction microscopy (STORM) have been used to look for the elusive lipid rafts. All these techniques (and combination of them) support the existence of nano-domains and the fundamental role played by cholesterol (2, 3).
Traditional Laurdan generalized polarization (GP) imaging visualizes lipid domains with different packing degree if their size is above the micrometer range. However, Laurdan GP imaging alone does not seem to be equally useful in the quest of a signature of membrane heterogeneity in vivo at the subdiffraction scale. In erythrocytes (4), HeLa (5), and Chinese hamster ovary (CHO) cells (6) Laurdan GP images show a plasma membrane homogeneous in GP value without macrodomains segregation. One case has been reported where macrodomains were visualized using GP imaging and 3D projection analysis in machrophages. However, as pointed out by the authors, this may be a particular case (7). Celli et al. added to the high spatial resolution of Laurdan GP imaging the high temporal resolution of scanning FCS. This combined methodology was successfully used to characterize the formation of submicrometric lipid domains in secondary lipid mixtures by detecting GP fluctuations at the transition temperature (8, 9). In this report we use this combined methodology to detect GP packing fluctuations in biological membranes.
Detecting Laurdan GP Fluctuations
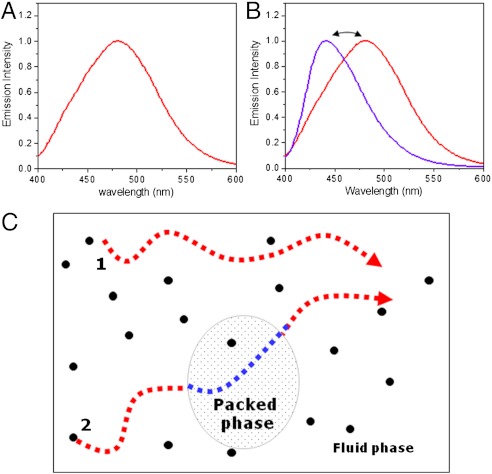
Laurdan, a membrane dye synthesized by Gregorio Weber in 1979, presents a large excited state dipole moment, which results in its ability to report the extent of water penetration into the bilayer surface due to the dipolar relaxation effect (10). Because water penetration has been correlated with lipid packing and membrane fluidity (11), Laurdan has been extensively used in membrane studies. A full discussion on the use and significance of GP Laurdan measurements (11, 12) and the combined use of FCS and Laurdan GP can be found in the literature (8, 9). The diagram in Fig. 1 explains the concept of Laurdan GP (Fig. 1 A and B) and the rational for using the Laurdan GP fluctuations measurements (Fig. 1C). The emission spectrum of Laurdan within a single phospholipid bilayer is centered at 490 nm when the lipids are in a disordered phase (Fig. 1A) and is shifted to the blue (around 440 nm) when the lipids are in a more packed phase (Fig. 1B). The GP function (13), GP = (I440 - I490)/(I440 + I490), gives a mathematically convenient and quantitative way to measure the emission shift.
Fig. 1.
Schematic of the Laurdan GP fluctuation measurements. (A) Emission spectra of Laurdan solubilized in DPPC small unilamellar vesicles at 50 °C). (B) Emission spectra of Laurdan solubilized in DPPC at 50 °C (red) and at 35 °C (blue) using excitation light at 340 nm. (C) Diagram showing the Laurdan molecules (small black dots) diffusing in a fluid phase (molecule 1) and crossing domains of a more tightly packed phase (molecule 2).
The combined use of FCS and Laurdan GP measures the fluctuations of the GP value in the membrane. Fig. 1C schematically explains the origin of these fluctuations. Laurdan molecules (small black dots) distribute homogeneously in the lipid bilayer and move freely. If a Laurdan molecule (molecule 1) moves in a homogeneous disordered phase (for example, the ld phase), its emission spectra (color) will not change; the response to membrane packing is the same along its trajectory. If the Laurdan molecule (molecule 2 in Fig. 1C) moves in a homogeneous fluid phase, but it crosses an area (domain) where the lipids are more packed, the molecule’s dipole will sense this difference and its spectra (color) will change when the molecule enters and leaves the domain. Changes in packing at the nanoscale have a minor effect on the average diffusion of Laurdan in the membrane (DLau parameter will not change), but the GP value fluctuates as the molecule enters and leaves the small domain. Consequently, in the scheme shown in Fig. 1C, fluctuations in GP value will be observed for molecule 2 but not for molecule 1.
Classical autocorrelation and cross-correlation analysis applied to the Laurdan intensity fluctuation measurements give information about molecules moving in and out of the illumination volume, but they cannot report changes in lipid packing at the nanoscale. Here we are not interested in the diffusion of Laurdan molecules, because the Laurdan fluorescence intensity per se is not informative of the lipid packing; instead we measure the fluctuation of the GP because the spectroscopic parameters report on the local packing of the membrane. If the GP is not changing, although the molecules could diffuse in and out of the excitation volume, we conclude that there is no difference in lipid packing. In our experiments we use a relatively large concentration of Laurdan so that regions of different packing can be “visualized” using fluctuation analysis as they move as nanometer-size objects in the membrane.
There are different regimes to consider:
domains much smaller than the point spread function (PSF); in this case, the amplitude of the GP correlation function should be proportional to the inverse of the number of domains; the relaxation time should depend on the mobility and size of the nano-domains;
domains comparable in size to the PSF; the GP fluctuations should tend to be slower but only few domains are present in the PSF, resulting in a larger value in the GP fluctuation amplitude;
very large domains; the GP fluctuations should be absent; however, the domains should become visible in the GP image.
We note that our approach of measuring the GP fluctuations is different than the usual FCS experiment in which the molecular fluctuations are measured and as such it has not been done before. In the conventional approach of looking at molecular fluctuations, the basic idea is to try to decrease the size of the PSF so that small domains carrying a specific marker can be visualized. Here, using a conventional-sized PSF, the number of independent nano-domains is inferred from the amplitude of the GP fluctuation. The diffusion of the entire domain is assumed to be slower than the diffusion of single Laurdan molecules. In our approach we don’t need to “anchor” the dye to the domain. On the contrary, the rapid diffusion of the dye in the membrane is beneficial. A crucial difference between measuring the diffusion of Laurdan alone and measuring the diffusion of GP domains is that we can detect packing micro-domain “directly” rather than resolving the diffusion of Laurdan in multiple components and measuring the fraction of molecules in these components (3, 14)
We observed GP fluctuations of different amplitude (GGP) and relaxation time (DGP) in membranes of live rabbit red blood cells (RRBC) and Chinese hamster ovary (CHO-K1) cells. We show that these GP fluctuations are due to diffusion of small structures (with different GP value than the background) through the illumination volume and may be related to functional rafts. On the basis of our observations we suggest a methodology to study domain size heterogeneity in live cells based on a GP fluctuation map obtained by the plot of the GP autocorrelation parameters GGP versus DGP.
Results
Laurdan GP Fluctuation in RRBC.
We performed experiments to test the existence of Laurdan GP fluctuations in the plasma membrane of RRBC at 37 °C. RRBC (8–9 μm in diameter; humans are 3–6 μm in diameter) were chosen because of their size and stability during the measurements. Only round, concave cells were chosen for the measurements. Equinocytes were excluded. As controls we used giant unilamellar vesicles (GUVs) of Palmitoyl oleoyl phosphatidyl choline (POPC) at 25 °C. At this temperature, far away from the reported Tm of -5 °C (15, 16), POPC presents a homogeneous liquid crystalline phase, and therefore no GP fluctuations are expected (an example is discussed in Fig. 1 for molecule 1).
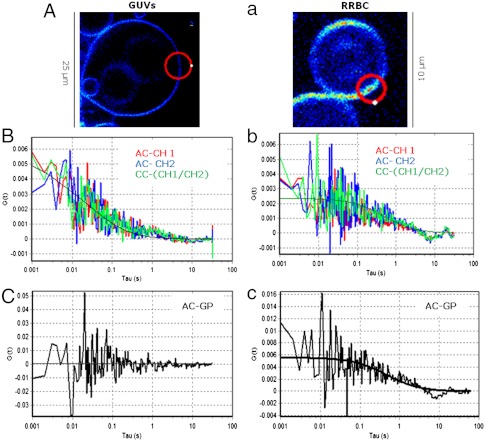
In this work we are using scanning FCS rather than single-point FCS. Two-photon SimFCS (sFCS), a method originally described by Berland (17), is particularly useful for working with membranes (18) because it measures the intensity along an orbit at several positions in the membrane without detectable photobleaching (8, 9). Representative results of applying this combined methodology are shown in Fig. 2 for POPC GUVs (Fig. 2 A–C) and RRBC (Fig. 2 a–c). One initial image was taken at the equatorial center of the GUV (Fig. 2A) or the RRBC (Fig. 2a). The orbit for the sFCS measurements was positioned in this image to cross twice the bilayer (red circle).
Fig. 2.
Representative results for Laurdan GP fluctuations for the control POPC at 25 °C (A–C) and RRBC at 37 °C (a–c). (A, a) Intensity images taken at the equatorial section show the location of the laser orbit (red circle) used to perform sFCS measurements. The orbit crosses twice the bilayer (see Methods), starts and finishes at the white point and moves clockwise. (B, b) Autocorrelation (AC) analysis applied to the independent channels (AC-CH1 in red and AC-CH2 in blue) together with the cross-correlation of the two channels (in green). (C, c) AC function applied to the pixels corresponding to the membrane on the GP carpet (AC-GP). Solid lines correspond to the fitting of the experimental data. For detailed explanations see Methods.
From the Laurdan intensity carpets (see Methods), the experimental autocorrelation (AC) for the independent channels (AC-CH1 and AC-CH2) and the cross-correlation (CC) of the two channels (CC-CH1-CH2) were obtained (Fig. 2B, b). Because it is the same molecule that produces fluctuations in the two channels, the individual autocorrelations and the cross-correlation curves overlap for the two systems as shown in Fig. 2B, b. This result shows that the cross-correlation of the two channels is not meaningful in these experiments. The experimental intensity AC curves for POPC were fitted using the equations for two diffusing species, and the average values for the DLau of Laurdan were equal to 4.5 + /-1.5 μm2/s and 0.08 + /-0.04 μm2/s (N = 64), the larger value is expected for Laurdan in POPC at 25 °C (Fig. 2B) and the second component we attributed to fluctuations produced by movements of the GUVs during the measurements. For Laurdan inside the RRBC at 37 °C we obtained a single value of 0.10 + /-0.04 μm2/s (N = 66). The errors are calculated using the standard deviation of measurements on different cells.
The Laurdan GP carpet contains the information about the fluctuations of the Laurdan GP value in the membrane which are sensitive to the local lipid packing. As expected for a homogeneous phase, the AC analysis of the GP carpet (see Methods) for POPC at 25 °C did not show autocorrelation (Fig. 2C). Instead, the analyses of the GP carpet corresponding to the RRBC membranes did show autocorrelation (Fig. 2c). The Laurdan GP fluctuations found in the RRBC membranes were not uniform. For the same cell at several locations and for different cells, the autocorrelation analysis shows fluctuations with different amplitude and apparent DGP.
We tested whether the GP fluctuations are due to the diffusion of nano-domains or rather to static fluctuations, for example, due to changes of Laurdan spectrum without a real motion. A common method to distinguish diffusion from static fluctuations is to systematically enlarge the PSF [see, for example, Didier Marguet et al. (14)]. If the relaxation is due to diffusion, the relaxation time becomes longer when the size of the PSF is increased. Instead static fluctuations should only decrease in amplitude when the PSF is enlarged without changing the relative time. A simple method (19) to enlarge the PSF in scanning FCS is to systematically average neighboring points along the orbit as to effectively broaden the PSF. We observed a decrease in DGP with pixel size (Fig. S1), which is the signature for the diffusion of nano-domains. The time constant obtained from the GP autocorrelations corresponds to the DGP of structures moving through the PSF.
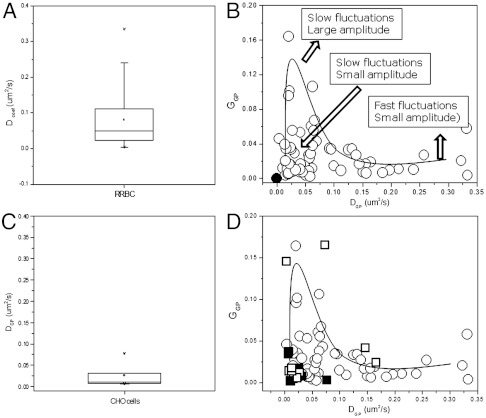
The parameters DGP and GGP for the GP analysis of all measurements in the RRBC are shown in Fig. 3 A and B. The total number of membranes measured was 64, 40 of which showed GP autocorrelation. Fig. 3A shows a large distribution of DGP, suggesting the existence of structures with different sizes. In Fig. 3B we plot GGP(0) versus DGP for all our data. For the cells not showing autocorrelation we assigned a (0,0) value. The plot of DGP versus GGP shows the heterogeneity of the GP fluctuations in the membrane of RRBC. Three cases can be distinguished: (i) fast fluctuations with small amplitude (high DGP and low GGP, (ii) slow fluctuations with large amplitude (low DGP high GGP), and (iii) slow fluctuations with low amplitudes (low DGP and GGP). Data were obtained from different cells and from different locations in the same cell. The same cell shows Laurdan GP fluctuations with different DGP and GGP. Because in the same cell the concentration of Laurdan should be homogeneous everywhere, the values of GGP and DGP for the GP fluctuations are largely independent on Laurdan concentration.
Fig. 3.
Laurdan GP fluctuation in RRBC (A and B) and CHO cells (C and D). Distribution of DGP (A and C). DGP versus GGP plots for the GP fluctuations obtained on RRBC (B and D, open circles) and CHO cells (D, closed squares). The black dot at (0,0) represents samples not showing autocorrelation. CHO cells incubated with rHDL for 2 h at 37 °C (D, open squares).
Laurdan GP Fluctuation in CHO-K1 Cells.
We applied the same methodology to test the existence of GP fluctuation in another cell type. CHO-K1 cells were grown to 70% confluence to allow the observation of single-cell membranes and kept at 37 °C during the measurements. We examined 15 cell membranes, 10 of which present autocorrelation. The average GP value for the membrane was 0.4, as reported before (6). The parameters DGP and GGP for the GP analysis of all the data are shown in Fig. 3 C and D. The DGP observed in control CHO cells (Fig. 3C) shows a narrower distribution than the one observed on RRBC (Fig. 3A), suggesting the existence of structures with more homogeneous sizes in this cell type. For comparison purposes in Fig. 3D the plot GGP (0) versus DGP show the data from CHO cells (closed square) and RRBC (open circles). The distribution of the fluctuations in control CHO are located in the area corresponding to slow fluctuations with small amplitude (Fig. 3D), suggesting a narrower size distribution of the domains in this cell type. To observe the effect of cholesterol in the distribution of the GP domains, we incubated CHO cells with rHDL particles for 2 h in order to remove cholesterol from the plasma membrane (16). After the treatment, the distribution of the fluctuations in CHO cells (Fig. 3D, open squares) changed in the direction of either slow fluctuations with large amplitudes or fast fluctuations with small amplitude, supporting the role of cholesterol to modify the size of the membrane domains.
Discussion
Functional lipids domains in vivo are conceived as tightly packed islands (sterol- and sphingolipid-enriched) immersed in a more fluid background. These domains of different sizes (10–200 nm) and highly dynamic (1) would create in the membrane heterogeneity in fluidity and composition. The focus of this work was to detect packing heterogeneity in the membrane of erythrocytes at the nanoscale using a methodology that combines two techniques: the high temporal resolution of sFCS and the sensitivity of Laurdan GP to membrane fluidity. In this approach we are not looking at clusters of proteins or the diffusion of single lipids, but we directly detect fluctuations in lipid packing.
For this study, we used a simple biological system, red blood cells. Earlier studies by Haverstick and Glaser on rabbit erythrocytes labeled with NBD-phospholipid (20, 21) using fluorescence digital imaging microscopy showed uneven distribution of the phospholipids in the membrane and attributed this heterogeneity to the presence of membrane domains of 1–2 μm in size. Fluorescent antibodies for CyaA (adenylate cyclase toxin) from Bordetella pertussis were used to identify rafts (regions rich in sphyingomyline and cholesterol easily disrupted by methyl-beta-cyclodextrin) as binding sites on sheep erythrocytes (22). Using Laurdan GP, imaging authors agreed on the observation that the plasma membrane of erythrocytes has similar fluidity than nucleated cells but does not show macrodomains separation in vivo. However the distribution of the Laurdan GP histogram suggested the existence of domains smaller than the pixel size (5).
Our results show that traditional autocorrelation analysis of Laurdan intensity fluctuations do not give information about lipid packing or domain existence. It gives information about the mobility of the Laurdan in the membrane as it moves in and out of the excitation volume. Our measurements indicate that Laurdan moves approximately 4 times slower in the erythrocyte membrane at 37 °C than in the POPC bilayer at 25 °C, a result that includes the effect of temperature and membrane composition.
The information about lipid packing domains in the membrane is given by the Laurdan GP fluctuations, detected only in the membranes of intact erythrocytes. Using the PSF scaling method analysis (19), we demonstrated the origin of the fluctuations being the diffusion of small structures with different Laurdan GP.
Diffusion and molecular sizes are related by the Stokes–Einstein equation. However, the application of this relationship is not possible when working with cells, where the mobility of the molecules is affected by other parameters such as cellular compartmentalization, binding of the protein to cellular structures, local viscosity, and the possible interactions with the intracellular matrix in the case of membrane proteins. For example, GFP expressed in cells show four times slower D than in solution (23). Proteins and protein complexes of different sizes show similar mobility (23). The DGP of the GP fluctuations found in the RRBC membranes falls in the same range of the values reported for proteins in membranes (23) and ribosomes (19); therefore, it is not possible to determine the size of the structure behind the GP fluctuations using only the DGP.
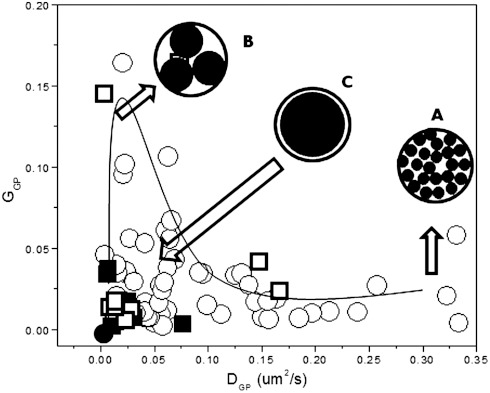
Considering the correlation plot of GGP and DGP and the relationship between GGP and the size of the PSF, we propose a possible scenario to explain the origin of Laurdan GP fluctuations. Fig. 4 shows the plot of GGP versus DGP for all measurements done in RRBC and CHO cells. This plot can be divided in four areas as shown in Fig. 3B.
Fig. 4.
Model for GP fluctuations in biological membrane. Nano-domains of different sizes will be present in the RRBC membrane: (A) Several small domains with radii in the range of approximately 25 nm, (B) big domains moving slowly with an average estimated radius of approximately 78 nm, and (C) even larger domains with estimated sizes between less than 300 nm and greater than 78 nm. Domains larger than the PSF will not show detectable fluctuations. Dotted circle represents the cross section of the PSF with a diameter of approximately 300 nm, and the black circled inside show the nano-domains responsible for the detected fluctuations.
Fast fluctuations with small amplitude: We propose that these conditions correspond to many small domains in the volume of observation. Using the average GGP value for these fluctuations and the gamma factor for the PSF intensity profile model used (γ = 0.3545) (24), we calculated an average of approximately 23 independent equal structures moving through the PSF. Using a simple geometrical calculation assuming circular illumination volume and circular structures of the same size passing through, we can estimate that these fluctuations are produced by structures with radii in the range of approximately 25 nm (25).
- Slow fluctuations with large amplitude GGP: Big domains move slowly. The estimated average number of equal circular structures in this case is approximately three because the GGP is large and the average estimated radius is about 78 nm (26).
- Slow fluctuations with small amplitudeGGP: These fluctuations will correspond to even larger domains, almost the size of the illumination volume (W0 = 300 nm) that slowly move through it; we can estimate a size for them between < 300 nm and > 78 nm.
No detectable fluctuations: This situation corresponds to membranes showing no GP fluctuations (38% in RRBC) and represented with a black point at (0,0) in the plot in Fig. 3B. This region of the plot corresponds to domains larger than the illumination volume (> 300 nm) that will not show fluctuations due to their size, although the Laurdan molecules are moving.
We postulate that the plasma membrane of live RRBC contains high-GP domains (tightly packed) moving (diffusing) in a background lipid phase with lower-GP (less packed). In the same cell, these domains can have different sizes and different mobility. We estimate the sizes ranging from > 300 nm to 25 nm.
The GP fluctuations found in the membranes of CHO cells are in the same temporal range of the ones founded in RRBC; however, the distribution is different. According to our model, the GP domains in CHO cells are in the range greater than 78 nm and less than 300 nm. Interestingly, our preliminary results show that cholesterol removal from the membrane decreases the size of the GP domains to a range between 78 and 25 nm. However, further studies are needed to relate the changes in domain size observed in this study with the mechanisms involved in cholesterol homeostasis in vivo. Several players including ABCA1 transporter have to be carefully considered.
Our results point to the existence in the plasma membrane of RRBC and CHO cells of structures with properties similar to the present definition of membrane raft: ”small (10–200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains” (1), suggesting that the GP fluctuations report here may be related to functional rafts in the plasma membrane of RRBC.
We propose the plot of DGP versus GGP as a way to study membrane heterogeneities in live cells. This map of Laurdan GP fluctuations may change under stimulus such as cholesterol removal; for example, the sizes of these domains may change, fuse, become smaller, or change packing (GP value). Different cells may have similar or different GP fluctuation map according to the organization of their membrane and the particular stage of the cell cycle. Using the approach presented here could allow observing changes in the distribution of this map in an easy, straightforward manner when cells are treated under different experimental conditions of biological or biophysical importance.
Material and Methods
Materials.
An amount of 6-lauroyl-2-(dimethylamino) naphthalene (LAURDAN) was purchased from Molecular Probes (Eugene, OR). Defibrinated rabbit blood with no additives or preservatives was obtained from the Colorado Serum Company. Erythrocytes were isolated by centrifugation and resuspended to the original hematocrit in PBS buffer (10 mM Na2PO4, pH 7.5) containing pH 147 mM NaCl and 3 mM KCl and kept at 37 °C. Samples were diluted to hematocrit 0.2% in PBS containing 0.4 μM Laurdan, incubated 15 min and observed under the microscope at 37 °C. CHO-K1 were seeded in MEM supplemented with penicillin/streptomycin solution (100 units/mL) and 10% FBS at 37 °C in a 5% CO2 atmosphere. Cells were grown until 70% confluence and Chol removal induced by incubation for 2 h with 96 Å rHDL (reconstituted HDL particles) 300 μg/mL for 2 h in serum-free medium (5). Next, cells were washed with PBS and incubated for 15 min at 37 °C with Laurdan to a final concentration 1 μM in Dulbecco modified Eagle medium (DMEM) with 2.5% FBS, washed, and kept in DMEM at the same temperature during the observation time in a thermostatized microscope stage.
Methods.
POPC GUVs were fabricated by the electroformation method, originally designed by Angelova et al. (12, 27). For the GP, FCS measurements fluctuations due to the movement of the GUV must be avoided. GUVs were formed in a specially designed chamber, where the platinum wires are in contact with the cover slide used to seal the chamber. In this configuration the liposome grow attached to the wire and to the glass, and their movement is restricted.
Laurdan GP Imaging/sFCS.
Laurdan GP images and the combined Laurdan GP/scanning FCS data were collected on a two-photon fluorescence microscope with a two-channel detection system previously described (28). A mode-locked titanium sapphire laser (Mira 900; Coherent) pumped by a frequency-doubled Nd: Vanadate laser (Verdi, Coherent) and set to 780 nm was used as the two-photon excitation source. The laser was guided by x-y galvanoscanner mirrors (Cambridge Technology) to achieve beam scanning in both x-y directions and circular orbit. A LD-Achroplan 60X long working distance water objective 1.2 NA (Olympus America Inc.) was used. The samples received from 0.5 to 1.5 mW of the excitation light. The fluorescence emission was observed through a broad band-pass filter with a pass band from 350 nm to 600 nm (BG39 filter, Chroma Technology, Brattleboro, VT). The fluorescence was split into two channels using a Chroma Technology 470DCXR-BS dichroic beam splitter in the emission path. Interference filters Ealing 440 ± 50 nm and Ealing 490 ± 50 nm were placed in the emission paths to isolate the two areas of the Laurdan emission spectrum, respectively. A two-channel detection system was attached and two miniature photomultiplier (R5600-P, Hamamatsu, Bridgewater, NJ) were used for simultaneously detection of the two channels in a photon counting mode. Corrections for the wavelength dependence of the emission detection system were accomplished through the comparison of the GP value of a known solution (Laurdan in DMSO) (29).
For the single-point FCS measurements, a 64 kHz sampling frequency was used. For the scanning FCS measurement, the center of the circular scanning path was selected from the fluorescence image. The data acquisition frequency was set at 64 kHz, with a 1-ms orbit period and a radius of 1.52 μm. Sixty-four data points, corresponding to 64 locations, were collected in each scanning orbit.
Data Acquisition.
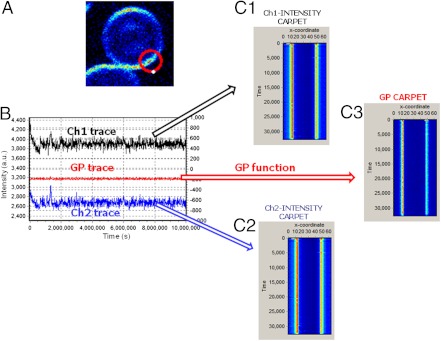
Fig. 5 shows a diagram for the acquisition and analysis of the sFCS data using the two-channel detection system. Fig. 5A shows the intensity image from one of the channels (CH1 at 440 nm) of a RRBC to indicate the circular orbit used for the sFCS measurements (red circle drawn on top of a RRBC). The orbit starts where the white point is located, and it moves clockwise crossing twice/period the RRBC membrane. The intensities from the two emission channels are saved as two independent traces (CH1 and CH2 traces in Fig. 5B).
Fig. 5.
Measurement and analysis of Laurdan GP fluctuations. (A) Intensity image of a rabbit erythrocyte showing the scanning orbit (red circle) of 1.52 um radius used in the circular scanning FCS data acquisition. Sixty-four points were acquired per ms (duration of the orbit). The white point on the orbit shows positions 0 and 64. The laser moves clockwise starting at point zero. (B) The Laurdan emission is recorded in two channels with interference filter center at 440 and 490 nm (blue and black traces). GP trace (red line) is generated using the traces and the GP formula (see Methods). (C) XY transformation of the raw scanning FCS intensity traces in intensity carpet (C1 and C2) and GP trace into GP carpet (C3). The x-position columns represent points along one circular scan (64), and the y-position rows represent successive scans every 1 ms.
Data Analysis.
Data were analyzed in two ways: (i) Classical sFCS analysis: From the independent traces (CH1 or CH2), data segments were transposed to form a fluorescence intensity matrix as a function of time known as an intensity-carpet (Fig. 5 C1, C2) (18, 19). Each vertical column in the carpet contains the intensity at 64 positions sampled every 1 ms. The two vertical bright lines observed in the intensity carpets C1 and C2 and in Fig. 5 correspond to the intensity of the Laurdan inside the erythrocyte membrane that have been crossed twice by the circular orbit of the laser as indicated in Fig. 5A. The analysis of these two bright columns contains information on the Laurdan intensity fluctuations. The autocorrelation analysis gives the DLau and the number of molecules of Laurdan in the membrane; (ii) sFCS/GP analysis: Intensity traces (Fig. 5B) are processed using the GP function [(CH1 - CH2)/(CH1 + CH2)] (9) and a GP trace is generated (Fig. 5B). Similarly to the process described previously a GP-carpet is created (Fig. 5C3). The two vertical lines corresponding to the membrane contain information on the GP fluctuations. Autocorrelation analysis applied to the vertical lines reports provide the DGP and the GGP, quantities related to the mobility and number of structures responsible for the GP fluctuations. Experimental autocorrelation functions were fit using a 3D-Gaussian intensity profile model (24). The point spread function (PSF) was calibrated using point FCS measurements of fluorescein in Tris pH > 9 and 25 °C assigning a D = 300 μm2/s. Typical w0 values were 0.29–0.35 μm. The SimFCS software (Laboratory for Fluorescence Dynamics) was used for all data analysis.
Laurdan GP Images.
GP images were generated using the same experimental setup used for sFCS measurements but directing the laser in a raster scanning motion. Two simultaneous mages (using the two emission channels) were obtained to generate the GP image by applying the GP formula pixel by pixel. The Laurdan GP value reported corresponds to the average of the Gaussian fitting of the histogram GP value v/s pixels obtained from the Laurdan GP image (12).
PSF Scaling Analysis for GP Fluctuations.
In this type of analysis, the point spread function (PSF) is artificially changed by binning adjacent pixels and then calculating the characteristic correlation time of the data averaged (19). The GP carpet (Fig. 5C3) is used for this analysis. In this representation, the positions corresponding to the membrane are analyzed, the center position is chosen and the program (sFCS) is set to bin three, five, seven, or nine adjacent columns to calculate the average diffusion coefficients after each binning operation. Because the orbit spans regions inside and outside the cell, data can be analyzed at different locations along one orbit as controls.
Supplementary Material
Acknowledgments.
S.S. and E.G. thank National Institutes of Health Grants P41-RRO3155 and P50-GM076516. M.A.T. thanks Agencia Nacional de Promoción Científica y Tecnológica, Argentina for Grants 2106-2008, Universidad Nacional de La Plata, for Grants M126 and M158, and Consejo Nacional de Investigaciones Científicas y Técnicas, PIP 112-200801-00953
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118288109/-/DCSupplemental.
References
- 1.Pike LJ. Rafts defined: A report on the keystone symposium on lipid rafts and cell function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson K, Mouritsen OG, Anderson RGW. Lipid rafts: At a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 3.He H-T, Marguet D. Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu Rev Phys Chem. 2011;62:417–436. doi: 10.1146/annurev-physchem-032210-103402. [DOI] [PubMed] [Google Scholar]
- 4.Smith SK, et al. Mechanisms by which intracellular calcium induces susceptibility to secretory phospholipase a2 in human erythrocytes. J Biol Chem. 2001;276:22732–22741. doi: 10.1074/jbc.M010880200. [DOI] [PubMed] [Google Scholar]
- 5.Sánchez SA, Tricerri MA, Ossato G, Gratton E. Lipid packing determines protein-membrane interactions: Challenges for apolipoprotein a-i and high density lipoproteins. Biochim Biophys Acta. 2010;1798:1399–1408. doi: 10.1016/j.bbamem.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaureguiberry MS, et al. Membrane organization and regulation of cellular cholesterol homeostasis. J Membr Biol. 2010;234:183–194. doi: 10.1007/s00232-010-9245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaus K, et al. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc Natl Acad Sci USA. 2003;100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celli A, Beretta S, Gratton E. Phase fluctuations on the micron-submicron scale in guvs composed of a binary lipid mixture. Biophys J. 2008;94:104–116. doi: 10.1529/biophysj.107.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celli A, Gratton E. Dynamics of lipid domain formation: Fluctuation analysis. Biochim Biophys Acta. 2010;1798:1368–1376. doi: 10.1016/j.bbamem.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber G, Farris FJ. Synthesis and spectral properties of a hydrophobic fluorescent probe: 2-dimethylamino-6-propionylnaphthalene. Biochemistry. 1979;18:3075–3078. doi: 10.1021/bi00581a025. [DOI] [PubMed] [Google Scholar]
- 11.Parasassi T, Gratton E. Membrane lipid domains and dynamics as detected by laurdan fluorescence. J Fluoresc. 1995;8:365–373. doi: 10.1007/BF00718783. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez SA, Tricerri MA, Gratton E. Interaction of high density lipoprotein particles with membranes containing cholesterol. J Lipid Res. 2007;48:1689–1700. doi: 10.1194/jlr.M600457-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Parasassi T, Stasio GD, D’Ubaldo A, Gratton E. Phase fluctuation in phospholipid membranes revealed by laurdan fluorescence. Biophys J. 1990;57:1179–1186. doi: 10.1016/S0006-3495(90)82637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenne P-F, et al. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 2006;25:3245–3256. doi: 10.1038/sj.emboj.7601214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee D-K, Wildman KH, Ramamoorthy A. Solid-state nmr spectroscopy of aligned lipid bilayers at low temperatures. J Am Chem Soc. 2004;126:2318–2319. doi: 10.1021/ja039077o. [DOI] [PubMed] [Google Scholar]
- 16.Sánchez SA, Tricerri MA, Ossato G, Gratton E. Lipid packing determines protein-membrane interactions: Challenges for apolipoprotein a-i and high density lipoproteins. Biochim Biophys Acta. 2010;1798:1399–1408. doi: 10.1016/j.bbamem.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berland KM, So PTC, Chen Y, Mantulin WW, Gratton E. Scanning two-photon fluctuation correlation spectroscopy: Particle counting measurements for detection of molecular aggregation. Biophys J . 1996;71:410–420. doi: 10.1016/S0006-3495(96)79242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruan Q, Cheng MA, Levi M, Gratton E, Mantulin WW. Spatial-temporal studies of membrane dynamics: Scanning fluorescence correlation spectroscopy (sfcs) Biophys J. 2004;87:1260–1267. doi: 10.1529/biophysj.103.036483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Marcos A, et al. Yeast ribosomal stalk heterogeneity in vivo shown by two-photon fcs and molecular brightness analysis. Biophys J. 2008;94:2884–2890. doi: 10.1529/biophysj.107.121822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers W, Glaser M. Characterization of lipid domains in erythrocyte membranes. Proc Natl Acad Sci USA. 1991;88:1364–1368. doi: 10.1073/pnas.88.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haverstick DM, Glaser M. Visualization of ca-induced phospholipid domains. Proc Natl Acad Sci USA. 1987;84:4475–4479. doi: 10.1073/pnas.84.13.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vojtavá J, Kofroňová O, Sěvo P, Benada O. Bordetella adenylate cyclase toxin induces a cascade of morphological changes of sheep erythrocytes and localizes into clusters in erythrocyte membranes. Microsc Res Tech. 2006;69:119–129. doi: 10.1002/jemt.20277. [DOI] [PubMed] [Google Scholar]
- 23.Ruan Q, Chen Y, Gratton E, Glaser M, Mantulin WW. Cellular characterization of adenylate kinase and its isoform: Two-photon excitation fluorescence imaging and fluorescence correlation spectroscopy. Biophys J. 2002;83:3177–3187. doi: 10.1016/S0006-3495(02)75320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller JD, Chen Y, Gratton E. Fluorescence correlation spectroscopy. Methods Enzymol. 2003;361:69–92. doi: 10.1016/s0076-6879(03)61006-2. [DOI] [PubMed] [Google Scholar]
- 25.Reis GE. Dense packing of equal circles within a circle. Math Mag. 1975;48:33–37. [Google Scholar]
- 26.Pirl U. Der mindestabstand von n in der einheitskreisscheibe gelegenen punkten. Math Nachr. 1969;40:111–124. [Google Scholar]
- 27.Angelova MI, Dimitrov DS. Liposome electroformation. Faraday Discuss Chem Soc. 1986;81:303–311. [Google Scholar]
- 28.So PTC, et al. Time resolved fluorescence microscopy using two-photon excitation. Bioimaging. 1995;3:49–63. [Google Scholar]
- 29.Sanchez SA, Bagatolli LA, Gratton E, Hazlett TL. A two-photon view of an enzyme at work: Crotalus atrox venom pla2 interaction with single-lipid and mixed-lipid giant unilamellar vesicles. Biophys J. 2002;82:2232–2243. doi: 10.1016/S0006-3495(02)75569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.