Abstract
Proteins with similar crystal structures can have dissimilar rates of substrate binding and catalysis. Here we used molecular dynamics simulations and biochemical analysis to determine the role of intradomain and interdomain motions in conferring distinct activation rates to two Gα proteins, Gαi1 and GPA1. Despite high structural similarity, GPA1 can activate itself without a receptor, whereas Gαi1 cannot. We found that motions in these proteins vary greatly in type and frequency. Whereas motion is greatest in the Ras domain of Gαi1, it is greatest in helices αA and αB from the helical domain of GPA1. Using protein chimeras, we show that helix αA from GPA1 is sufficient to confer rapid activation to Gαi1. Gαi1 has less intradomain motion than GPA1 and instead displays interdomain displacement resembling that observed in a receptor–heterotrimer crystal complex. Thus, structurally similar proteins can have distinct atomic motions that confer distinct activation mechanisms.
Heterotrimeric G proteins are molecular switches that are activated in response to extracellular stimuli including hormones, light, and neurotransmitters. In animals, the G-protein heterotrimer is activated by cell-surface receptors that trigger the Gα subunit of the heterotrimer to release GDP and bind GTP. GTP binding induces conformational changes in three small switch regions that result in heterotrimer dissociation (1). The free subunits then relay signals by activating or inhibiting downstream effectors. G-protein signaling is terminated after Gα hydrolyzes GTP and the heterotrimer reassociates. Thus, Gα proteins serve as timing devices that determine the duration of signaling.
Gα proteins from different organisms and subclasses share nearly identical structural features (2–4). However, G proteins exhibit a large spectrum of nucleotide exchange and hydrolysis rates. Basal nucleotide exchange in Gαq, for example, is essentially undetectable without a receptor (5), whereas the Gα protein (GPA1) from the plant Arabidopsis thaliana exchanges nucleotides at a pace of at least 4/min (6). Thus, GPA1 serves as a counterexample to slowly exchanging animal proteins and provides an opportunity to compare the molecular basis for receptor-dependent and -independent signaling.
The Gα subunit is composed of two domains connected by two short linker regions: (i) a domain that resembles the monomeric G protein Ras and (ii) an all α-helical domain unique to heterotrimeric G proteins. The guanine nucleotide binds at the interface of the two domains, but nucleotide-binding residues and switch regions are contained within the Ras domain and linker regions. The recently solved cocrystal structure of a G-protein heterotrimer with an activated receptor shows a large receptor-induced displacement of the Gα helical domain relative to the Ras domain (7). It is unclear whether this interdomain rearrangement is the cause or the consequence of nucleotide release. Nonetheless this work shows that both intradomain and interdomain rearrangements are required for G-protein activation.
Here we investigated the role of intradomain and interdomain motion in conferring the different activation properties of two structurally similar G proteins. Toward this end, we used molecular dynamics (MD) simulations and essential dynamics (ED) analyses to identify motions in G proteins that accompany nucleotide exchange. By this method we determined that the helical domains of GPA1 and Gαi1 rotate away from the Ras domains, consistent with the motion in Gαs induced by the β2 adrenergic receptor (7). However, intradomain and interdomain motions in GPA1 and Gαi1 were differentially influenced by the bound nucleotide and activation state. We tested predictions generated from MD simulations with protein chimeras and identified a small region from GPA1 sufficient to confer rapid nucleotide exchange to Gαi1. Together these approaches revealed distinct motions in GPA1 and Gαi1 that account for their distinct activation mechanisms. More broadly these analyses reveal atomic determinants of timing mechanisms in G proteins.
Results
GPA1 and Gαi1 Are Structurally Similar.
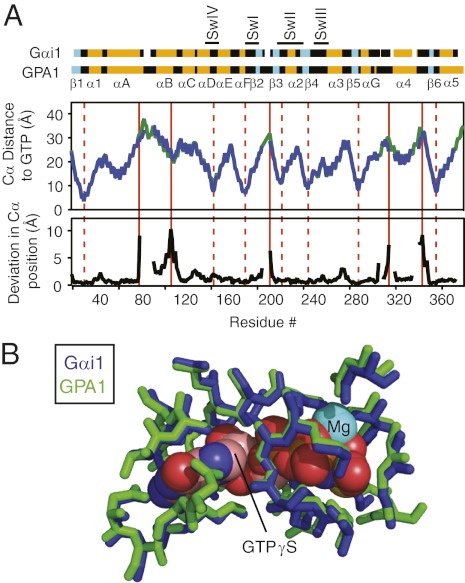
We chose to study GPA1 because basal nucleotide exchange is >100-fold faster than that of Gαi1 under optimal conditions for each enzyme (6, 8). Despite large differences in activation rates, GPA1 and Gαi1 crystal structures (9, 10) show remarkable similarity, with an average root-mean-square deviation of only 1.4 Å in core residue Cα atoms (Fig. 1A). Cα positions are most similar for residues closest to the nucleotide-binding pocket (Fig. 1A), and GPA1 and Gαi1 side chain atoms are in nearly identical positions surrounding the guanine nucleotide (Fig. 1B). In other words, structural comparison did not reveal the determinants of the disparate activation properties for GPA1 and Gαi1. Because intradomain and interdomain rearrangements are known to accompany G-protein activation (1, 7), we next analyzed these proteins for differences in atomic motions.
Fig. 1.
GPA1 and Gαi1 have similar crystal structures. (A) (Top) Secondary structure and domain architecture of Gαi1 and GPA1 residues used in MD simulations. Switch regions I–IV (“Sw”) are marked with lines. (Middle) Distance of each Cα from the center of the guanine nucleotide (GTPγS) is plotted for GPA1 (2XTZ, green) and Gαi1 (1GIA, blue). (Bottom) Deviation in Cα positions in GPA1 and Gαi1 is plotted against residue number. Solid red lines mark residues that have large deviations in Cα position and are distant from the nucleotide; dashed red lines mark residues that have small deviations in Cα position and are close to the nucleotide. (B) Superimposition of residues from GPA1 (2XTZ, green) and Gαi1 (1GIA, blue) within 4 Å of the guanine nucleotide.
Molecular Dynamics Simulations.
We conducted all-atom MD simulations to assess the effect of bound nucleotide and switch conformation on motions required for nucleotide exchange. We calculated average protein structures and used fluctuations of Cα residues relative to the average structure as a measure of dynamic motion. We also calculated covariance matrices that show how residues move relative to one another to identify differences in collective and correlated motions within the Gα proteins. Next we used ED to identify individual modes of motion (vibrational modes) that would permit nucleotide exchange and compared relative frequencies of these motions in GPA1 and Gαi1. Finally, we tested predictions from MD simulations with purified protein chimeras, GTP binding assays, and biophysical measurements of protein dynamics.
MD Simulations Reveal Unique Regions Sensitive to Bound Nucleotide.
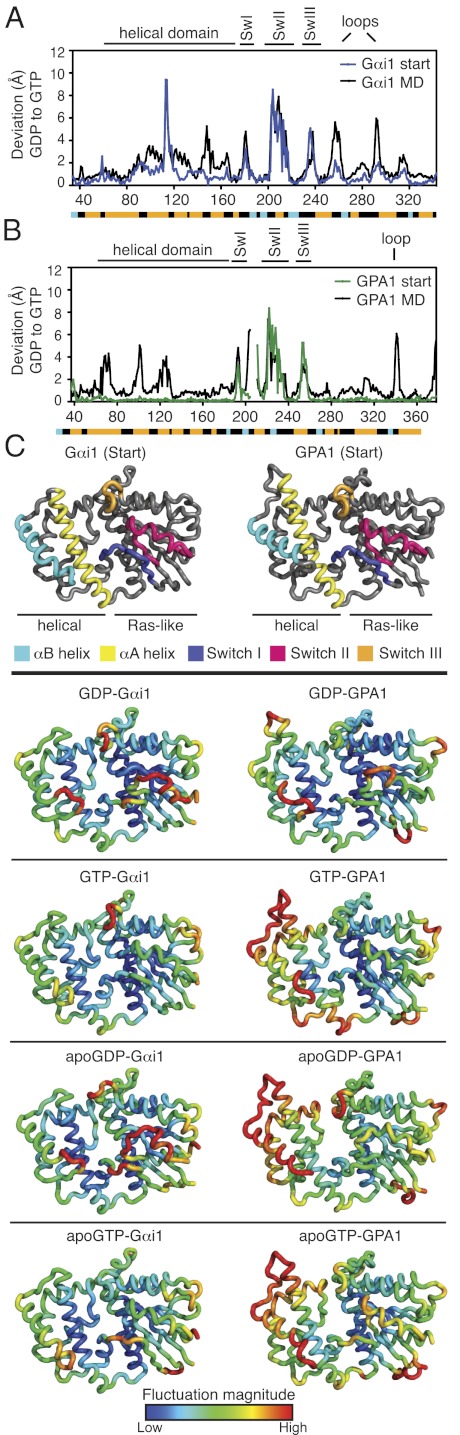
On the basis of crystal structures, Gα proteins have four small regions [switches I–III and a region that lies upstream of switch I sometimes called “switch IV” (residues 142–151 in Gαi1 and Fig. 1)] that differ in conformation between inactive (GDP-bound) and active (GTP-bound) Gα states (11–13). However, crystal structures represent just one or a few possible conformations of a protein, and this conformation is influenced by packing constraints in the crystal lattice. MD simulations build on information from crystal structures to model protein movements in solution. To identify regions that differ between the inactive and active forms of Gαi1 in solution, we calculated deviations in GDP- and GTP-bound Gα proteins from the average structures from our MD simulations. As with crystal structures, the MD approach also identified switches I–IV as differing between active and inactive states (Fig. 2 A and B). However, additional regions changed average conformation in Gαi1, including much of the helical domain and two surface loops in the Ras domain (Fig. 2A). Likewise, several regions in addition to the switches changed average conformation in GPA1, including a large portion of the helical domain and the α4-β6 loop in the Ras domain (Fig. 2B). Notably, these nucleotide-sensitive regions and the differences between GPA1 and Gαi1 were not evident in earlier crystallographic studies.
Fig. 2.
Localized fluctuations in GPA1 and Gαi1 are differentially affected by bound nucleotides. (A) Differences in Cα positions, comparing GDP-Gαi1 and GTP-Gαi1 determined from crystal structures (blue) or the average structures from MD simulations (black). (B) Same as A but with GPA1 (green). Note that the GDP-GPA1 input structure is derived from a model described in ref. 20. (C) (Upper) Regions of Gα proteins discussed in the text are colored on MD simulation input structures of GPA1 and Gαi1. (Lower) Average structures from MD simulations are colored according to the magnitude of fluctuation of each residue. Color scale indicates fluctuation magnitude.
Intradomain Motion in Gαi1 Is Localized Mainly to Switch Regions.
To reveal differences in GPA1 and Gαi1 that underlie their distinct activation mechanisms, we determined how each residue was influenced by switch conformation and bound nucleotides. Fig. 2C shows average structures from each simulation colored according to fluctuation magnitudes for each Cα residue. For GDP-Gαi1, we found that fluctuations were greatest in areas within the switch regions in the Ras domain, which are known to undergo conformational changes (Fig. 2C). We also found large fluctuations in an area with unknown function (helix αB, Fig. 2C). GTP diminished motion in helix αB (Fig. 2C) even though this helix is >17 Å from the guanine nucleotide at the closest point (Fig. 1A). This region was also suggested to differ in conformation between active and inactive states of transducin (14). Together these results suggest communication between the Gαi1 nucleotide-binding pocket and distant residues within the helical domain.
Intradomain Motion in Gαi1 Depends on Switch Conformation and Bound Nucleotide.
The active and inactive states of the Gα protein differ in at least two ways. First, GTP binding is accompanied by conformational changes in the switch regions (1). Second, the contacts between protein and nucleotide differ for GDP and GTP. To differentiate between the effect of a change in switch conformation and that of a change of nucleotide on dynamic motion in Gα proteins, we compared motion in two ligand-free forms of Gαi1. Apo enzymes were generated from both the GDP-bound (“apoGDP”) and the GTP-bound (“apoGTP”) structures, and thus each differed in switch conformations only. Simulations showed large differences in apoGDP-Gαi1 and apoGTP-Gαi1 fluctuations localized to switches II and III (Fig. 2C), suggesting that the switch conformation alone (without the nucleotide present) influenced dynamic motion in Gαi1.
Switch conformation accounts for some, but not all of the differences in motion between GDP- and GTP-bound Gαi1. To determine how motion was affected by the presence of guanine nucleotide, we compared simulations with Gαi1 that had the same switch conformation, but either contained or lacked the guanine nucleotide. ApoGDP-Gαi1 and GDP-Gαi1 had similar fluctuations, but apoGTP-Gαi1 and GTP-Gαi1 had differences throughout the protein. The presence of GTP reduced motion in switches I and II (Fig. 2C and Fig. S1), but GTP increased motion in switch III (up to 2.7 Å). Together these simulations suggest that the conformations of the switch regions, as well as the presence of the guanine nucleotide, influence dynamic motion in Gαi1.
Intradomain Motion in GPA1 Is Predominantly in the Helical Domain.
Dynamic motion in GPA1 differed from that in Gαi1 in many ways. The most apparent difference was that fluctuations were largest throughout the helical domain of GPA1 compared with the large differences found in the switch regions of Gαi1 (Fig. 2C). These differences were particularly evident in αA and αB of the helical domain (Fig. 2C and Fig. S1). Also in contrast to Gαi1, neither the switch conformation nor the presence of GTP had major effects on fluctuation magnitudes in GPA1 (Fig. 2C and Fig. S1). Finally, the presence of GDP generally had little effect on Gαi1, whereas GDP stabilized the GPA1 helical domain (Fig. 2C and Fig. S1). Collectively our findings show that structurally similar proteins can have strikingly different dynamic properties, and these properties vary according to protein conformation and bound ligand.
G-Protein Activation State Affects Collective Movements of Residues in Gα Proteins.
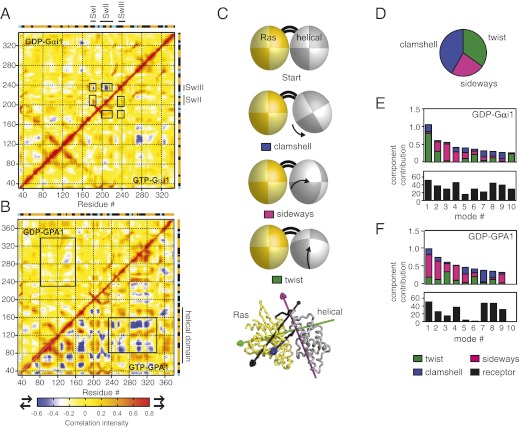
Having found differences in localized movements (fluctuations in individual residues) in GPA1 and Gαi1, we next identified differences in collective movements of pairs of Gα residues. Correlated motion plots generated from MD simulations show how atoms move relative to each other. Motions can be positively correlated (in the same direction), anticorrelated (in the opposite direction), or uncorrelated. Correlated (red) and anticorrelated (blue) movements in switch regions of GDP-Gαi1 suggested coordinated switch movement in Gαi1 that was not evident in GTP-Gαi1 or GDP-GPA1 (Fig. 3 A and B). Correlated motion plots also revealed how the Ras domain and the helical domain moved as collective units relative to each other: GTP-bound GPA1 displayed strong anticorrelated motion between the Ras and helical domains, consistent with frequent domain separation in the presence of GTP (Fig. 3B). Domain separation was less frequent in other simulations (GDP-Gαi1, GTP-Gαi1, and GDP-GPA1). For both GPA1 and Gαi1, we observed strong anticorrelated motions between Ras and helical domains in apo-enzyme simulations, consistent with frequent domain separation in the absence of guanine nucleotides (Fig. S1). Together these results suggest that collective movements in GPA1 and Gαi1 are differentially influenced by the activation state of the protein.
Fig. 3.
Gαi1 and GPA1 display different collective and two-domain motions. (A) Correlation matrix for pairs of residues in GDP-Gαi1 and GTP-Gαi1. Red indicates positive correlation; blue indicates anticorrelation; yellow indicates uncorrelated motion. Correlated and anticorrelated motions between switch regions are boxed. (B) Same as A, but with GPA1. Anticorrelated motion between the Ras and helical domains is boxed. (C) (Upper) Schematics of three variants of two-domain motion involving movement of the helical domain (gray ellipsoid) relative to the Ras domain (yellow ellipsoid), connected by domain linkers (black lines). (Lower) Axes of rotation for each variant are indicated on a Gα structure cartoon. (D) Contribution of two-domain motions depicted in C to the receptor-stimulated displacement of the Gαs helical domain as observed in 3SN6 (7). (E) (Upper) Contribution of two-domain motions depicted in C to displacement of the helical domain observed in eigenmodes from simulations with GDP-Gαi1, with each eigenmode scaled by its contribution to the total fluctuation. (Lower) Similarity of motion in each eigenmode to the receptor-stimulated displacement of the Gαs helical domain (7), with a maximum of 90 reflecting identical movements. (F) Same as E, but with GDP-GPA1.
GPA1 and Gαi1 Exhibit Different Frequencies of Two-Domain Motions.
Several reports suggest that the Ras and helical domains separate to allow nucleotide exchange. The crystal structure of the β2 adrenergic receptor–Gs heterotrimer complex reveals a large receptor-stimulated rotational displacement of the helical domain relative to the Ras domain (7). Likewise, our previous work suggests that the helical domain of the self-activating GPA1 protein frequently dissociates from the Ras domain (9). On the basis of these recent observations, we compared two-domain motions in GPA1 and Gαi1. We used DynDom3D (15) to identify the predominant motions involving two clusters of residues (one helical domain cluster and one Ras domain cluster). This analysis identified two-domain motions in almost all of the top 10 modes for simulations with GPA1 and Gαi1 (Fig. S2). DynDom3D also identified the axes of rotation of the helical domain relative to the Ras domain. We analyzed these axes of rotation for three nearly orthogonal components that we termed “twist,” “sideways,” and “clamshell” (see models in Fig. 3C and prototype motions in Movies S1, S2, S3, and S4). Each of these variants of motion could provide an exit route for the bound guanine nucleotide, and each component contributed to the Gα helical domain displacement observed in the cocrystal complex of the β2 adrenergic receptor with the Gs heterotrimer (Fig. 3D, calculated from ref. 7). For additional data and discussion on two-domain motions of GDP-bound Gs and Gβγ-bound Gαi1, see SI Text and Fig. S2. The twist variant was the dominant component in the top modes for GDP-Gαi1, but the sideways component was dominant in GDP-GPA1 (Fig. 3 E and F, Upper). In other words, our analyses suggest that both GPA1 and Gαi1 have frequent interdomain motions, but the types of two-domain motions vary greatly when comparing these two proteins.
We also analyzed the top modes from simulations for receptor-like two-domain motion. We used DynDom3D to calculate the receptor-induced axis of rotation of the helical domain relative to the Ras domain shown in reported crystal structures (7, 16). Both proteins displayed interdomain rearrangements resembling those stimulated by the receptor (Fig. 3 E and F, Lower), suggesting that receptors activate G proteins by enhancing the frequency or magnitude of motions that are intrinsic to the Gα protein. Compared with GPA1, Gαi1 displayed more receptor-like interdomain motion, and the two Gα proteins differed in the effects of nucleotides on their two-domain motions (Fig. S2). Collectively our analyses suggest that differences in intrinsic two-domain motions may underlie distinct activation mechanisms for the plant and animal Gα proteins.
Helix αA from GPA1 Promotes Fast G-Protein Activation.
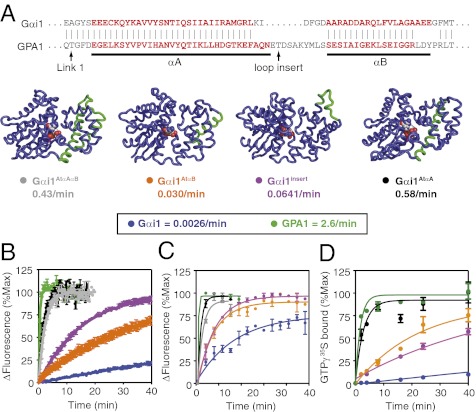
The results from our MD simulations showed that dynamic motion in helices αA and αB differed between GPA1 and Gαi1 and thus may account for differences in their basal nucleotide exchange rates. To test this prediction experimentally, we constructed a protein chimera with helices αA and αB from Gαi1 replaced with the corresponding helices from GPA1 (Fig. 4A, Gαi1AtαAαB). We purified this protein and measured its nucleotide exchange rate in a fluorescence assay (GTP-bound Gα proteins have higher intrinsic fluorescence than GDP-bound G proteins) (17). Consistent with a previous study (6), wild-type GPA1 had a spontaneous rate of nucleotide exchange that was fast relative to that of wild-type Gαi1 (Fig. 4B). However, the rate of GTP binding to Gαi1AtαAαB (0.43/min) was 160-fold faster than that of GTP binding to Gαi1 (0.0026/min). This rate of GTP binding nearly matched that of GPA1 (2.6/min). In other words, the αA-αB region from GPA1 was sufficient to confer rapid activation to Gαi1. To identify more precisely the region that conferred rapid nucleotide exchange, we substituted helix αA, the loop between helix αA and αB, or helix αB from GPA1 into Gαi1 (Fig. 4A and Table S1). Of these substitutions, we found that helix αA from GPA1 conferred the largest (220-fold) increase in Gαi1 nucleotide exchange (Fig. 4B and Table S1). In comparison, the seven-residue loop insert after αA of GPA1 conferred a 25-fold increase in Gαi1 nucleotide exchange, and helix αB from GPA1 conferred only a 10-fold increase in nucleotide exchange to Gαi1. We observed similar differences in nucleotide exchange assays with a fluorescently tagged guanine nucleotide (Fig. 4C and Table S1) and a radioactive nucleotide (Fig. 4D and Table S1). The single-turnover hydrolysis rate of the chimera with the fastest exchange rate (Gαi1AtαA) was measured at 0.38/min (Fig. S3A), similar to that of Gαi1 at 0.32/min and dissimilar to that of GPA1 (9). Thus, this substitution increased the exchange rate without affecting hydrolysis. We also found, with stability measurements (18), that helix αA from GPA1 conferred to Gαi1 the GPA1 property of thermal lability (9) (Fig. S3B). Reciprocal chimeras, substituting the αA and/or αB helices of GPA1 with those from Gαi1, were expressed poorly and could not be characterized.
Fig. 4.
Helix αA from GPA1 is sufficient to activate Gαi1. (A) (Upper) Comparison of primary Gα sequences from helices αA and αB. (Lower) Cartoon representation of models of protein chimeras used in these experiments and nucleotide exchange rates measured in B. (B) The change in intrinsic fluorescence (excite 284 nm, emit 340 nm) for the indicated Gα protein (400 nM) was measured after addition of GTPγS (10 μM). (C) The increase in fluorescence (excite 502 nm, emit 511 nm) of BODIPYFL-GDP (100 nM) was measured after addition of the indicated Gα protein (300 nM). (D) [γ-35S]GTPγS bound to Gα protein (400 nM) was measured over time. All experiments are average and SEM for at least two individual experiments.
Our results suggest that αA is sufficient to confer dynamic behavior and rapid basal activation to the Gαi1 protein. More broadly, they highlight the importance of considering vibrational modes as determinants of Gα protein properties. Although the crystal structures of GPA1 and Gαi1 are nearly identical, our analyses shows that a subset of comparable regions of these proteins has strikingly different dynamics (SI Text and Fig. S4), and these same regions confer distinct activation properties. Moreover, these results reveal an unexpected role for “action at a distance” in regulating G-protein signal initiation.
Discussion
Heterotrimeric G proteins are present in a wide variety of organisms and they exist in multiple distinct subclasses. Although nearly identical in structure, Gα subunits in particular exhibit remarkable diversity of function (2–4, 9). Recent analysis identified residues in Gα proteins that confer differences in effector interactions (19). However, an unanswered question was how Gα proteins acquired such a broad range of activation properties. Whereas animal G proteins have slow rates of nucleotide exchange, the Gα protein from A. thaliana self-activates (6).
Here we show that two prototype Gα proteins, one from animals and one from plants, have distinct intradomain and interdomain motions. Our previous crystallographic and biochemical analyses suggested a potential role for the larger helical domain in controlling basal nucleotide exchange rates (9). Here we found that helices αA and αB from GPA1 are more dynamic than the homologous helices from Gαi1. Follow-up experimental analysis using chimeric proteins established that the αA helix is largely responsible for the differences in activation. These results were particularly surprising given that helix αA is so distant from regions involved in receptor coupling, effector activation, nucleotide binding, and hydrolysis.
Thus our results suggest that localized dynamics in helix αA allow receptor-independent nucleotide release in plant Gα proteins. Moreover our findings, together with the recent crystal structure of a receptor–G-protein complex (7), reveal a distinct mechanism whereby the enhancement of two-domain motions allows receptor-dependent nucleotide release in animal Gα proteins. We propose that intradomain and interdomain motions evolved throughout G-protein divergence and led to the distinct activation mechanisms in plants and animals. More broadly, our results demonstrate the utility of MD simulations for elucidating structure–function relationships and highlight the importance of considering dynamic motion as a determinant of protein activities.
Materials and Methods
Structures Used for MD Analyses.
Structural coordinates for GTPγS-GPA1 (2XTZ), GNPPNP-Gαi1 (1CIP), and GDP-Gαi1 (1GP2) were obtained from www.pdb.org. GDP-GPA1 was modeled as described before (20). For consistency, PDB files were edited to delete N-terminal helix residues. Single-atom replacements converted nonhydrolyzable nucleotides (GTPγS or GNPPNP) to GTP (S or N replaced with O). Simulations included residues R32–N347 from Gαi1 and residues H37–L382 from GPA1 as well as guanine nucleotides and Mg2+ (for GTP-Gα simulations). The helical domain was defined as residues Y61–V179 from Gαi1 and residues F66–V191 from GPA1. Simulations and data analysis methods are described in SI Text.
Protein Purification and Measurement of Nucleotide Exchange Rates.
Standard methods were used for the purification of His-tagged Gα proteins as well as measurements of intrinsic fluorescence, binding to BODIPYFL-GDP, and binding to [γ35S]GTPγS, as detailed in SI Text.
Supplementary Material
Acknowledgments
We thank John Sondek for sharing equipment, Dan Isom for technical advice, and Jeff Duffy and Daisuke Urano for experimental assistance. This work was supported by National Institutes of Health Grants GM073180, GM080739 (to H.G.D,), and GM065989 and by National Science Foundation Grants MCB-0723515 and MCB-0718202 (to A.M.J.). We also thank the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02-05er15671 (to A.M.J.) for funding the technical support used here.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202943109/-/DCSupplemental.
References
- 1.Sprang SR. G protein mechanisms: Insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 2.Wall MA, et al. The structure of the G protein heterotrimer Gi α 1 β 1 γ 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura A, et al. Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc Natl Acad Sci USA. 2010;107:13666–13671. doi: 10.1073/pnas.1003553107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambright DG, et al. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 5.Berstein G, et al. Reconstitution of agonist-stimulated phosphatidylinositol 4,5-bisphosphate hydrolysis using purified m1 muscarinic receptor, Gq/11, and phospholipase C-β 1. J Biol Chem. 1992;267:8081–8088. [PubMed] [Google Scholar]
- 6.Johnston CA, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen SG, et al. Crystal structure of the β(2) adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linder ME, Ewald DA, Miller RJ, Gilman AG. Purification and characterization of Go α and three types of Gi α after expression in Escherichia coli. J Biol Chem. 1990;265:8243–8251. [PubMed] [Google Scholar]
- 9.Jones JC, et al. The crystal structure of a self-activating Gα protein reveals its distinct mechanism of signal activation. Sci Signal. 2011;4:ra8. doi: 10.1126/scisignal.2001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman DE, et al. Structures of active conformations of Gi α 1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 11.Noel JP, Hamm HE, Sigler PB. The 2.2 A crystal structure of transducin-α complexed with GTP γ S. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 12.Lambright DG, Noel JP, Hamm HE, Sigler PB. Structural determinants for activation of the α-subunit of a heterotrimeric G protein. Nature. 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 13.Mixon MB, et al. Tertiary and quaternary structural changes in Gi α 1 induced by GTP hydrolysis. Science. 1995;270:954–960. doi: 10.1126/science.270.5238.954. [DOI] [PubMed] [Google Scholar]
- 14.Ceruso MA, Periole X, Weinstein H. Molecular dynamics simulations of transducin: Interdomain and front to back communication in activation and nucleotide exchange. J Mol Biol. 2004;338:469–481. doi: 10.1016/j.jmb.2004.02.064. [DOI] [PubMed] [Google Scholar]
- 15.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 16.Sunahara RK, Tesmer JJ, Gilman AG, Sprang SR. Crystal structure of the adenylyl cyclase activator Gsα. Science. 1997;278:1943–1947. doi: 10.1126/science.278.5345.1943. [DOI] [PubMed] [Google Scholar]
- 17.Higashijima T, et al. The effect of activating ligands on the intrinsic fluorescence of guanine nucleotide-binding regulatory proteins. J Biol Chem. 1987;262:752–756. [PubMed] [Google Scholar]
- 18.Isom DG, Marguet PR, Oas TG, Hellinga HW. A miniaturized technique for assessing protein thermodynamics and function using fast determination of quantitative cysteine reactivity. Proteins. 2010;79:1034–1047. doi: 10.1002/prot.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temple BR, Jones CD, Jones AM. Evolution of a signaling nexus constrained by protein interfaces and conformational states. PLoS Comput Biol. 2010;6:e1000962. doi: 10.1371/journal.pcbi.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones JC, Temple BR, Jones AM, Dohlman HG. Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. J Biol Chem. 2011;286:13143–13150. doi: 10.1074/jbc.M110.190355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.