Abstract
Many RNA-processing events in the cell nucleus involve the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex, which contains the poly(A) polymerase Trf4p, the Zn-knuckle protein Air2p, and the RNA helicase Mtr4p. TRAMP polyadenylates RNAs designated for processing by the nuclear exosome. In addition, TRAMP functions as an exosome cofactor during RNA degradation, and it has been speculated that this role involves disruption of RNA secondary structure. However, it is unknown whether TRAMP displays RNA unwinding activity. It is also not clear how unwinding would be coordinated with polyadenylation and the function of the RNA helicase Mtr4p in modulating poly(A) addition. Here, we show that TRAMP robustly unwinds RNA duplexes. The unwinding activity of Mtr4p is significantly stimulated by Trf4p/Air2p, but the stimulation of Mtr4p does not depend on ongoing polyadenylation. Nonetheless, polyadenylation enables TRAMP to unwind RNA substrates that it otherwise cannot separate. Moreover, TRAMP displays optimal unwinding activity on substrates with a minimal Mtr4p binding site comprised of adenylates. Our results suggest a model for coordination between unwinding and polyadenylation activities by TRAMP that reveals remarkable synergy between helicase and poly(A) polymerase.
Keywords: ATP, surveillance, kinetics, decay, metabolism
The Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex is involved in nuclear RNA surveillance, 3′-end processing of rRNA, small nucleolar RNAs (snoRNAs), and snRNAs, and in gene silencing and chromatin maintenance (1–3). The TRAMP complex consists of three subunits that are highly conserved in eukaryotes: a noncanonical poly(A) polymerase (Trf4p or Trf5p in Saccharomyces cerevisiae), a Zn-knuckle protein (Air2p or Air1p), and the RNA helicase Mtr4p (4–7). TRAMP assists RNA degradation by the nuclear exosome, most notably by appending short (∼4–5 nt) oligo(A) tails at the 3′ ends of RNAs slated for exosome-mediated degradation (4, 5, 8–10). In addition, it has been speculated that TRAMP enables the nuclear exosome to efficiently degrade structured RNA through unwinding activity associated with Mtr4p (1, 2, 11–13).
However, RNA helicase activity has not been demonstrated for TRAMP. Though Mtr4p alone has been shown to unwind RNA duplexes in vitro (12, 13), it is not known whether binding of Trf4p/Air2p abolishes, decreases, increases, or otherwise alters this helicase activity. This question is important for TRAMP function, because Mtr4p and Trf4p operate with opposite polarities. Mtr4p only unwinds duplexes with a 3′ unpaired region, i.e., with a 3′ to 5′ polarity (12, 13). Trf4p polyadenylates the 3′ end of RNA and thus possesses 5′ to 3′ polarity (4, 5). How unwinding and polyadenylation with opposite polarities are coordinated in one complex is not readily apparent.
Moreover, Mtr4p controls the lengths of poly(A) tails appended by TRAMP (9). This role of Mtr4p requires ATP, but does not involve unwinding (9). Instead, Mtr4p binds to the 3′ end of the RNA, detects the number of 3′-terminal adenosines, and, in response, modulates ATP affinity and adenylation rate constants of Trf4p (9). These observations suggest an intricate functional interplay between helicase and polymerase during polyadenylation.
Here we show that TRAMP possesses robust unwinding activity that is also directed by complex functional interplay between Mtr4p and Trf4p/Air2p. Using recombinant S. cerevisiae TRAMP, we find that Trf4p/Air2p significantly stimulates the unwinding activity of Mtr4p. However, this stimulation does not depend on ongoing polyadenylation. Nonetheless, polyadenylation by Trf4p enables Mtr4p to unwind substrates that it otherwise cannot separate. Our data show optimal unwinding activity of TRAMP on substrates that contain just a minimal binding site for Mtr4p, preferably consisting of adenylates. Together with previous results, our findings suggest a model for the coordination between unwinding and polyadenylation by TRAMP that highlights remarkable synergy between helicase and polymerase activities with opposite polarities.
Results
Unwinding Activity of Mtr4p Is Stimulated in the TRAMP Complex.
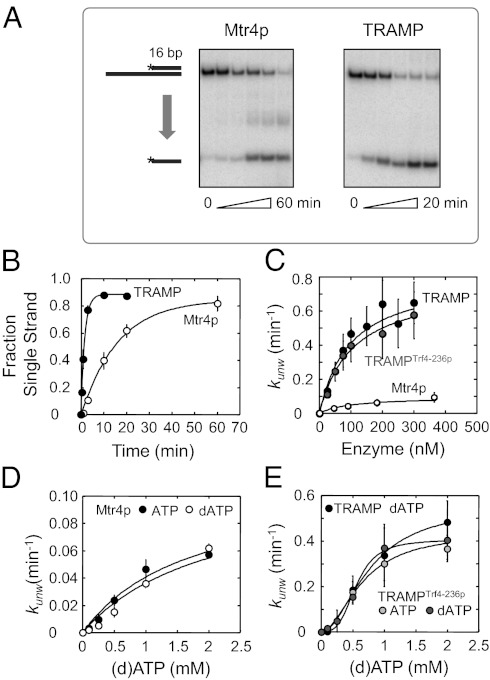
To examine RNA unwinding activity by the TRAMP complex, we performed pre–steady-state unwinding reactions with a substrate containing a 16-bp duplex and a 3′ single-stranded extension of 25 nt (Fig. 1A). This substrate had been previously used for unwinding measurements by Mtr4p (13). To preclude complications in data analysis and interpretation caused by simultaneous polyadenylation, we conducted unwinding reactions with dATP, which promotes Mtr4p-driven unwinding but not polyadenylation by Trf4p (Fig. 1A).
Fig. 1.
The unwinding activity of Mtr4p is stimulated in the TRAMP complex. (A) Representative PAGE for unwinding reaction (RNA: 16-bp duplex with 3′ 25-nt single-stranded region, 0.5 nM, 2 mM equimolar dATP-Mg2+) by 200 nM Mtr4p (Left) and 200 nM TRAMP (Right). Aliquots were removed at 1, 3, 10, 20, and 60 min for Mtr4p, and at 0.5, 1, 3, 10, and 20 min for TRAMP. (B) Time courses for unwinding reactions with Mtr4p (○) and TRAMP (●). Conditions were as in A. Data show averages from three independent experiments; error bars represent one SD. Curves represent best fits to the integrated first-order rate law, yielding observed rate constants (kobs). For Mtr4p, kobs, unw = 0.06 ± 0.01 min−1; for TRAMP, kobs, unw = 0.59 ± 0.06 min−1. (C) Dependence of unwinding rate constants (2 mM dATP-Mg2+) on enzyme concentrations for Mtr4p (open circles), WT TRAMP (filled black circles), and TRAMPTrf4-236p (filled gray circles). Rate constants were determined from multiple independent reactions; error bars represent one SD. Curves represent the best fit to the binding isotherm, kunw = kmax, E [E]/([E] + K1/2, E). [E], enzyme concentration. For Mtr4p, kmax, Mtr4p = 0.090 ± 0.003 min−1, K1/2, Mtr4p = 105 ± 36 nM. For WT TRAMP, kmax, TR = 0.84 ± 0.10 min−1, K1/2, TR = 94 ± 31 nM. For TRAMPTrf4-236p, kmax, TR(m) = 0.76 ± 0.06 min−1, K1/2, TR(m) = 105 ± 20 nM. (D) Dependence of unwinding rate constants on ATP and dATP concentrations for Mtr4p (800 nM). Rate constants were determined from multiple reactions; error bars represent one SD. Curves represent the best fit to a binding isotherm, kunw = kmax, dATP [dATP]/([dATP] + K1/2, dATP). With ATP, kmax, ATP = 0.11 ± 0.03 min−1, K1/2, ATP = 1.74 ± 0.75 mM. With dATP, kmax, dATP = 0.11 min−1, K1/2, dATP = 2.08 ± 0.27 mM. (E) Dependence of unwinding rate constants on ATP and dATP concentrations for TRAMPTrf4-236p and WT TRAMP (both at 300 nM). Rate constants were determined from multiple reactions; error bars represent one SD. Curves represent the best fit to the Hill equation kunw = kmax, dATP [dATP]n/([dATP]n + (K1/2, dATP)n). For WT TRAMP, kmax, dATP = 0.56 ± 0.04 min−1, K1/2, dATP = 0.77 ± 0.08 mM, n = 1.9 ± 0.2. For TRAMPTrf4-236p with dATP, kmax, dATP = 0.40 ± 0.01 min−1, K1/2, dATP = 0.56 ± 0.03 mM, n = 3.4 ± 0.7. For TRAMPTrf4-236p with ATP, kmax, ATP = 0.43 ± 0.03 min−1, K1/2, ATP = 0.65 ± 0.08 mM, n = 1.9 ± 0.3.
Unwinding was seen with Mtr4p alone (Fig. 1A), consistent with previous data (13). No unwinding was observed with only Trf4p/Air2p. The complete TRAMP complex unwound the substrate faster than Mtr4p at identical enzyme concentrations (Fig. 1 A and B). Unwinding rate constants extrapolated to enzyme saturation were approximately ninefold higher for TRAMP than for Mtr4p (Fig. 1C). Functional substrate affinities of TRAMP and Mtr4p (K1/2) did not differ significantly (Fig. 1C).
The use of dATP in the unwinding reactions did not alter reaction parameters for Mtr4p alone, compared with ATP (Fig. 1D). To measure parameters with dATP and ATP for TRAMP, we generated TRAMP with Trf4p mutations (Trf4-236p) in the polymerase active site that abolish polyadenylation activity (5). TRAMPTrf4-236p displayed unwinding activity highly similar to WT TRAMP with dATP (Fig. 1C). The activity was virtually unchanged with ATP (Fig. 1E). The data also revealed that the stimulation of the unwinding activity of Mtr4p by Trf4p/Air2p does not require an intact active site in the poly(A) polymerase.
Both WT TRAMP and TRAMPTrf4-236p displayed a slightly higher affinity for dATP/ATP [K1/2(dATP) = 0.74 ± 0.04 mM] compared with Mtr4p alone [K1/2(dATP) = 2.08 ± 0.27 mM; compare Fig. 1D with Fig. 1E]. This increase in dATP/ATP affinity contributes to the observed stimulation of Mtr4p by Trf4p/Air2p (Fig. 1C). Notably, dATP/ATP binding by TRAMP and TRAMPTrf4-236p was cooperative (n = 1.9 ± 0.2; Fig. 1E). No clear cooperativity was seen for Mtr4p (n = 1.5 ± 0.1; Fig. 1D). These observations raise the possibility that unwinding by TRAMP involves dATP binding to both the helicase and the polymerase, although no polyadenylation took place. Collectively, our data show robust unwinding activity by TRAMP and reveal that Trf4p/Air2p stimulates the helicase activity of Mtr4p by increasing the ATP affinity and the strand-separation rate constant (kunwmax), without significantly changing RNA affinity of Mtr4p.
Unwinding Rate Constants by Mtr4p and TRAMP Depend on Duplex Length.
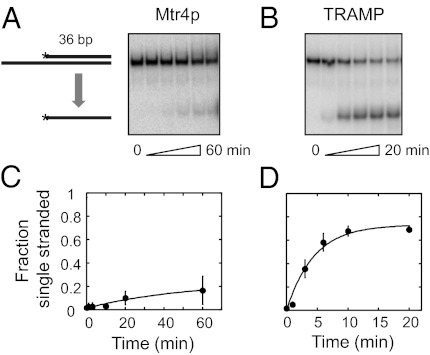
We next examined the impact of duplex length on the unwinding rate constants for TRAMP and Mtr4p using a substrate with a 36-bp duplex region (Fig. 2). Both Mtr4p and TRAMP unwound the substrate, but TRAMP showed significantly higher activity (Fig. 2). Unwinding of this duplex by Mtr4p is notable, despite the modest reaction amplitude, because other RNA helicases, especially members of the DEAD-box family, do not separate duplexes of this length to an appreciable degree (14, 15). Both Mtr4p and TRAMP also facilitated strand annealing, which accounts for the lower reaction amplitudes of the 36-bp duplex, compared with the reaction with the 16-bp complex (Fig. S1). Unwinding rate constants for both TRAMP and Mtr4p were roughly fourfold lower for the 36-bp substrate than for the 16-bp substrate (Fig. 2 C and D). Sensitivity of unwinding rate constants to duplex length and stability is a hallmark of DEAD-box RNA helicases, which unwind duplexes in a nonpolar fashion (16–20). In contrast, the viral RNA helicases of the NS3/NPH-II family, which unwind duplexes with strict polarity, show comparably small or no effects of duplex length or stability on unwinding rate constants (21, 22).
Fig. 2.
Mtr4p and TRAMP unwind a 36-bp RNA duplex. (A) Representative PAGE for unwinding reaction (RNA: 36-bp duplex with 3′ 25-nt single-stranded region, 0.5 nM, 2 mM equimolar dATP-Mg2+) by 200 nM Mtr4p. Aliquots were removed at 1, 3, 10, 20, and 60 min. (B) Representative PAGE for unwinding reaction (RNA and conditions as in A) by 200 nM TRAMP. Aliquots were removed at 0.5, 1, 3, 10, and 20 min. (C) Time course for unwinding of the 36-bp duplex RNA by Mtr4p (conditions as in A). Data show averages from three independent experiments; error bars indicate one SD. For curve fitting, see Fig. S1 (observed unwinding rate constant kobs, unw = 0.02 ± 0.02 min−1). (D) Time course for unwinding of the 36-bp duplex RNA by TRAMP (conditions as in A). Data show averages from three independent experiments; error bars indicate one SD. For curve fitting, see Fig. S1 (kobs, unw = 0.22 ± 0.05 min−1).
Duplex Unwinding by Mtr4p and TRAMP Requires an RNA Loading Strand.
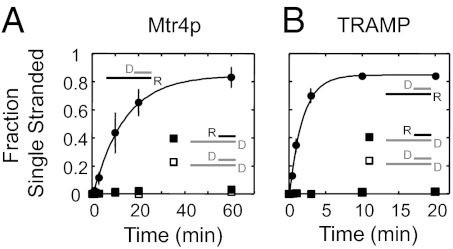
To further probe the extent to which duplex unwinding by Mtr4p and TRAMP resembled unwinding by the nonpolar DEAD-box RNA helicases, we examined unwinding of RNA/DNA hybrids. DEAD-box helicases separate DNA/RNA hybrids, regardless of which strand is DNA (19, 20, 23). In contrast, RNA helicases that act with defined polarity, such as viral NS3/NPH-II RNA helicases or the DEAH/RHA helicase Prp22p, usually require the loading strand to be RNA (24, 25). Both Mtr4p and TRAMP readily unwound the hybrid substrate with the RNA loading strand, but not the substrate with a DNA loading strand (Fig. 3). These characteristics clearly differ from DEAD-box helicases (15).
Fig. 3.
Duplex unwinding by Mtr4p and TRAMP requires an RNA bottom strand. Time courses of unwinding reactions of DNA/RNA hybrid duplexes by Mtr4p (A) and TRAMP (B) (conditions as in Fig. 1). In the 16-bp RNA duplex with the 25-nt single-stranded region (Fig. 1A), top, bottom, or both RNA strands (R) were replaced by DNA (D), as indicated. Duplexes with DNA bottom strands were not unwound. Mtr4p unwound the substrate with the DNA top strand at kobs, unw = 0.07 ± 0.01 min−1; TRAMP at kobs, unw = 0.54 ± 0.07 min−1.
The data thus show that duplex unwinding by Mtr4p and TRAMP shares major similarities with the unwinding mode of polar NS3/NPH-II or DEAH/RHA helicases. However, the duplex length dependence of unwinding rate constants resembles DEAD-box proteins. Together, these observations suggest that unwinding by Mtr4p and TRAMP involves binding of the helicase to the duplex/single-stranded region in a defined, polar fashion. However, it is not clear whether strand separation involves translocation over many steps, as in NS3/NPH-II RNA helicases (26). We were not able to experimentally evaluate processivity or translocation in the unwinding reaction for either Mtr4p or TRAMP, because no unwinding activity could be detected under single-cycle reaction conditions with excess scavenger RNA. Presumably, Mtr4p and TRAMP dissociate from the substrate significantly faster than they separate the duplex.
Simultaneous Unwinding and Polyadenylation Reveals Minimal Overhang Length for Strand Separation by TRAMP.
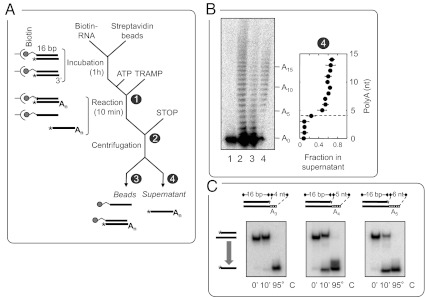
Having defined basic characteristics of the unwinding reaction by TRAMP in the absence of polyadenylation, we next examined duplex unwinding with ongoing polyadenylation. Continuous adenylation creates a population of substrates with increasing single-stranded extensions at the 3′ end (9). It was therefore important to first probe whether and how the number of appended adenylates affected the ability of TRAMP to unwind duplexes. To this end, we simultaneously monitored unwinding and polyadenylation, using the 16-bp duplex with a single unpaired nucleotide at the 3′ end (Fig. 4A). The single unpaired nucleotide is necessary for efficient polyadenylation, but does not support unwinding in the absence of polyadenylation (9). The duplex was immobilized on streptavidin beads via a biotin moiety at the 3′ end of the top strand. Duplex unwinding released adenylated loading strands into the supernatant, whereas the species not unwound remained on the beads (Fig. 4A). The distribution of unwound vs. not unwound species was then analyzed on denaturing PAGE (Fig. 4B).
Fig. 4.
Simultaneous polyadenylation and unwinding reaction by TRAMP reveals the oligo(A) length required for duplex unwinding. (A) Experimental design for simultaneous polyadenylation and unwinding reaction. A 16-bp duplex with a single unpaired nucleotide at the 3′ terminus was immobilized via a biotin moiety (gray dot; asterisk marks the radiolabel) on streptavidin beads. The Reaction was started by adding TRAMP (300 nM final, 2 mM ATP-Mg2+). After 10 min, the sample was centrifuged, and beads and supernatant were separated and then analyzed on denaturing PAGE. The circled numbers refer to samples and controls analyzed in B. (B) Representative denaturing PAGE (Left) and quantification of unwinding for each RNA species (Right). Lane 1: RNA before reaction; lane 2: RNA after reaction and before centrifugation; lane 3: RNA on beads sample after centrifugation; lane 4: RNA in supernatant after centrifugation. The number of adenines appended (A0 … An) is indicated. The plot (Right) shows the fraction of unwound duplex for individual adenylated species (signals from lane 4 of the PAGE, divided by the sum of signals from lanes 3 and 4 for each species). The sum of signals in lanes 3 and 4 faithfully represents the distribution of species in the unsedimented RNA in lane 2 (Fig. S3). The dashed line indicates A4. (C) Unwinding reactions (10 min, conditions as in Fig. 1) for 16-bp substrates with defined overhangs of three, four, and five adenylates (A3–A5, 4- to 6-nt overhang) corresponding to RNAs in B. The lane marked 95 °C shows heat-denatured duplexes.
We observed exclusively species with at least four adenylates in the supernatant and a concurrent decline of the fraction of species with four and more adenylates in the duplex (Fig. 4B). This result indicates a minimum of five unpaired nucleotides (one protruding nucleotide + A4) to be required for unwinding. Virtually identical results were obtained by examining polyadenylation and unwinding reactions simultaneously without immobilization (Fig. S2). This control confirmed that the observed minimum of five unpaired nucleotides was not caused by the experimental setup involving substrate immobilization.
To further verify the overhang length required for unwinding, we generated three 16-bp duplexes with defined unpaired extensions of 4 (U + A3), 5 (U + A4), and 6 (U + A5) nt and measured unwinding of these substrates by TRAMP without polyadenylation (Fig. 4C). Little unwinding was seen for the substrate with the 4-nt extension, whereas significant unwinding was observed for the substrate with a 5-nt extension, and even more strand separation for the substrate with the 6-nt extension (Fig. 4C). These observations are clearly consistent with the results obtained when measuring unwinding and polyadenylation simultaneously, and thus confirm that efficient duplex unwinding by TRAMP requires five unpaired nucleotides. Collectively, the data define a minimal overhang length for unwinding by TRAMP and indicate that this minimal overhang length is independent of the actual process of polyadenylation.
TRAMP and Mtr4p Preferentially Unwind Substrates with Short Poly(A) Tails.
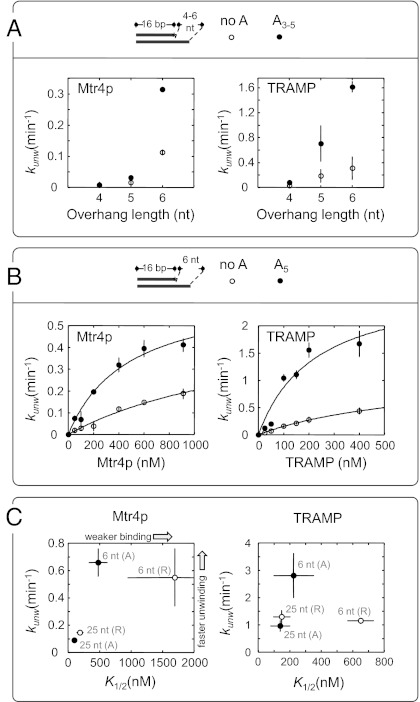
The minimal overhang length coincides roughly with the length of poly(A) tails where Mtr4p starts to inhibit polyadenylation by Trf4p/Air2p (9). We therefore tested whether TRAMP and Mtr4p also preferred adenylates in unwinding substrates with minimal overhangs. We compared unwinding rate constants for substrates with unpaired 4-, 5-, and 6-nt extensions containing adenylates (U + A3–5) to unwinding rate constants of substrates with the same overhang length without adenylates (Fig. 5A). For both, Mtr4p and TRAMP unwinding rate constants increased with the number of nucleotides in the overhang. For a given overhang length, the substrate with adenylates was unwound with a higher rate constant than the substrate without adenylates (Fig. 5A). The similarities between Mtr4p and TRAMP suggest that Mtr4p is involved in the detection of overhang length and adenine bases.
Fig. 5.
Mtr4p and TRAMP preferably unwind substrates with short single-stranded regions containing adenylates. (A) Unwinding of 16-bp duplexes (Fig. 1) with 4- to 6-nt single-stranded overhangs containing adenosines (●) or a sequence without adenosines (○) by Mtr4p (400 nM; Left) and TRAMP (400 nM; Right). Observed unwinding rate constants (kunw) are averages from three independent measurements (2 mM dATP-Mg2+; other conditions as in Fig. 1); error bars represent the SD. (B) Observed unwinding rate constants for a 16-bp duplex with a 6-nt single-stranded overhang containing adenosines (●) or a sequence without adenosines (○) as a function of Mtr4p (Left) and TRAMP (Right) concentration. Conditions were as in A, observed unwinding rate constants (kunw) are averages from multiple independent measurements; error bars represent the SD. Curves represent best fits to the binding isotherm, kunw = kmax, E [E]/([E] + K1/2, E). [E], enzyme concentration; K1/2, functional affinities; kmax, E, unwinding rate constant at enyzme saturation. Obtained values were as follows: Mtr4p, A-rich substrate; kmax, Mtr4p = 0.66 ± 0.10 min−1; K1/2, Mtr4p = 477 ± 145 nM; Mtr4p, the non-A substrate, kmax, Mtr4p = 0.55 ± 0.21 min−1, K1/2, Mtr4p = 1,697 ± 908 nM; TRAMP, A-rich substrate; kmax, TR = 2.81 ± 0.81 min−1; K1/2, TR = 225 ± 126 nM; TRAMP, non-A substrate; kmax, TR = 1.15 ± 0.10 min−1; K1/2, TR = 657 ± 81 nM. (C) Functional affinities and unwinding rate constants at enzyme saturation for substrates with 6-nt and 25-nt overhangs containing adenosines (A) or a sequence without adenosines (R) for Mtr4p (Left) and TRAMP (Right).
To investigate the basis for the adenylate preference, we determined unwinding rate constants for the two substrates with 6-nt extensions at increasing concentrations of Mtr4p and TRAMP (Fig. 5B). These substrates were chosen because substrates with shorter overhangs could not be saturated with enzyme. Both Mtr4p and TRAMP displayed higher functional affinity (K1/2) for the substrate with the adenylate tail, compared with the substrate with the tail of a different sequence (Fig. 5B). In addition, an adenylate tail increased the unwinding rate constant at enzyme saturation (kmax, TR = 2.81 ± 0.81 min−1), compared with a tail with a different sequence (kmax, TR = 1.15 ± 0.10 min−1). No significant effect on the unwinding rate constant at enzyme saturation was seen for Mtr4p (Fig. 5B).
The preference for adenylates in substrates with 6-nt overhangs prompted us to examine whether similar differences were seen for substrates with longer overhangs. We compared unwinding rate constants and functional affinities for two substrates with 25-nt overhangs, one with an adenylate tail and the other with a tail of a different sequence (Fig. 5C). For both Mtr4p and TRAMP, a slightly higher functional affinity was measured for the 25-nt substrate with adenylates compared with the substrate without adenylates in the overhang (Fig. 5C). No significant difference was seen in the unwinding rate constant at enzyme saturation for TRAMP (Fig. 5C). However, for Mtr4p unwinding rate constants at enzyme saturation were significantly lower for the substrates with 25-nt overhangs than for the substrates with 6-nt overhangs (Fig. 5C). For TRAMP, the unwinding rate constant at enzyme saturation for the substrate with the 6-nt overhang containing adenylates was significantly higher than for all other substrates (Fig. 5C). Collectively, these data reveal a preference of TRAMP and Mtr4p for substrates with short overhangs that contain adenylates.
Discussion
In this study, we have demonstrated that TRAMP displays robust ATP-driven RNA unwinding activity. Trf4p/Air2p stimulates the inherent unwinding activity of the RNA helicase Mtr4p through increases in ATP affinity and in the rate constant for strand separation. For substrates with short (6 nt), but not with longer (25 nt), overhangs, TRAMP also increases the RNA affinity of Mtr4p. The stimulation of Mtr4p by Trf4p/Air2p argues that TRAMP, rather than Mtr4p alone, is the preferred agent for duplex unwinding.
Trf4p/Air2p modulates multiple reaction parameters of Mtr4p, indicating that Trf4p/Air2p are not simply RNA binding adaptors for Mtr4p. Instead, the alteration of ATP affinity and unwinding rate constant of Mtr4p by Trf4p/Air2p suggest an intricate functional coupling between helicase and Trf4p/Air2p. This observation expands the scope of functional coupling between helicase and polymerase that is seen during polyadenylation by TRAMP, where Mtr4p modulates Trf4p activity, notably also by altering ATP affinity and adenylation rate constants of Trf4p (9).
Notwithstanding the functional coupling between Mtr4p and Trf4p, the stimulation of Mtr4p in the TRAMP complex does not require ongoing polyadenylation by Trf4p. Similarly, the modulation of polyadenylation by Mtr4p does not require unwinding (9). These observations indicate that functional coupling between Mtr4p and Trf4p does not require polyadenylation and unwinding to occur simultaneously. This arrangement might prevent potentially detrimental effects caused by the opposite polarities of the two activities.
Our data show that TRAMP is able to unwind duplexes with several helical turns (Fig. 2). Although duplexes of this length are not thought to occur in cellular RNA, unwinding of extended RNA duplexes has been seen with certain other RNA helicases, such as Prp22 of the DEAH/RHA family (25), or the viral HCV NS3 and NPH-II (21, 22). Like Mtr4p, these enzymes unwind RNA duplexes in a strictly polar fashion. TRAMP and Mtr4p further share with Prp22 the requirement for RNA in the loading strand (Fig. 3). These findings suggest common functional features in Ski2-like and DEAH/RHA helicases, consistent with structural conservation in the C termini of both helicase families, and the presence of a conserved β-hairpin between the helicase motifs Va and VI in Ski2-like, DEAH/RHA, and viral NS3/NPH-II helicases (14, 27–34). However, given that no activity was seen in single-cycle experiments with both Mtr4p and TRAMP, it would be premature to extrapolate from the available data that Mtr4p/TRAMP unwind duplexes by processive translocation. Single-molecule approaches might be required to probe processivity of Mtr4p and TRAMP during the unwinding reaction.
A striking feature of the unwinding activity of Mtr4p/TRAMP is the preference for short (6 nt) unpaired overhangs (Fig. 5). Other RNA helicases show either increasing unwinding activity with increasing overhang length or little sensitivity to the overhang length beyond a critical number of nucleotides (19, 35). The minimal number of nucleotides required for unwinding by Mtr4p/TRAMP coincides with the Mtr4p binding-site size (11, 31), and thus most likely reflects binding of Mtr4p to the 3′ tail. Binding of Mtr4p in the immediate vicinity of the duplex potentially promotes previously proposed contacts of the Arch/KOW domain with the duplex (31).
On substrates with short overhangs, both Mtr4p and TRAMP display higher affinity for adenylates, compared with other nucleotides (Fig. 5). Preferred binding to adenylates during the unwinding reaction correlates with higher affinity of Mtr4p for oligo(A) in equilibrium binding experiments (11). TRAMP, but not Mtr4p alone, unwinds substrates with short adenylate tails faster than substrates with short tails of other sequences (Fig. 5). This finding suggests that binding of Mtr4p to short adenylate tails in the context of TRAMP elicits further modulation of Mtr4p by Trf4p/Air2p. Substrates with longer overhangs (25 nt) are unwound slower by TRAMP and Mtr4p, compared with substrates with short overhangs, and the difference between functional affinities for adenylates vs. other sequences is far less pronounced in substrates with long overhangs (Fig. 5). We speculate that longer overhangs facilitate binding of Mtr4p farther away from the duplex, possibly preventing optimal positioning of TRAMP/Mtr4p for unwinding.
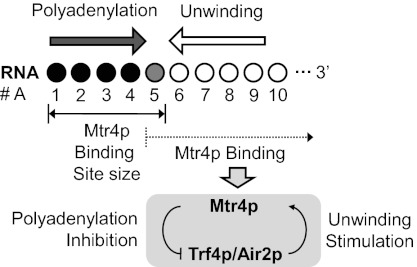
Taken together, the results from this and previous studies (9) suggest a model for the coordination between polyadenylation and unwinding by TRAMP (Fig. 6). Central to this coordination is the binding of Mtr4p to single-stranded 3′ extensions. Mtr4p can only fully bind when the 3′ extensions contain at least 5–6 nucleotides, corresponding to the Mtr4p binding site size (31). Substrates with smaller extensions are polyadenylated by TRAMP until the minimal binding site for Mtr4p is formed. As shown previously, Mtr4p binding to four or more 3′ adenylates inhibits polyadenylation by Trf4p (9). Thus, TRAMP inherently disfavors generation of substrates that are not optimal for unwinding. It was also shown that TRAMP dissociates frequently from the RNA during the polyadenylation process, thereby enabling Mtr4p to repeatedly interrogate the number of 3′ terminal adenylates (9).
Fig. 6.
Coordination between polyadenylation and unwinding activities in TRAMP. ○, number of added adenylates; ●, minimal number of adenylates involved in Mtr4p binding. See text for further explanation.
This model for coordination between unwinding and polyadenylation activities highlights synergy between polyadenylation and unwinding activities in the TRAMP complex. The model further supports the notion that TRAMP is inherently optimized to append only a few adenylates to its targets. Despite coordination between polyadenylation and unwinding, the two activities do not absolutely depend on each other. TRAMP polyadenylates substrates that are not unwound, and although TRAMP preferably unwinds RNAs with short adenylate tails, it does unwind substrates without adenylates in the single-stranded extensions. We speculate that this accommodation of a large variety of potential RNA substrates may be important for the processing of the multitude of biological TRAMP targets (10).
Materials and Methods
Materials.
Recombinant TRAMP complex was purified from reconstituted components expressed in E. coli, as described previously (9). Mutagenesis of TRF4 to create trf4-236 has been described previously (36). Recombinant TRAMPTrf4-236p was purified using procedures identical to those used for WT TRAMP reconstitution. Recombinant Mtr4p was expressed and purified as described previously (13).
RNAs were purchased from Dharmacon. Sequences are listed in SI Materials and Methods. RNA oligonucleotides were 5′ radiolabeled with T4 polynucleotide kinase, followed by purification on denaturing PAGE. The duplexes were generated by annealing top strand (R16, R36) to its corresponding bottom strand, followed by purification on nondenaturing PAGE (37).
Unwinding Reactions.
Unwinding reactions were performed at 30 °C in a temperature-controlled heating block in a buffer containing 40 mM Mops (pH 6.5), 100 mM NaCl, 0.5 mM MgCl2, 5% glycerol (vol/vol), 0.01% Nonidet P-40 (vol/vol), 2 mM DTT, and 0.7 U/μL Protector RNase Inhibitor (Roche). Before the reaction, radiolabeled duplex RNA (0.5 nM final concentration) was incubated for 5 min with the indicated concentration of TRAMP. Reactions were started by addition of equimolar dATP (or ATP) and MgCl2. After the reaction start, aliquots were removed at times indicated, and the reaction was stopped by addition of an equal volume of 1% SDS, 0.5 mM EDTA, 20% glycerol, and dye markers. Samples were applied to 15% nondenaturing PAGE, and duplex and single-stranded RNAs were separated by electrophoresis at 15 V/cm. Gels were dried, bands were visualized on a Storm PhosphorImager (GE Healthcare) and quantified using ImageQuant software (GE Healthcare). Unwinding rate constants were calculated as described (37).
Simultaneous Measurement of Polyadenylation and Unwinding Reactions on Streptavidin Beads.
R16-bio/R17 duplex (50 fmol) was incubated with 5 μL of Streptavidin UltraLink Resin (Thermo Scientific) in unwinding reaction buffer (200 μL) for 1 h at 4 °C. After incubation, the resin was washed twice with reaction buffer until no radiation could be detected in the wash, and resuspended to a 50% slurry. Reaction buffer (10 μL) containing 2 mM equimolar ATP and MgCl2 was added to the resin, followed by incubation for 5 min on a temperature-controlled heating block at 30 °C. TRAMP (300 nM final) was then added to initiate the reaction. At 10 min after reaction start, an equal volume of stop buffer (1% SDS, 50 mM EDTA) was added to terminate the reaction, the sample was centrifuged, and the supernatant was removed. Samples were applied to a 15% denaturing PAGE. Gels were dried, and individual bands were visualized on a Storm PhosphorImager (GE Healthcare) and quantified using ImageQuant software (GE Healthcare).
Supplementary Material
Acknowledgments
We thank Sukanya Srinivasan (Case Western Reserve University) for help with TRAMP preparations, and members of the E. J. laboratory for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grants GM067700 (to E.J.) and GM069949 (to J.T.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201085109/-/DCSupplemental.
References
- 1.Anderson JT, Wang X. Nuclear RNA surveillance: No sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 2.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: The link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779:239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. J Cell Sci. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- 4.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 5.Vanácová S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Lubas M, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia H, et al. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein J, Ballin JD, Patterson DN, Wilson GM, Toth EA. Unique properties of the Mtr4p-poly(A) complex suggest a role in substrate targeting. Biochemistry. 2010;49:10357–10370. doi: 10.1021/bi101518x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein J, Patterson DN, Wilson GM, Toth EA. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, a 3′→5′ helicase partner of the nuclear exosome. J Biol Chem. 2008;283:4930–4942. doi: 10.1074/jbc.M706677200. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder P, Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 16.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 17.Jankowsky E. RNA helicases at work: Binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 21.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 22.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers GW, Jr, Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 24.Shuman S. Vaccinia virus RNA helicase. Directionality and substrate specificity. J Biol Chem. 1993;268:11798–11802. [PubMed] [Google Scholar]
- 25.Tanaka N, Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- 26.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 27.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson RN, et al. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pena V, et al. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell. 2009;35:454–466. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, et al. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009;16:731–739. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walbott H, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–134. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J Biol Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 36.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.