Abstract
Axon regeneration in the central nervous system normally fails, in part because of a developmental decline in the intrinsic ability of CNS projection neurons to extend axons. Members of the KLF family of transcription factors regulate regenerative potential in developing CNS neurons. Expression of one family member, KLF7, is down-regulated developmentally, and overexpression of KLF7 in cortical neurons in vitro promotes axonal growth. To circumvent difficulties in achieving high neuronal expression of exogenous KLF7, we created a chimera with the VP16 transactivation domain, which displayed enhanced neuronal expression compared with the native protein while maintaining transcriptional activation and growth promotion in vitro. Overexpression of VP16-KLF7 overcame the developmental loss of regenerative ability in cortical slice cultures. Adult corticospinal tract (CST) neurons failed to up-regulate KLF7 in response to axon injury, and overexpression of VP16-KLF7 in vivo promoted both sprouting and regenerative axon growth in the CST of adult mice. These findings identify a unique means of promoting CST axon regeneration in vivo by reengineering a developmentally down-regulated, growth-promoting transcription factor.
Keywords: gene therapy, spinal cord injury, adeno associated virus, laser capture microdissection
Axon regeneration generally fails after injury to the CNS, preventing substantial recovery. CNS regeneration is limited in part by the presence of extrinsic inhibitory molecules at the injury site, and also because adult CNS neurons possess an intrinsically low capacity for axon growth compared with embryonic or peripheral nervous system (PNS) neurons (1–3). Intrinsic regenerative capacity appears to be particularly low in the corticospinal tract (CST), an essential motor control pathway and important therapeutic target in humans (4). For instance, CST axons have shown mixed responses when inhibitory signals are neutralized (5, 6), and regenerate minimally or not at all into growth permissive tissue grafts that support some regeneration from propriospinal and brainstem neurons (7–10).
The intrinsic molecular mechanisms that limit CNS axon growth remain unclear, but likely reflect a suboptimal pattern of regenerative gene expression (1, 11). In sensory neurons, overexpression of GAP43 and CAP23 or transcription factors, including ATF3, STAT3, or ID2, have produced modest gains in spinal regeneration (12–15). In the visual system, overexpression of p300 modestly increases growth (16), and knockdown of PTEN and SOCS3 results in substantial regeneration (17, 18). In the CST, overexpression of the neurotrophin receptor TrkB enables regeneration into subcortical BDNF-secreting tissue grafts, and, notably, knockout of PTEN evokes regeneration in the spinal cord (19, 20). The regenerative response evoked by these gene manipulations, including PTEN knockout, remains incomplete in terms of both the distance traveled by regenerating axons and the percent of neurons that respond to treatment. These findings illustrate the critical need to develop additional tools to enhance the intrinsic growth state of CNS neurons.
We recently identified the Krüppel-like factors (KLFs), a 17-member family of zinc finger transcription factors, as regulators of CNS axon regeneration (21, 22). Knockout of one family member, KLF4, promotes axon regeneration by retinal ganglion cells (RGCs) in vivo, demonstrating that KLF family members can act as neuron-intrinsic suppressors of axon growth. In contrast to KLF4, KLF7 increases axon growth when overexpressed in cultured neurons. KLF7 is expressed in CNS neurons during developmental periods of axon growth, and down-regulated as neurons mature (22, 23). Neurons that successfully regenerate, including zebrafish RGCs and peripherally injured mammalian DRGs, up-regulate KLF7 in response to axotomy (24, 25). Here we explore whether increasing KLF7 activity can enhance regenerative ability in the adult CNS. We first found that through an unknown mechanism that relies on its N-terminal domain, KLF7 protein expression is suppressed in mature neurons even after attempted overexpression. To overcome this suppression, we engineered a fusion protein with the VP16 viral transcriptional activation domain (26). In dissociated neurons, VP16-KLF7 effectively promotes neurite outgrowth while displaying enhanced expression compared with wild-type KLF7, and in cortical slice cultures, VP16-KLF7 expression overcomes an age-dependent decline in axon growth after injury. Moreover, adult CST neurons fail to reexpress KLF7 after injury, and forced expression of VP16-KLF7 promotes regeneration of CST axons after spinal cord injury. These data provide an example of a reengineered intrinsic growth program that results in robust CST regeneration in the spinal cord.
Results
Expression of KLF7 Is Suppressed by an N-terminal Sequence.
KLF7 and the closely related KLF6 enhance neurite outgrowth when overexpressed in cultured RGCs or cortical neurons (21, 22), but their potential for promoting axon growth in the adult CNS remains unexplored. We focused on KLF7, based on its larger effects in vitro (22). We directly visualized exogenously expressed KLF7 with an N-terminal EGFP tag (EGFP-KLF7). Luciferase assays confirmed EGFP-KLF7’s ability to activate transcription from the p21/CIF promoter, a previously identified target (27) (Fig. S1). As expected, EGFP-KLF7 localized to the nucleus of transfected cortical neurons (Fig. S2) and significantly increased neurite lengths compared with an EGFP control (Fig. 1 A and C). Unexpectedly, however, EGFP-KLF7 was detected in only ∼4% of neurons (Fig. 1B), compared with 41% for EGFP. Expression of KLF7 in cultured neurons was not increased by using smaller epitope tags, such as Flag or myc, or by attaching tags to the C terminus, suggesting that poor expression of KLF7 was not an artifact of tag size or location. Other KLF family members are subject to rapid degradation under the control of posttranslational modifications and protein–protein interactions; notably, the stability of both KLF5 and of KLF6 (which has high sequence homology to KLF7) is increased by deleting N-terminal motifs (28, 29). We found that deletion of the KLF7 N terminus, a previously identified 76-aa acidic transactivation domain, increased KLF7 expression more than threefold (Fig. 1B). This truncated KLF7 localized appropriately to the nucleus (Fig. S2), but no longer increased neurite length, and, as expected, lacked the ability to activate the p21 promoter (Fig. 1 C and D and Fig. S1) (30). Thus, N-terminal domains of KLF7 reduce neuronal expression, but also appear to be required for transcriptional activation and promotion of neurite outgrowth.
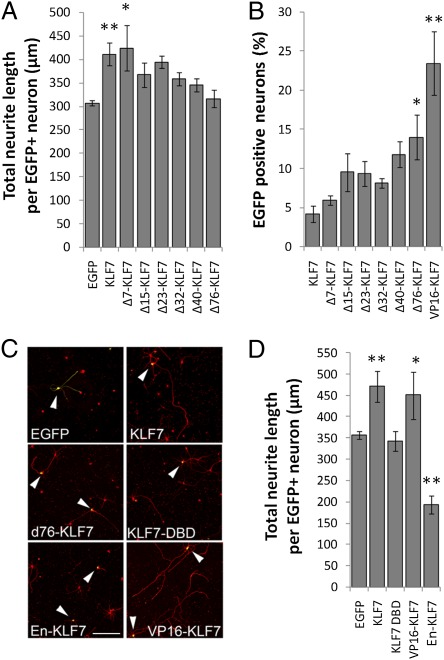
Fig. 1.
Structure/function analysis of KLF7 identifies VP16-KLF7 as an effectively expressed growth enhancer. Cortical neurons were transfected with EGFP-tagged KLF7 constructs, cultured for 3 d, and stained for neuron-specific βIII tubulin. Neurite lengths and EGFP expression were quantified by automated image analysis (Cellomics ArrayScan VTI). (A) EGFP-KLF7 expression significantly increased neurite length, and this effect was blocked by removal of the N-terminal 76-aa acidic activation domain. (B) The percentage of neurons with detectable EGFP-KLF7 expression was increased by deletion of 76 N-terminal amino acids, and was highest in a construct in which the endogenous N-terminal domain was replaced with VP16. (C and D) Wild-type EGFP-KLF7 or EGFP-VP16-KLF7 significantly increased neurite lengths, whereas EGFP-En-KLF7 significantly decreased neurite lengths. The DNA binding domain of KLF7 alone did not affect neurite outgrowth. For each treatment, >200 individual neurons from three replicate experiments were analyzed. Error bars are SEM. **P < 0.01 vs. EGFP (A and D) or KLF7 (C), ANOVA, Dunnett’s post hoc comparison. (Scale bar: 50 μm.)
VP16-KLF7 Promotes Neurite Outgrowth and Is Effectively Expressed.
Poor expression of KLF7 could limit its usefulness as a potential tool for increasing axon growth. We therefore explored partial deletion of the N-terminal domain as a strategy to enhance expression while maintaining growth promotion. In a series of truncation mutants, however, we found that significant growth promotion was lost upon deletion of 15 or more N-terminal amino acids, whereas KLF7 expression was not significantly increased until 76 amino acids were deleted (Fig. 1 A and B). As an alternative, we replaced the endogenous KLF7 N terminus with the viral VP16 transactivation domain, a widely used and clinically approved motif that promotes strong transcriptional activation through recruitment of basal transcription factors and acetyl transferases (26, 31–34). Luciferase assays confirmed activation of the p21 promoter by VP16-KLF7 (Fig. S1). Transfection of cortical neurons with EGFP-tagged VP16-KLF7 resulted in a fivefold increase in the number of EGFP+ neurons compared with wild-type EGFP-KLF7 (Fig. 1B), consistent with suppression of KLF7 expression by endogenous N-terminal motifs. Furthermore, VP16-KLF7 localized to the nucleus and increased neurite lengths similarly to wild-type KLF7 (Fig. S2 and Fig. 1 C and D). In contrast, replacement of the endogenous KLF7 N terminus with an Engrailed transcriptional repression domain suppressed the p21 promoter and decreased neurite lengths by 45% (Fig. S1 and Fig. 1 C and D) (35). These data establish a tight correlation between transcriptional activation and neurite growth promotion by KLF7, and identify transcriptionally active VP16-KLF7 as an effective stimulator of neurite outgrowth that is expressed more effectively than exogenous wild-type KLF7.
Overexpression of VP16-KLF7 Overcomes the Age-Dependent Loss of Axon Growth in Cortical Slice Culture.
KLF7 expression in the CNS declines during the postnatal period when neurons lose regenerative capacity (22, 23, 36). Can continued KLF7 activity abrogate the developmental loss of axon growth ability? We developed a slice culture assay to address this question because we found that cortical neurons, as they age postnatally, become increasingly difficult to maintain in dissociated culture. Bilateral slices of P5 cortex were unilaterally labeled with AAV8 viral particles expressing EGFP (AAV8-EGFP), then transected at the midline and cultured in apposition to an unlabeled contralateral half slice. Growth of EGFP+ axons across the midline was robust (∼10,000 axons) in slices aged only 1 d in vitro (DIV), but declined 88% when transections were performed at 8 DIV, even when 8 DIV slices were provided with contralateral recipient tissue aged only 1 DIV (Fig. 2J). This decline thus reflects changes in the ability of labeled neurons to extend axons, rather than changes in the recipient growth environment. KLF7 expression in cortical slices, as assessed by qRT-PCR, declined by 83% between 1 and 8 DIV (Fig. 2I). Thus, both axon growth across the midline and KLF7 expression in postnatal cortical slices declined with age in culture.
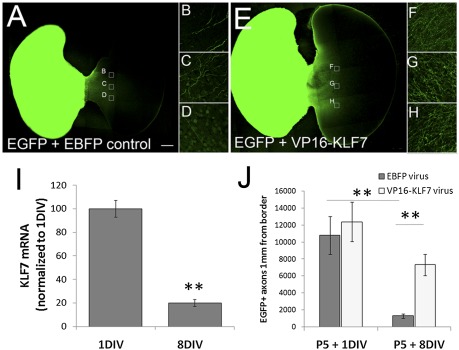
Fig. 2.
VP16-KLF7 attenuates the age-dependent decrease in axon growth in transected cortical slice cultures. Bilateral cortical slices connected by the corpus callosum were prepared from P5 rats and treated with a mixture of viral particles encoding AAV8-EGFP and either AAV8-EBFP (control) or AAV8-VP16-KLF7. After 1 or 8 DIV, the hemispheres were separated and paired with unlabeled P5 recipient tissue; axon growth was evaluated 7 d later. Slices aged 8 DIV and treated with AAV8-VP16-KLF7 (E–H) show enhanced axon growth compared with control (A–D). (I) KLF7 expression decreased ∼80% between 1 and 8 DIV. Three replicate slices from three experiments were analyzed. **P < 0.01, paired t test, mean ± SEM. (J) Axon growth from cortical slices declined about eightfold between 1 and 8 DIV. Application of VP16-KLF7 did not increase axon growth after injury at 1 DIV but increased axon growth about sixfold at 8 DIV. Error bars are SEM. **P < 0.01, ANOVA with Tukey’s post hoc comparison. Four replicate slices in two experiments were analyzed. (Scale bar: 500 μm.)
To test whether transcriptional activation by KLF7 can prevent the developmental loss of axon growth, we prepared viral particles encoding Flag-EBFP (control), Flag-KLF7, or Flag-VP16-KLF7. To identify transduced neurons, mCherry was coexpressed using a 2A linker, producing two separate proteins from a single transcript (Fig. S3) (37). In preliminary experiments we encountered significant difficulty in effectively delivering wild-type KLF7 using AAV8. First, despite repeated attempts with various tagged viral plasmids (KLF7-2A-mCherry, Flag-KLF7), titers were consistently two orders of magnitude lower than for EBFP or VP16-KLF7. Second, when AAV8-KLF7 was applied to cortical slices, the flag or 2A tags were barely detectable, in contrast to robust expression of tagged AAV8-EBFP and AAV8-VP16-KLF7. The mechanism(s) responsible for the low expression of AAV8-KLF7 and its potential relationship to the low expression in transfected cortical neurons remains unclear, but precluded the use of wild-type KLF7 in these experiments. We therefore proceeded with AAV8-VP16-KLF7, which was expressed more efficiently than wild type, and was equally effective in promoting neurite outgrowth in vitro. In cortical slices, AAV8-VP16-KLF7 had no effect on the robust growth seen at 1 DIV, but caused a fivefold increase in axon growth across the midline at 8 DIV (Fig. 2 A, E, and J). Thus, overexpression of VP16-KLF7 promoted axon growth selectively in older cortical neurons.
Overexpression of VP16-KLF7 Enhances CST Axon Growth in the Injured Spinal Cord.
Zebrafish RGCs and mammalian dorsal root ganglion (DRG) neurons, which are regeneration competent, up-regulate KLF7 mRNA in response to axonal injury (24, 25). In contrast, we detected no change in KLF7 gene expression after cervical axotomy of adult CST neurons when these neurons were purified by laser capture microdissection (LCM; Fig. 3 A and B) and analyzed by qRT-PCR (Fig. 3C). As a control for detection of KLF7 we measured twofold higher expression of KLF7 in LCM-harvested cortex at P1 (Fig. 3C) (36). Thus, unlike neurons capable of successful regeneration, adult CST neurons do not up-regulate KLF7 expression after spinal axotomy (38).
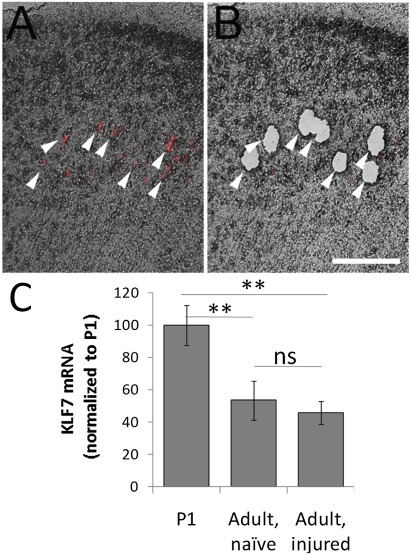
Fig. 3.
KLF7 expression in CST neurons is insensitive to cervical axotomy. CST neurons were retrogradely labeled and subjected to either sham injury or cervical dorsal transection. (A) Fluorescently labeled CST neurons in a transverse section of frontal cortex (arrows). (B) After laser capture microdissection, CST neurons were selectively gathered. (C) CST neurons were collected from adult naïve or spinally injured animals. qRT-PCR detected a 50% decrease in KLF7 expression compared with P1 cortex, independent of injury. n = 4 animals in each treatment. Error bars are SEM. **P < 0.01, ANOVA with Tukey’s post hoc comparison. (Scale bar: 100 μm.)
Can increased KLF7 activity promote growth in adult CST neurons in vivo? We first used unilateral pyramidotomy to test VP16-KLF7’s ability to promote compensatory sprouting by unlesioned axons (6). One week before injury, the left sensorimotor cortex of adult mice was coinjected with AAV8-EGFP and either AAV8-EBFP-2A-mCherry (control) or AAV8-VP16-KLF7-2A-mCherry at a 1:2 ratio; >95% of EGFP-labeled neurons coexpressed mCherry in cell bodies (Fig. 4 B–D). Two weeks after injection, EGFP brightly labeled axons in the internal capsule, the left medullary pyramid, and the right spinal CST (Fig. 4 A and E–K). As expected, lesion of the right (nontransduced) pyramid largely deprived the left side of the spinal cord of CST input; complete lesions were verified by the unilateral loss of PKCγ immunoreactivity in the left CST tract (Fig. S4) (6). CST axons in control animals displayed minimal contralateral sprouting (Fig. 5 A and C), and this sprouting response was significantly enhanced by VP16-KLF7 treatment (Fig. 5 B and C). These results demonstrate that following unilateral denervation of the spinal cord, overexpression of VP16-KLF7 increases contralateral sprouting of uninjured CST axons.
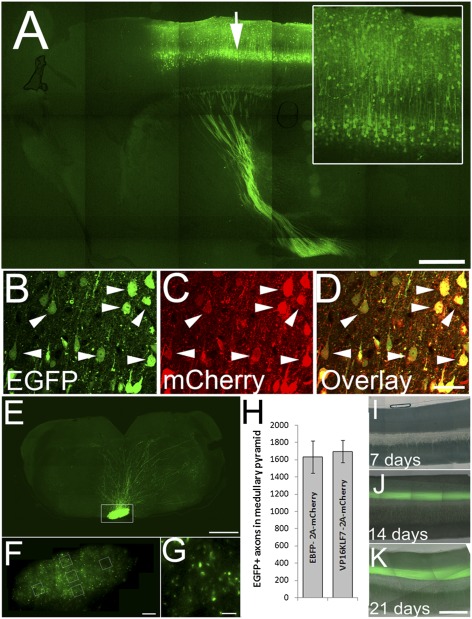
Fig. 4.
AAV8-EGFP identifies transduced CST neurons. (A) Sagittal brain section 4 wk after injection with AAV8-EGFP (arrow). Neuronal somata and dendrites (Inset) and descending axons are brightly labeled with EGFP. (B–D) AAV8-EGFP and AAV8-EBFP-2A-mCherry were coinjected at a 1:2 ratio. Four weeks later, nearly all EGFP+ cell bodies also express mCherry (arrowheads). (E–G) At 9 wk after cortical injection of AAV8-EGFP, the pyramidal tract is visible in horizontal sections of the medulla (box, E), and at higher magnification individual axons are clearly visible (F and G). Axon counts were sampled (boxes, F), and the total number of labeled CST axons was extrapolated from the cross-sectional area of the pyramid. (H) There was no significant difference in average axon counts between control (EBFP) and VP16KLF7-injected animals (n = 9 control, 10 VP16-KLF7 animals; mean ± SEM). (I–K) Midsagittal sections of cervical spinal cord 7, 14, and 21 d after viral injections. Error bars are SEM. A, E, F, and I–K are composites of multiple fields of view. (Scale bars: A and E, 500 μm; D, 30 μm; F, 100 μm; G, 10 μm; K, 250 μm.)
Fig. 5.
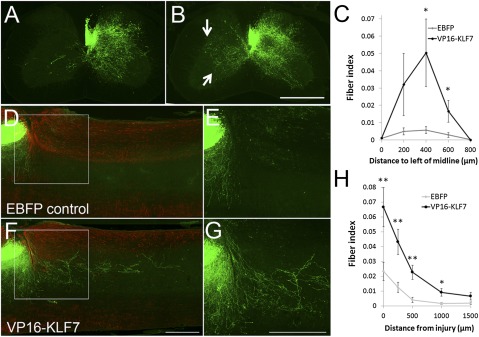
Overexpression of VP16-KLF7 promotes CST axon sprouting and regeneration in the injured spinal cord. (A and B) Transverse sections of spinal cord 4 wk after unilateral right pyramidotomy and 5 wk after coinjection of AAV8-EGFP and AAV8-EBFP (A) or AAV8-VP16-KLF7 (B) into motor cortex. Contralateral EGFP+ sprouts are abundant in VP16-KLF7–treated animals (arrow, B). Quantification (C) shows significantly elevated sprouting in VP16-KLF7 animals up to 600 μm beyond the midline. *P < 0.05, paired t test, n = 9 in each group. (D–G) Sagittal sections of cervical spinal cord 8 wk after dorsal hemisection and 9 wk after coinjection of AAV8-EGFP and AAV8-EBFP (D and E) or AAV8-VP16-KLF7 (F and G) into motor cortex. GFAP (red) shows gliosis around the injury site. EGFP+ axonal profiles (green) are visible caudal to the injury in VP16-KLF7–treated animals but not controls. (H) Quantification of EGFP+ profiles shows a significant increase up to distances of 1 mm caudal to the injury. Error bars are SEM. *P < 0.05, paired t test, n = 9 (control), 10 (VP16KLF7). A, B, D, and F are composites of multiple fields of view. (Scale bars: 500 μm.)
Finally, to test the ability of VP16-KLF7 to promote regeneration from injured CST axons, virus was injected into motor cortex and 1 wk later, before detectable expression of EGFP in the spinal cord (Fig. 4I), transduced CST axons on the right side of the spinal cord were severed by dorsal hemisection. Eight weeks later, growth of EGFP-labeled axons in the distal spinal cord was quantified by a blinded observer, with counts normalized to the number of EGFP+ CST fibers in the medullary pyramids. In control animals, few EGFP+ fibers were detected distal to the injury, likely reflecting trace amounts of spontaneous CST regeneration (39) (Fig. 5 D, E, and H). Mice treated with VP16-KLF7 had significantly more CST axons distal to the injury site (Fig. 5 F–H). VP16-KLF7 expression did not affect the number of labeled axons in the pyramids (Fig. 4H), ruling out potentially confounding effects of the efficiency of viral transduction or intensity of EGFP expression. Distal growth was concentrated near the main CST tract on the medial-lateral axis, and examination of a complete sagittal series of spinal cords, as well as horizontal sections from a second cohort of animals, confirmed complete severing of dorsolateral CST tracts and an absence of labeled ventral CST axons, ruling out spared minor tracts as the source (Figs. S5 and S6). The second cohort of animals also confirmed the regeneration-promoting ability of VP16-KLF7 (Fig. S6). New growth did not appear to emerge directly from the injured ends of axons. Thus, it is most likely that VP16-KLF7 promotes “regenerative sprouting,” that is, regenerative growth that arises from collateral branching by injured axons (39). These data demonstrate that manipulating the activity of KLFs in fully adult neurons can increase sprouting and regenerative ability, and provide an example of effective growth promotion by viral particle-mediated gene delivery.
Discussion
KLF7 expression correlates with regenerative capacity across neuronal age, species, and cellular subtype, and is important both for the development of cortical efferent axons in mammals and for regenerative axon growth in the zebrafish (22–24, 40). Our findings show that adult mammalian CST neurons, which regenerate poorly and developmentally down-regulate KLF7, fail to increase KLF7 expression in response to spinal axotomy. Interestingly, in RGCs another KLF, KLF4, is developmentally up-regulated and acts to inhibit axonal regeneration (22). We now demonstrate that VP16-KLF7 promotes axon regeneration in the adult CST, supporting a model in which transcriptional activity by KLF family members can regulate axon growth both positively and negatively across diverse CNS populations.
We also demonstrate that the molecular biology of KLF7 regulation of axon growth has an intriguing complexity. First, the N-terminal activation domain of KLF7 is required for transcriptional activation and growth promotion (Fig. 1A), although it could also possess a nontranscriptional activity that drives neurite extension. The viral VP16 activation domain restores growth promotion by N-truncated KLF7, and the Drosophila Engrailed repression domain switches KLF7 from growth promotion to inhibition. The fact that two evolutionarily divergent domains, with well-characterized and opposite effects on transcription, give opposite effects on neurite outgrowth when fused to KLF7’s DNA binding domain strongly favors transcriptional activation as KLF7’s mechanism of action. Second, N-terminal sequences keep expression low in neurons even after attempted overexpression, possibly by targeting KLF7 protein for degradation (28, 29). We developed VP16-KLF7 as a means to enhance expression, but cannot rule out the possibility that VP16-KLF7 could activate nonphysiological targets, which would confound conclusions regarding endogenous KLF7. Previous comparisons of VP16 fusion proteins to wild type have shown broad but imperfect similarity in transcriptional targets, often due to elevated activation of targets by VP16 chimeras (32–34, 41). These precedents, combined with the fact that VP16-KLF7 and wild-type KLF7 share growth-promoting properties in vitro, increase the likelihood that VP16-KLF7 and wild-type KLF7 enhance axon growth through a common mechanism in vivo. Overall, with these caveats in mind, our data support a model in which activation of KLF7 target genes promotes neurite outgrowth, and repression of these target genes inhibits neurite outgrowth.
What might these KLF7 target genes be? p21, a KLF7 target (Fig. S1) (27), can stimulate axon growth by blocking RhoA signaling (42). KLF7 also binds and activates the promoters of regeneration-associated genes, including TrkA and TrkB receptors, GAP-43, and the L1CAM adhesion molecule (43–45). Interestingly, CST neurons normally up-regulate growth-associated proteins only in response to cortical, but not spinal, axotomy (38). We speculate that overexpression of VP16-KLF7 may circumvent this distance requirement by activating growth-associated genes and enabling a more effective regenerative response to distant axotomy.
A critical question for future study is whether axons stimulated to regenerate by VP16-KLF7 can form appropriately targeted synapses and lead to functional recovery (5, 20), or whether additional interventions might be necessary to achieve behavioral improvements. Indeed, similar to previous work (20), VP16-KLF7 appears to evoke a regenerative response from a minority of transduced neurons. To boost the number of injured neurons that participate in the regenerative response and the distance they regenerate, further engineering of VP16-KLF7 might be necessary, as might a combinatorial approach with other molecular manipulations. Therapeutic combination targets could include extracellular inhibitory molecules (5, 46), stimulation by neurotrophins or cytokines (47, 48), and a wide range of regeneration-associated transcription factors (e.g., cJun, ATF3, Sox11, c/EBP) (49) or effector molecules (e.g., SPIRRA, Arginase), in addition to those previously discussed (50, 51). VP16-KLF7 stands out as being effective in promoting CST regeneration when delivered to adult wild-type animals, and precedent exists for the clinical use of a VP16 chimera (31). KLF7 thus represents a powerful and potentially therapeutically relevant means by which to promote regenerative capacity in adult CNS neurons, motivating future exploration of optimal combinations of VP16-KLF7 with existing strategies.
Methods
Detailed methods are available in SI Methods.
Cloning and Virus Construction.
EGFP-tagged KLF7 mutants: full-length KLF7, truncation mutants, or VP16/Engrailed domains fused to the KLF7 zinc finger and adjacent nuclear localization signal were inserted 3′ to EGFP in pAAV-MCS (Stratagene). 2A-mCherry constructs: A 2A peptide sequence (37) was inserted 5′ to mCherry in a pAAV-MCS backbone, and then EBFP (Addgene plasmid 14891). KLF7 or VP16-KLF7 were inserted 5′ to the 2A sequence. Viral particles were produced at the Miami Project Viral Vector Core with titers of 2.5–3 × 1014 viral particles per milliliter.
Luciferase Assays.
293T cells were cultured in supplemented DMEM and cotransfected at a 3:1 ratio with EGFP-tagged KLF constructs and a reporter plasmid with 970 bp of the p21 promoter, or scrambled control sequence, upstream of Renilla luciferase (SwitchGear Genomics). Luminescence was quantified 24 h posttransfection following cell lysis and addition of luciferase substrate.
Neuronal Cell Culture.
Neurons were cultured as described previously (21). Briefly, rat cortical neurons from postnatal day 3 pups were dissociated, transfected with EGFP-tagged constructs, and plated on poly-d-lysine/laminin-coated 24-well plates at 10,600 cells per well in supplemented, glial-enriched neurobasal media. After 3 d, neurons were fixed and stained for neuron-specific βIII-tubulin, and then imaged using an ArrayScan VTI HCS Reader (Cellomics). Neurite lengths were quantified for all EGFP+ neurons using the Neuronal Profiling Algorithm.
Cortical Slice Culture.
Viral particles were applied to slices of P5 rat cortices comprising both hemispheres connected by the corpus callosum. Sections were rinsed 24 h later and either immediately or 7 d later, transected along the midline and paired with contralateral P5 slices that had not been treated with viral particles. After 7 d, confocal microscopy quantified EGFP+ profiles that extended >1 mm from the border at all Z-planes.
Laser Capture Microdissection and qRT-PCR Analysis of KLF7 Expression.
Cholera toxin subunit B (CTB)-Alexa Fluor 555 was injected into the C3/C4 dorsal column of 8-wk-old female C57/BI6 mice. One week later, labeled cells were collected from cryosections into extraction buffer using a Laser Capture Microscope (Leica LMD6000), and RNA was extracted. 18S and KLF7 and primer efficiencies were calculated in a dilution series as described; KLF7 primers recognize all known isoforms (22). Using 18S as a reference gene, KLF7 abundance for each sample was normalized to the average threshold cycle (CT) of KLF7 at P1 using the Plaffl equation (52). In slice cultures, KLF7 abundance was calculated similarly, with all samples normalized to the average KLF7 CT at 1 DIV.
Spinal Cord Injury Experiments.
A total of 0.5 μL of AAV8 viral particles were injected into sensorimotor cortex of 8-wk-old female C57/Bl6 mice. Pyramidotomies were performed 1 wk postinjection as previously described (6), and 4 wk postinjury, EGFP+ axon counts in the denervated gray matter were normalized to total axons in the medulla. For spinal injuries, 1 wk postinjection the right dorsal quadrant of the spinal cord was transected at C4/C5 using a custom-built Vibraknife (53) set to a depth of 0.9 mm and extending 1 mm to the left of the midline and beyond the right edge of the spinal cord. Eight weeks postinjury, sections of brain, medulla, and spinal cord were prepared. Immunohistochemistry used anti-GFAP antibodies (DAKO; 1:500). In all sagittal sections of right (injured) spinal cord and the first section to the left of the midline, a virtual line was drawn at the site of injury, and EGFP+ profiles were counted at 0, 250, 500, 1,000, or 1,500 μm caudal, then normalized to axon counts in the medullary pyramids (fiber index) (20).
Supplementary Material
Acknowledgments
We thank Y. Martinez, O. Franch, Y. Shi, Pingping Jia, and Anthony Oliva for technical assistance. This work was supported by the James and Esther King Biomedical Research Program Grant JEK09KW-05 (to J.L.B.); the Craig H. Neilsen Foundation (M.G.B.); US Army Grant W81XWH-05-1-0061 (to M.G.B., V.P.L., and J.L.B.); National Institutes of Health Grants HD057521 (to V.P.L.), NS059866 (to J.L.B.), and NEI EY020913 and P30-EY014801 (to J.L.G.); the American Health Assistance Foundation (J.L.G.); Research to Prevent Blindness (J.L.G.); the Buoniconti Fund; and the Walter G. Ross Distinguished Chairs in Developmental Neuroscience (V.P.L.) and in Ophthalmic Research (J.L.G.).
Footnotes
Conflict of interest statement: J.L.G., V.P.L., J.L.B., and M.G.B have filed a patent pertaining to KLF-mediated neural repair.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120684109/-/DCSupplemental.
References
- 1.Sun F, He Z. Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr Opin Neurobiol. 2010;20:510–518. doi: 10.1016/j.conb.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huebner EA, Strittmatter SM. Axon regeneration in the peripheral and central nervous systems. Results Probl Cell Differ. 2009;48:339–351. doi: 10.1007/400_2009_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science. 2002;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- 4.Lemon RN, Griffiths J. Comparing the function of the corticospinal system in different species: Organizational differences for motor specialization? Muscle Nerve. 2005;32:261–279. doi: 10.1002/mus.20333. [DOI] [PubMed] [Google Scholar]
- 5.Cafferty WB, Duffy P, Huebner E, Strittmatter SM. MAG and OMgp synergize with Nogo-A to restrict axonal growth and neurological recovery after spinal cord trauma. J Neurosci. 2010;30:6825–6837. doi: 10.1523/JNEUROSCI.6239-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JK, et al. Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron. 2010;66:663–670. doi: 10.1016/j.neuron.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearse DD, et al. cAMP and Schwann cells promote axonal growth and functional recovery after spinal cord injury. Nat Med. 2004;10:610–616. doi: 10.1038/nm1056. [DOI] [PubMed] [Google Scholar]
- 8.Xu XM, Zhang SX, Li H, Aebischer P, Bunge MB. Regrowth of axons into the distal spinal cord through a Schwann-cell-seeded mini-channel implanted into hemisected adult rat spinal cord. Eur J Neurosci. 1999;11:1723–1740. doi: 10.1046/j.1460-9568.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- 9.Richardson PM, Issa VM, Aguayo AJ. Regeneration of long spinal axons in the rat. J Neurocytol. 1984;13:165–182. doi: 10.1007/BF01148324. [DOI] [PubMed] [Google Scholar]
- 10.Hollis ER, 2nd, Lu P, Blesch A, Tuszynski MH. IGF-I gene delivery promotes corticospinal neuronal survival but not regeneration after adult CNS injury. Exp Neurol. 2009;215:53–59. doi: 10.1016/j.expneurol.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi F, Gianola S, Corvetti L. Regulation of intrinsic neuronal properties for axon growth and regeneration. Prog Neurobiol. 2007;81:1–28. doi: 10.1016/j.pneurobio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Bomze HM, Bulsara KR, Iskandar BJ, Caroni P, Skene JH. Spinal axon regeneration evoked by replacing two growth cone proteins in adult neurons. Nat Neurosci. 2001;4:38–43. doi: 10.1038/82881. [DOI] [PubMed] [Google Scholar]
- 13.Yu P, et al. Inhibitor of DNA binding 2 promotes sensory axonal growth after SCI. Exp Neurol. 2011;231:38–44. doi: 10.1016/j.expneurol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bareyre FM, et al. In vivo imaging reveals a phase-specific role of STAT3 during central and peripheral nervous system axon regeneration. Proc Natl Acad Sci USA. 2011;108:6282–6287. doi: 10.1073/pnas.1015239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaub P, et al. The histone acetyltransferase p300 promotes intrinsic axonal regeneration. Brain. 2011;134:2134–2148. doi: 10.1093/brain/awr142. [DOI] [PubMed] [Google Scholar]
- 17.Sun F, et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park KK, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollis ER, 2nd, Jamshidi P, Löw K, Blesch A, Tuszynski MH. Induction of corticospinal regeneration by lentiviral trkB-induced Erk activation. Proc Natl Acad Sci USA. 2009;106:7215–7220. doi: 10.1073/pnas.0810624106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K, et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat Neurosci. 2010;13:1075–1081. doi: 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackmore MG, et al. High content screening of cortical neurons identifies novel regulators of axon growth. Mol Cell Neurosci. 2010;44:43–54. doi: 10.1016/j.mcn.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore DL, et al. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laub F, et al. Developmental expression of mouse Krüppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev Biol. 2001;233:305–318. doi: 10.1006/dbio.2001.0243. [DOI] [PubMed] [Google Scholar]
- 24.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Zou H, Ho C, Wong K, Tessier-Lavigne M. Axotomy-induced Smad1 activation promotes axonal growth in adult sensory neurons. J Neurosci. 2009;29:7116–7123. doi: 10.1523/JNEUROSCI.5397-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 27.Smaldone S, Laub F, Else C, Dragomir C, Ramirez F. Identification of MoKA, a novel F-box protein that modulates Krüppel-like transcription factor 7 activity. Mol Cell Biol. 2004;24:1058–1069. doi: 10.1128/MCB.24.3.1058-1069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Zhou Z, Guo P, Dong JT. Proteasomal degradation of the KLF5 transcription factor through a ubiquitin-independent pathway. FEBS Lett. 2007;581:1124–1130. doi: 10.1016/j.febslet.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez E, Aburjania N, Priedigkeit NM, DiFeo A, Martignetti JA. Nucleo-cytoplasmic localization domains regulate Krüppel-like factor 6 (KLF6) protein stability and tumor suppressor function. PLoS ONE. 2010;5:e12639. doi: 10.1371/journal.pone.0012639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto N, et al. Cloning the cDNA for a new human zinc finger protein defines a group of closely related Krüppel-like transcription factors. J Biol Chem. 1998;273:28229–28237. doi: 10.1074/jbc.273.43.28229. [DOI] [PubMed] [Google Scholar]
- 31.Rajagopalan S, et al. Use of a constitutively active hypoxia-inducible factor-1alpha transgene as a therapeutic strategy in no-option critical limb ischemia patients: Phase I dose-escalation experience. Circulation. 2007;115:1234–1243. doi: 10.1161/CIRCULATIONAHA.106.607994. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Lazar MA. Differential gene regulation by PPARgamma agonist and constitutively active PPARgamma2. Mol Endocrinol. 2002;16:1040–1048. doi: 10.1210/mend.16.5.0825. [DOI] [PubMed] [Google Scholar]
- 33.Tang DQ, et al. Reprogramming liver-stem WB cells into functional insulin-producing cells by persistent expression of Pdx1- and Pdx1-VP16 mediated by lentiviral vectors. Lab Invest. 2006;86:83–93. doi: 10.1038/labinvest.3700368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirai H, Tani T, Kikyo N. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int J Dev Biol. 2010;54:1589–1596. doi: 10.1387/ijdb.103194hh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han K, Manley JL. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 37.de Felipe P, et al. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Mason MR, Lieberman AR, Anderson PN. Corticospinal neurons up-regulate a range of growth-associated genes following intracortical, but not spinal, axotomy. Eur J Neurosci. 2003;18:789–802. doi: 10.1046/j.1460-9568.2003.02809.x. [DOI] [PubMed] [Google Scholar]
- 39.Steward O, et al. Regenerative growth of corticospinal tract axons via the ventral column after spinal cord injury in mice. J Neurosci. 2008;28:6836–6847. doi: 10.1523/JNEUROSCI.5372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laub F, et al. Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol Cell Biol. 2005;25:5699–5711. doi: 10.1128/MCB.25.13.5699-5711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang C, et al. Gene expression profiles in human cardiac cells subjected to hypoxia or expressing a hybrid form of HIF-1 alpha. Physiol Genomics. 2002;8:23–32. doi: 10.1152/physiolgenomics.00058.2001. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka H, et al. Cytoplasmic p21(Cip1/WAF1) enhances axonal regeneration and functional recovery after spinal cord injury in rats. Neuroscience. 2004;127:155–164. doi: 10.1016/j.neuroscience.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 43.Kajimura D, Dragomir C, Ramirez F, Laub F. Identification of genes regulated by transcription factor KLF7 in differentiating olfactory sensory neurons. Gene. 2007;388:34–42. doi: 10.1016/j.gene.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 44.Lei L, et al. The zinc finger transcription factor Klf7 is required for TrkA gene expression and development of nociceptive sensory neurons. Genes Dev. 2005;19:1354–1364. doi: 10.1101/gad.1227705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kingsbury TJ, Krueger BK. Ca2+, CREB and krüppel: A novel KLF7-binding element conserved in mouse and human TRKB promoters is required for CREB-dependent transcription. Mol Cell Neurosci. 2007;35:447–455. doi: 10.1016/j.mcn.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradbury EJ, et al. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 47.Cafferty WB, et al. Conditioning injury-induced spinal axon regeneration fails in interleukin-6 knock-out mice. J Neurosci. 2004;24:4432–4443. doi: 10.1523/JNEUROSCI.2245-02.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grill R, Murai K, Blesch A, Gage FH, Tuszynski MH. Cellular delivery of neurotrophin-3 promotes corticospinal axonal growth and partial functional recovery after spinal cord injury. J Neurosci. 1997;17:5560–5572. doi: 10.1523/JNEUROSCI.17-14-05560.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore DL, Goldberg JL. Multiple transcription factor families regulate axon growth and regeneration. Dev Neurobiol. 2011;71:1186–1211. doi: 10.1002/dneu.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci. 2002;22:1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai D, et al. Arginase I and polyamines act downstream from cyclic AMP in overcoming inhibition of axonal growth MAG and myelin in vitro. Neuron. 2002;35:711–719. doi: 10.1016/s0896-6273(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 52.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang YP, et al. Dural closure, cord approximation, and clot removal: Enhancement of tissue sparing in a novel laceration spinal cord injury model. J Neurosurg. 2004;100(4 Suppl Spine):343–352. doi: 10.3171/spi.2004.100.4.0343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.