Abstract
DegP, a member of the highly conserved HtrA family, performs quality-control degradation of misfolded proteins in the periplasm of Gram-negative bacteria and is required for high-temperature survival of Escherichia coli. Substrate binding transforms DegP from an inactive oligomer containing two trimers into active polyhedral cages, typically containing four or eight trimers. Although these observations suggest a causal connection, we show that cage assembly and proteolytic activation can be uncoupled. Indeed, DegP variants that remain trimeric, hexameric, or dodecameric in the presence or absence of substrate still display robust and positively cooperative substrate degradation in vitro and, most importantly, sustain high-temperature bacterial growth as well as the wild-type enzyme. Our results support a model in which substrate binding converts inactive trimers into proteolytically active trimers, and simultaneously leads to cage assembly by enhancing binding of PDZ1 domains in one trimer to PDZ2′ domains in neighboring trimers. Thus, both processes depend on substrate binding, but they can be uncoupled without loss of biological function. We discuss potential coupling mechanisms and why cage formation may have evolved if it is not required for DegP proteolysis.
Keywords: macromolecular assembly, periplasmic degradation, protein quality control, stress survival
Efficient proteolytic removal of misfolded and/or damaged proteins is essential for intracellular protein-quality control, but degradation of the wrong proteins can waste cellular resources and destroy essential proteins. Thus, controlling the activity and specificity of intracellular proteases is critical for maintaining viability. The DegP protease is a member of the HtrA family, functions in the periplasm of Gram-negative bacteria, is essential for survival of Escherichia coli at high temperatures, and its expression is increased substantially following heat shock or other stresses that result in protein misfolding (1–6).
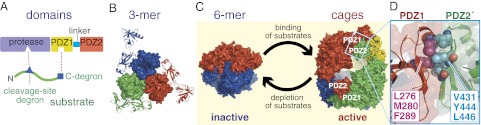
The fundamental structural unit of DegP is a trimer (7), which is stabilized by contacts between trypsin-like protease domains, each of which contains a conventional Ser-His-Asp catalytic triad and has two attached PDZ domains (Fig. 1 A and B). In the absence of substrate, two DegP trimers stack face-to-face in an inactive hexamer with malformed active sites (Fig. 1C, Left) (7). Intriguingly, when protein substrates are added, DegP assembles into cage-like polyhedrons (Fig. 1C) that can contain 4, 6, 8, or more trimers (8–10). Packing between PDZ1 domains in one trimer and PDZ2′ domains in neighboring trimers stabilize these cages (Fig. 1D). The active sites are inside these cages and can degrade unfolded substrates that were trapped by assembly or subsequently diffuse into the chamber through openings at the cage vertices.
Fig. 1.
Domain and oligomeric structures of DegP. (A) DegP subunits contain a protease domain and two PDZ domains. Simple model substrates contain a C-terminal degron that binds to PDZ1 and a cleavage-site degron that binds to the active site of the protease domain. (B) A DegP trimer is the fundamental unit of assembly. This trimer is stabilized by packing between protease domains, which are shown in surface representation and colored individually. The PDZ1 and PDZ2 domains are on the periphery of the trimer and are shown in cartoon representation in the same color as the attached protease domain. The trimer shown consists of subunits A, B, and C from PDB structure 3OTP (10). (C) Upon addition of substrate, DegP is transformed from a proteolytically inactive hexamer to larger cages containing 4, 6, 8, or more trimers (8–10). Each trimer in the hexamer (1KY9) and the dodecamer (3OTP) shown is displayed in surface representation and is a different color. After the substrate is degraded, cages dissociate back to inactive hexamers (10). (D) Close-up view of PDZ1–PDZ2′ contact, emphasizing PDZ1 residues (L276, M280, and F289) and PDZ2′ residues (V431, Y444, and L446) that pack together in the hydrophobic interface (shown in Corey–Pauling–Koltun representation).
The best model substrates for DegP contain a hydrophobic C-terminal residue, which binds in a pocket in the PDZ1 domain, and a hydrophobic residue at the P1 position of the scissile peptide bond, which binds in the active-site cleft (Fig. 1A) (11, 12). The linked binding of both the cleavage-site and C-terminal degrons in a substrate contribute to stabilization of DegP cages (10). Proteolytic cleavage uncouples these degrons, and once the intact substrate concentration has been reduced to a sufficiently low level, cages disassemble and revert to the inactive hexamer (10). This synchronization of substrate-dependent proteolytic activation and cage assembly suggests an obligatory relationship. However, the importance of cage formation for proteolysis is brought into question by the observation that certain DegP variants form trimers in vitro but retain some proteolytic activity (13, 14).
Here, we probe the relationship between DegP cage formation and activity in vitro and in vivo. We find that wild-type DegP and a variant, which is trimeric in the presence or absence of substrate, cleave a model substrate in a positively cooperative manner with similar steady-state kinetic parameters. Robust proteolytic activity was also observed for DegP variants that are hexameric or dodecameric in a substrate-independent fashion. Importantly, wild-type DegP and the obligate trimeric, hexameric, and dodecameric variants all support E. coli survival at heat-shock temperatures equally well. Thus, cage formation is not required for this biological activity. Our results show that allosteric activation of DegP proteolysis by substrate binding is an intrinsic property of the trimer. We also show that substrate binding enhances binding of PDZ1 domains in one trimer to PDZ2′ domains in neighboring trimers, explaining why substrate binding normally leads to cage assembly as well as allosteric activation.
Results
Cage-Defective DegP Variant.
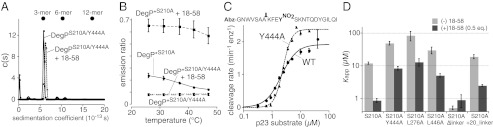
Assembly of DegP cages containing 12, 18, 24, or more subunits is most easily assayed in DegP variants containing the S210A active-site mutation; is stimulated by peptide and protein substrates; and requires binding of the PDZ1 domains in one DegP trimer to the PDZ2′ domains in neighboring DegP trimers (Fig. 1) (8–10). Previously, we found that introducing the Y444A substitution into the PDZ2 portion of the cage interface in DegPS210A/Y444A resulted in a variant that ran as a hexamer in gel-filtration chromatography at 10 °C (10), both in the absence and presence of a model substrate derived from residues 18–58 of lysozyme (henceforth called 18–58). When we tested the oligomeric form of DegPS210A/Y444A by sedimentation-velocity experiments performed at 20 °C, the protein was predominantly trimeric irrespective of the absence or presence of 18–58 (Fig. 2A). Thus, the Y444A mutation prevents substrate-stabilized cage formation under these conditions. By contrast, 18–58 induces formation of cages of DegPS210A that are predominantly dodecameric as assayed by gel filtration, sedimentation, and the crystal structure of an 18–58•DegPS210A complex (10).
Fig. 2.
Cage assembly is not required for proteolysis or cooperative substrate binding. (A) DegPS210A/Y444A (30-μM monomer equivalent) was predominantly trimeric in the absence or presence of the 18–58 substrate (33 μM) in sedimentation velocity centrifugation experiments. The expected sedimentation positions for trimers, hexamers, and dodecamers are shown as dots (10). (B) Donor and acceptor dye-labeled DegP*S210A or DegP*S210A/Y444A (1 μM total monomer equivalent) plus or minus 18–58 (60 μM) were incubated at different temperatures, and the FRET emission ratio was determined. For DegP*S210A with substrate, FRET was high at all temperatures, indicating formation of stable cages. For DegP*S210A without substrate, FRET was substantially lower and decreased as the temperature was increased. Cages may be marginally stable at low but not high temperatures in the absence of substrate; alternatively, the FRET decrease might correspond to partial dissociation of hexamers. FRET was low under all conditions for DegP*S210A/Y444A, as expected if this variant cannot form cages and is predominantly trimeric. (C) Initial rates of DegP or DegPY444A (0.2-μM monomer equivalent) cleavage of different concentrations of the p23 substrate at 25 °C were determined and fitted to the Hill form of a Michaelis–Menten equation (rate = Vmax⋅[S]H/(KMH + [S]H). The sequence of the p23 substrate sequence is shown above the plot. Abz represents a 2-aminobenzoic acid fluorophore, and YNO2 represents a 3-nitrotyrosine quencher. Error bars are averages ±1 SD (n = 3). (D) Apparent equilibrium dissociation constants (Kapp) of flC18–58 binding to different variants of inactive DegPS210A were determined by titrating protein against a fixed concentration of the substrate (50 or 25 nM), measuring changes in fluorescence anisotropy, and fitting the data to a hyperbolic binding equation. Each variant was assayed in the absence or presence of one-half molar equivalent of nonfluorescent 18–58. Bars represent the error of fitting.
DegP*S210A is a variant in which the wild-type cysteines are mutated to serines and the N296C mutation provides a site for labeling with fluorescent dyes (10). When one batch of DegP*S210A is labeled with a donor dye and another batch with an acceptor dye, mixing in the presence of substrate results in cage formation that can be monitored by increased FRET. Using this assay, DegP*S210A + 18–58 showed high FRET, indicative of cage formation, at temperatures between 25 and 47 °C (Fig. 2B). As expected, the FRET signal for a mixture of donor- and acceptor-labeled DegP*S210A was much lower in the absence of 18–58 and decreased even more at higher temperatures. The latter effect may reflect dissociation of some metastable cages that form at low temperature in the absence of substrate. Importantly, DegP*S210A/Y444A gave a very low FRET signal at each temperature whether 18–58 was present or absent (Fig. 2B). Thus, multiple assays under a variety of conditions show that the Y444A mutation prevents or greatly destabilizes cage formation.
Cage Assembly Is Not Required for Proteolysis or Cooperative Substrate Binding.
To determine if the inability to form cages affects proteolytic activity, we determined initial rates of DegP and DegPY444A cleavage of different concentrations of a two-degron substrate with 23 residues (p23) in which cleavage at a single site increases fluorescence by separating a fluorophore and quencher (Fig. 2C). The proteolytic activities of both enzymes were similar. Fitting the DegP data to the Hill form of the Michaelis–Menten equation gave a Vmax of 1.9 min−1⋅enzyme−1, an apparent KM of 2.1 μM, and a Hill constant of 1.6. For DegPY444A, these values were 2.4 min−1⋅enz−1, 2.4 μM, and 2.9. Thus, substrate cleavage by both enzymes was positively cooperative, with comparable maximal rates and apparent KM values. We conclude that cage formation is not required for the intrinsic proteolytic activity of DegP or for cooperative interactions with substrates. Note, however, that apparent KMs are complex functions of the strength of substrate binding to the oligomers present as well as the energetic costs of coupled conformational changes from the inactive to the active conformation and, in the case of wild-type DegP, the cost of the dodecamer assembly reaction. Thus, substrate could bind more tightly to dodecamers (as required if substrate binding drives assembly), but apparent KM values could be similar if this difference was offset by higher energetic costs of active dodecamer assembly.
To assay substrate binding in the absence of cleavage, we used fluorescence anisotropy to monitor the binding of DegPS210A or DegPS210A/Y444A to fluorescently labeled 18–58 (flC18–58). In this assay, positive cooperativity, a hallmark of allosteric enzymes, can be observed by improved binding to the fluorescent substrate in the presence of subsaturating amounts of nonfluorescent 18–58 (10). We found that DegPS210A/Y444A bound flC18–58 more tightly in the presence of 0.5 equivalents of nonfluorescent substrate, but binding under both conditions was weaker than observed for the DegPS210A parent (Fig. 2D) (10). We constructed and assayed two additional variants that destabilize the PDZ1 (L276A) or PDZ2 (L446A) portions of the cage interface. Compared with DegPS210A, the DegPS210A/L276A and DegPS210A/L446A variants showed a reduced ability to form substrate-stabilized cages (Fig. S1). Moreover, DegPS210A/L276A and DegPS210A/L446A resembled DegPS210A/Y444A in terms of 18–58 binding (Fig. 2D); in each case, binding was weaker than for wild-type DegP but retained positive cooperativity. These results support our conclusion that cage formation is not required for cooperative substrate binding.
The 18–58 substrate bound more tightly to DegPS210A than to DegPS210A/Y444A (Fig. 2D), but the p23 substrate was cleaved with similar apparent KMs by DegP and DegPY444A (Fig. 2C). These variations may arise because of differences in the sequences of the two substrates, the enzymes used (S210A background for binding assays), or the different assay conditions. For example, the substrate titration experiments were performed using 0.2 μM DegP, whereas half-maximal binding of DegPS210A to a fixed substrate concentration occurred at an enzyme concentration of ∼0.6 μM. This ∼threefold change in concentration would increase the stability of the dodecamer relative to the hexamer and could account for the tighter apparent binding in the protein titration assay.
Binding of Isolated PDZ2 Blocks Assembly and Enhances Substrate Binding.
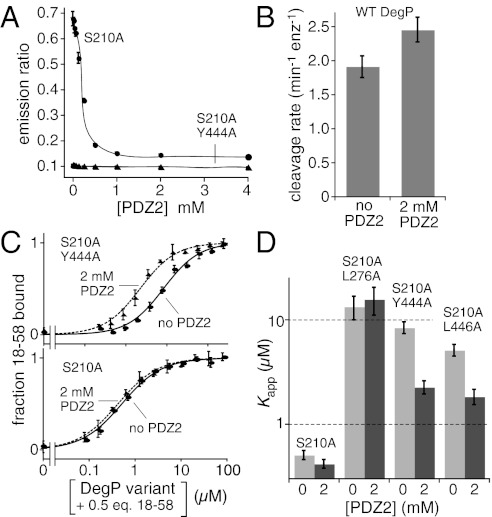
Because the PDZ2 domain plays an essential role in cage formation, we tested if isolated PDZ2 domains could inhibit cage assembly using the DegP* FRET assay. Indeed, when we assembled donor dye- and acceptor dye-labeled DegP*S210A cages in the presence of 18–58 and then added increasing amounts of the purified PDZ2 domain, the FRET signal was reduced in a dose-dependent manner and eventually reached a baseline level expected for complete cage disassembly (Fig. 3A). By contrast, when cage-defective donor and acceptor dye-labeled DegP*S210A/Y444A trimers were mixed with 18–58, the FRET signal remained low in the absence or presence of PDZ2 (Fig. 3A). Thus, high concentrations of the isolated PDZ2 domain can block DegP cage assembly.
Fig. 3.
PDZ1–PDZ2′ interactions enhance substrate binding. (A) Donor and acceptor dye-labeled DegP*S210A or DegP*S210A/Y444A (1 μM total monomer equivalent) were mixed with 18–58 (20 μM) in the presence of different concentrations of the isolated PDZ2 domain, and a FRET emission ratio was calculated. Increasing PDZ2 resulted in disassembly of DegP*S210A cages but no change in the assembly status of DegP*S210A/Y444A. Error bars are averages ±1 SD (n = 3). (B) Rates of DegP (0.2 μM) cleavage of the p23 substrate (40 μM) in the absence and presence of 2 mM PDZ2. Error bars are averages ±1 SD (n = 3). (C) Addition of 2 mM PDZ2 strengthened flC18–58 binding by DegPS210A/Y444A (Upper) but not by DegPS210A (Lower) as assayed by changes in fluorescence anisotropy. In both experiments, nonfluorescent 18–58 was present at half the concentration of DegP subunits. The curves are fits to a hyperbolic equation. Error bars are averages ±1 SD (n = 3). (D) The presence of 2 mM PDZ2 decreased Kapp for flC18–58 binding to DegPY444A and DegPL446A but not to wild-type DegP or DegPL276A, which contains a mutation in the PDZ1 domain that weakens PDZ2′ binding. Bars represent the error of fitting to hyperbolic binding curves, as shown in C.
We assayed cleavage of a high concentration of the p23 substrate by wild-type DegP in the presence of 2 mM PDZ2 to prevent cage formation. The cleavage rate in this experiment was ∼2.4 min−1⋅enz−1 (Fig. 3B). In the absence of added PDZ2, the cleavage rate was ∼1.9 min−1⋅enz−1. These results support our conclusion that cage assembly is not required for high-efficiency DegP proteolysis.
Because mutations that prevent or destabilize PDZ1–PDZ2′ interactions between trimers weaken 18–58 binding, we reasoned that binding of the isolated PDZ2 domain might increase the substrate affinity of DegPS210A/Y444A by mimicking PDZ1–PDZ2′ interactions that are normally made in cages. This result was observed, and binding was ∼fourfold tighter in the presence of 2 mM PDZ2 (Fig. 3C, Upper). By contrast, addition of 2 mM PDZ2 did not change the affinity of DegPS210A for 18–58 (Fig. 3C, Lower). We interpret this observation as indicating that interactions made by binding of the isolated PDZ2 domain have no net effect because they simply replace comparable interactions in wild-type cages. In support of this model, we found that 2 mM PDZ2 also stabilized 18–58 binding by DegPS210A/L446A, which contains a different cage-destabilizing mutation in PDZ2, but did not stabilize binding by DegPS210A/L276A, which contains a mutation in PDZ1 and thus cannot interact stably with added PDZ2 (Fig. 3D). The Fig. 3 C and D experiments were performed in the presence of 0.5 equivalents of unlabeled 18–58, but similar results were observed without added unlabeled substrate (Fig. S2 A and B). Thus, PDZ1–PDZ2′ binding results in stronger 18–58 binding to DegP even when the PDZ2′ domain is not part of a neighboring trimer in a cage.
Effects of the PDZ1–PDZ2 Linker on Assembly, Substrate Binding, and Proteolysis.
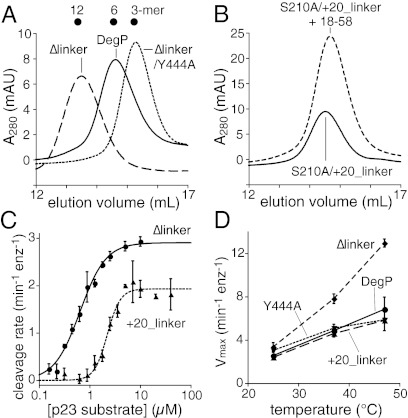
In each DegP subunit, a linker consisting of residues 357–368 connects the PDZ1 and PDZ2 domains. These linkers were ordered in a crystal structure of a symmetric DegP24 cage but were largely disordered in a structure of an asymmetric DegP12 cage, suggesting that changes in linker conformation provide flexibility in cage geometry (9, 10). Iwanczyk et al. (15) constructed a DegPΔlinker variant (Δ357–364) and demonstrated that it eluted as a dodecamer in gel filtration and sedimentation equilibrium experiments without added substrate. We confirmed that DegPΔlinker chromatographed as a dodecamer (Fig. 4A) and also determined that addition of the 18–58 substrate did not change its elution position. However, the DegPΔlinker/Y444A variant eluted as a trimer (Fig. 4A), establishing that the DegPΔlinker dodecamer, like the wild-type dodecamer, is stabilized by PDZ2-dependent interactions. DegPS210A/Δlinker and DegPS210A bound the flC18–58 substrate with comparable apparent affinities in the presence of 0.5 equivalents of unlabeled substrate (Fig. 2D).
Fig. 4.
Variants with shorter or longer PDZ1–PDZ2 linkers show altered assembly but maintain robust proteolytic activity. (A) DegP, DegPΔlinker, and DegPΔlinker/Y444A (25-μM loading concentration) eluted from a Superose 6 gel-filtration column at positions expected for a hexamer, dodecamer, and trimer, respectively. (B) DegPS210A/+20_linker (25-μM loading concentration) eluted as a hexamer in gel filtration in the absence and presence of 18–58 (38-μM loading concentration). (C) Concentration dependence of the rates of cleavage of the p23 substrate by DegPΔlinker and DegP+20_linker (0.2- and 0.1-μM monomer equation, respectively). The curves are fits to the Hill form of the Michaelis–Menten equation. Error bars are averages ±1 SD (n = 3). (D) Temperature dependence of the rate of cleavage of the p23 substrate (60 μM) by DegP, DegPY444A, DegPΔlinker, and DegP+20_linker (0.5-μM monomer equivalent).
To determine the effects of lengthening the PDZ1–PDZ2 linker, we inserted 20 residues between residues Q356 and S357 in wild-type and S210A backgrounds. Interestingly, DegPS210A/+20_linker eluted as a hexamer both with and without added substrate (Fig. 4B). The binding of flC18–58 to DegPS210A/+20 was still positively cooperative, but binding in the presence of 0.5 equivalents of unlabeled 18–58 was somewhat weaker than observed for the cages of wild-type DegP or DegPΔlinker (Fig. 2D).
Although their oligomeric states are apparently fixed, DegPΔlinker and DegP+20_linker cleaved the p23 substrate in positively cooperative reactions with Hill constants of 1.7 and 3.3, Vmax values of 2.9 and 1.9 min−1⋅enz−1, and apparent KM values of 0.6 and 2.2 μM, respectively (Fig. 4C). Thus, linker length plays an important role in substrate-dependent cage assembly but plays a relatively modest role in determining the steady-state kinetics of substrate cleavage. Moreover, these results show that preformed hexamers and dodecamers still display positively cooperative interactions with the p23 substrate. To test if temperature caused substantial changes in the proteolytic activities of DegP, DegPY444A, DegPΔlinker, and DegP+20_linker, we assayed cleavage of a single high concentration of the p23 substrate at 25, 37, and 47 °C (Fig. 4D). In each instance, the cleavage rate increased as the temperature was increased. Thus, all of these enzymes would be expected to be active proteases at heat-shock temperatures.
Cage Assembly Is Not Required for High-Temperature Cell Survival.
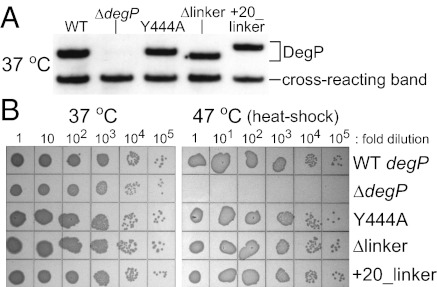
To determine how DegP mutations affect biological function, we constructed otherwise isogenic bacterial strains in which the Y444A, Δlinker, and +20_linker mutations were recombined into the chromosomal degP gene. Western blots confirmed that each mutant protein was expressed at a level similar to wild-type DegP (Fig. 5A). To test cell viability, cultures were grown to log phase at 37 °C, and serial 10-fold dilutions were spotted onto LB agar plates and incubated overnight at 37 or 47 °C (Fig. 5B). Strikingly, the degPY444A, degP+20_linker, and degPΔlinker strains all grew as well as the wild-type strain at both temperatures (Fig. 5B), even though the first two variants do not assemble into cages, and the third forms permanent dodecameric cages. Thus, reversible cage formation does not seem to be an important determinant of high-temperature growth. By contrast, a strain with an in-frame deletion of the DegP coding sequence (ΔdegP) was inviable at 47 °C but grew normally at 37 °C (Fig. 5B).
Fig. 5.
Assembly of DegP cages is not required for high-temperature cell survival. (A) E. coli encoding wild-type DegP, DegPY444A, DegPΔlinker, or DegP+20_linker at the standard chromosomal locus expressed similar levels of DegP protein. Strains were grown to log phase in LB broth at 37 °C, and equivalent numbers of cells were lysed and separated by SDS/PAGE. DegP was identified by Western blotting using an anti-DegP antibody. A ΔdegP strain expressed no DegP. All strains expressed comparable levels of a cross-reacting lower molecular-weight protein. (B) Strains expressing wild-type DegP, DegPY444A, DegPΔlinker, or DegP+20_linker exhibited similar overnight growth on LB agar plates at 37 and 47 °C. The ΔdegP strain grew normally at 37 °C but was inviable at 47 °C.
Discussion
Previous studies have shown that substrate binding drives assembly of DegP cages and activates proteolysis, suggesting a causal relationship (8–10). However, Spiess et al. (13) and Jomaa et al. (14) showed that deletion of the DegP PDZ2 domain results in trimers that retain some proteolytic activity. Our results confirm this observation in that we found that a trimeric variant, DegPY444A, degrades a model substrate in a positively cooperative fashion with apparent KM and Vmax values similar to wild-type DegP. The Y444A mutation in PDZ2 affects packing interactions that normally occur in cages between the PDZ2 domains of one trimer and the PDZ1 domains of a neighboring trimer. We also found that addition of high concentrations of the isolated PDZ2 domain could disassemble wild-type cages without eliminating proteolysis. Thus, cage formation is not a prerequisite for proteolytic activation, and the molecular mechanisms that drive these reactions must be different, albeit energetically coupled in the wild-type enzyme.
DegS is a DegP paralog that contains a protease domain and one PDZ domain and functions as a stable trimer (16–18). Substrates bind and activate DegS in a positively cooperative reaction that involves a transition from a proteolytically inactive trimer to an active trimer and is fit well by the concerted Monod–Wyman–Changeux model of allostery (19, 20). Because our results establish that trimeric DegPY444A also shows positive cooperativity in substrate binding and proteolysis, we assume that this trimer undergoes a similar transition between an inactive conformation that predominates in the absence of substrate and an active conformation that is stabilized by substrate binding.
How is substrate binding normally linked to assembly of DegP cages? From an energetic perspective, we found that the 18–58 substrate binds more tightly to wild-type DegP, which can form cages, than to DegPY444A, which cannot form cages. Moreover, addition of high concentrations of the isolated PDZ2 domain increased the 18–58 affinity of the DegPY444A trimer ∼fourfold. These results indicate that docking of exogenous PDZ2 domains with the PDZ1 domains of DegPY444A stabilizes a trimer conformation with higher substrate affinity. Thermodynamic linkage then ensures that substrate binding to a trimer will also stabilize tighter binding of its PDZ1 domains to exogenous PDZ2 domains, providing the driving force for substrate-dependent assembly of DegP cages (Fig. 6). An important aspect of this model is that geometric constraints imposed by the structure of the trimer prevent its PDZ2 domains from binding to the PDZ1 domains of the same trimer in a way that mimics trimer–trimer interactions within cages. Although a fourfold stabilization for each trimer is a relatively small value, it could be amplified substantially by the presence of multiple trimers in a cage. Indeed, DegP trimers appear to assemble into a range of different geometrically allowed cage- or bowl-like structures in the presence of different substrates and/or membranes (8–10, 21).
Fig. 6.
A model for linkage between substrate binding and cage assembly. Cages assemble when substrate binding enhances the interaction between PDZ1 domains in one trimer and PDZ2′ domains in neighboring trimers. Although the mechanisms of cage assembly and proteolytic activation are separable, both depend upon substrate binding and thus appear to be coupled in wild-type DegP.
Two potential structural mechanisms might link substrate binding to enhanced PDZ1–PDZ2′ interactions. The first mechanism is a direct contact of substrate residues between the cleavage-site degron and C-terminal degron in one trimer with a PDZ2′ domain in a neighboring trimer. A contact of this type appeared to be present in one subunit of the 18–58 bound dodecamer (Fig. S3A) (10). The second mechanism is an allosteric change in the structure of PDZ1 upon binding to the C terminus of a substrate or to a PDZ2′ domain. Indeed, modest changes in PDZ1 structure are observed between substrate-bound cages and substrate-free structures (Fig. S3B) (9), although it is difficult to know if these conformational changes would affect the binding reactions. We note, however, that allosteric coupling between the peptide-binding site and the opposite face of PDZ domains has been proposed based upon patterns of sequence covariation (22).
In DegP variants, the length of the linker between the PDZ1 and PDZ2 domains of each DegP subunit can modulate the stability of higher oligomers. A DegP mutant with a substantially longer PDZ1–PDZ2 linker formed hexamers in the presence or absence of substrate, whereas a mutant with a very short linker formed dodecamers in the presence or absence of substrate. Both of these linker variants had robust proteolytic activity and showed cooperative activation by substrates. These results reinforce our conclusion that the mechanisms that drive substrate-dependent formation of wild-type cages and result in substrate-dependent proteolytic activation are separable. From an evolutionary perspective, altering the length or flexibility of the PDZ1–PDZ2 linker might provide a way to tune the coupling between substrate binding and the assembly of cages or other oligomers.
Multiple studies have shown that DegP is required for survival of E. coli at high temperatures or when misfolded envelope proteins are overexpressed at lower temperatures (4, 5, 23, 24). Importantly, we found that strains expressing DegPY444A, DegP+20_linker, or DegPΔlinker from the chromosome were as effective as wild-type DegP in supporting growth at 47 °C. In vitro, these mutants form stable trimers, hexamers, or dodecamers, respectively, and thus the ability of wild-type DegP to form cages in a reversible substrate-dependent manner or to convert from an inactive hexamer to an active cage does not appear to be an important factor in survival at 47 °C. The DegPY444A, DegP+20_linker, and DegPΔlinker mutants all displayed robust and cooperative proteolysis in vitro, suggesting that this activity is an important aspect of their biological roles in combating protein-folding stress. Indeed, proteolytically inactive DegPS210A did not support growth at 47 °C but supported some growth at 42 °C, as expected from the previous work (13, 25).
Why has DegP evolved the ability to form cages in a substrate-dependent fashion if these cages are not required for biological function as assayed by high-temperature growth? There are several possibilities. First, cages may provide additional fitness and thus a strong selective advantage under stress conditions that we have not tested or are difficult to reproduce in laboratory cultures. Second, if cages provided even a marginal growth benefit at high temperatures or under other conditions, then this small advantage exponentially magnified over thousands of generations could provide a strong selective pressure. For example, cages might suppress proteolysis of improper substrates under some stress conditions, providing a fitness benefit. Third, DegP cages may facilitate additional biological activities. For example, degPS210A supports growth at 42 °C better than a ΔdegP allele, a property that has been proposed to result from chaperone activity (13) or sequestration of toxic misfolded proteins (26, 27). Finally, DegP cages may allow more efficient degradation of large protein substrates, even though this activity is not required for high-temperature survival.
Materials and Methods
DegP variants, the 18–58 model substrate, and flC18–58 were prepared as described (10). The +20_linker variant carries an additional 20 residues (ASGAGGSEGGGSEGGTSGAT) between Q356 and S357. The p23 substrate was synthesized by the Massachusetts Institute of Technology Biopolymer Laboratory or CHI Scientific, Inc. and was purified by HPLC; its concentration was determined by absorption at 381 nm (ε = 2,200 M−1⋅cm−1).
Gel filtration chromatography, FRET, fluorescence anisotropy, and analytical ultracentrifugation experiments were performed as described (10) with minor modifications. Sedimentation velocity experiments were carried out at 20 °C and 30,000 rpm using an An-50 Ti rotor in a Beckman XL-I analytical ultracentrifuge (Biophysical Instrumentation Facility, Massachusetts Institute of Technology). Data were fitted with SEDFIT (28) to a model for continuous sedimentation coefficient distribution with a single floating frictional ratio. Kinetic assays for the p23 cleavage were performed with excitation at 320 nm and emission at 435 nm (cutoff filter at 420 nm) using a SpectraMax M5 microplate reader (Molecular Devices). Each reaction was started with the addition of wild-type DegP or variants and terminated by addition of excess wild-type DegP (∼25 μM) to determine fluorescence values associated with complete substrate cleavage.
Different degP mutations were introduced into E. coli strain W3110 (wild-type) using a scarless lambda-RED–mediated recombineering method, in which the wild-type degP gene is replaced by two successive recombination steps (29) (details are available in SI Materials and Methods and Table S1). The ΔdegP strain constructed bore an in-frame deletion of only protein-coding sequence. The sequences of all chromosomal degP alleles were confirmed by DNA sequencing (GENEWIZ) following PCR amplification using flanking primers.
For Western blotting, cells expressing DegP variants were grown to log phase (OD600 ∼0.6) in LB at 37 °C. A 1-mL sample of the culture was harvested, resuspended with OD600/2 mL of SDS sample buffer, heated at 100 °C for 15 min, and centrifuged at 16,000 × g for 10 min. Samples (5 μL) were separated by SDS/PAGE and transferred to PVDF membrane (Bio-Rad). The membrane was sequentially incubated in protein-free T20 Blocking buffer (Thermo Scientific), anti-DegP antibody (30,000:1 dilution), and anti-rabbit IgG-HRP (30,000:1 dilution; GE Healthcare), and then blotted with SuperSignal West Pico reagent (Thermo Scientific). To test viability, cells were at 37 °C grown to OD600 ∼0.2, and dilutions of 10, 102, 103, 104, and 105 were prepared using LB broth. A 5-μL sample of each dilution was spotted onto LB agar plates and incubated at 37 °C and 47 °C for ∼11 h.
Supplementary Material
Acknowledgments
We thank D. Barthelme, A. Olivares, D. Pheasant, A. de Regt, and J. Sohn for helpful discussions and assistance. Anti-DegP antibody was a generous gift from T. Silhavy. Support for this work was provided by National Institutes of Health Grant AI-16892.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204791109/-/DCSupplemental.
References
- 1.Pallen MJ, Wren BW. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–221. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- 2.Clausen T, Kaiser M, Huber R, Ehrmann M. HTRA proteases: Regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol. 2011;12:152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- 3.Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: A sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–10067. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipinska B, Fayet O, Baird L, Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strauch KL, Johnson K, Beckwith J. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J Bacteriol. 1989;171:2689–2696. doi: 10.1128/jb.171.5.2689-2696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duguay AR, Silhavy TJ. Quality control in the bacterial periplasm. Biochim Biophys Acta. 2004;1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J, et al. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc Natl Acad Sci USA. 2008;105:11939–11944. doi: 10.1073/pnas.0805464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krojer T, et al. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 10.Kim S, Grant RA, Sauer RT. Covalent linkage of distinct substrate degrons controls assembly and disassembly of DegP proteolytic cages. Cell. 2011;145:67–78. doi: 10.1016/j.cell.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krojer T, et al. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci USA. 2008;105:7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krojer T, Sawa J, Huber R, Clausen T. HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat Struct Mol Biol. 2010;17:844–852. doi: 10.1038/nsmb.1840. [DOI] [PubMed] [Google Scholar]
- 13.Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 14.Jomaa A, et al. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J Bacteriol. 2007;189:706–716. doi: 10.1128/JB.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwanczyk J, et al. Role of the PDZ domains in Escherichia coli DegP protein. J Bacteriol. 2007;189:3176–3186. doi: 10.1128/JB.01788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 17.Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 18.Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 19.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 20.Sohn J, Sauer RT. OMP peptides modulate the activity of DegS protease by differential binding to active and inactive conformations. Mol Cell. 2009;33:64–74. doi: 10.1016/j.molcel.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Q-T, et al. Bowl-shaped oligomeric structures on membranes as DegP’s new functional forms in protein quality control. Proc Natl Acad Sci USA. 2009;106:4858–4863. doi: 10.1073/pnas.0811780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 23.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7(12B):2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 24.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 25.Skórko-Glonek J, Wawrzynów A, Krzewski K, Kurpierz K, Lipińska B. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene. 1995;163:47–52. doi: 10.1016/0378-1119(95)00406-v. [DOI] [PubMed] [Google Scholar]
- 26.Misra R, CastilloKeller M, Deng M. Overexpression of protease-deficient DegP(S210A) rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J Bacteriol. 2000;182:4882–4888. doi: 10.1128/jb.182.17.4882-4888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CastilloKeller M, Misra R. Protease-deficient DegP suppresses lethal effects of a mutant OmpC protein by its capture. J Bacteriol. 2003;185:148–154. doi: 10.1128/JB.185.1.148-154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis JH, Baker TA, Sauer RT. Small-molecule control of protein degradation using split adaptors. ACS Chem Biol. 2011;6:1205–1213. doi: 10.1021/cb2001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.