Abstract
Cisplatin (cis-diamminedichloroplatinum) and related compounds cause DNA damage and are widely used as anticancer agents. Chemoresistance to cisplatin treatment is due in part to translesion synthesis by human DNA polymerase η (hPol η). Here, we report crystal structures of hPol η complexed with intrastrand cisplatin-1,2–cross-linked DNA, representing four consecutive steps in translesion synthesis. In contrast to the generally enlarged and nondiscriminating active site of Y-family polymerases like Dpo4, Pol η is specialized for efficient bypass of UV–cross-linked pyrimidine dimers. Human Pol η differs from the yeast homolog in its binding of DNA template. To incorporate deoxycytidine opposite cisplatin–cross-linked guanines, hPol η undergoes a specific backbone rearrangement to accommodate the larger base dimer and minimizes the DNA distortion around the lesion. Our structural analyses show why Pol η is inefficient at extending primers after cisplatin lesions, which necessitates a second translesion DNA polymerase to complete bypass in vivo. A hydrophobic pocket near the primer-binding site in human Pol η is identified as a potential drug target for inhibiting translesion synthesis and, thereby, reducing chemoresistance.
Keywords: chemotherapy, inhibitor, translesion DNA synthesis, yeast polymerase η
Cisplatin [cis-diamminedichloroplatinum (II)] was discovered to be a potent antitumor agent in 1969 (1) and was approved for treatment of testicular and ovarian cancer within 10 years. Currently, cisplatin and related platinum-anticancer drugs are used to treat a variety of sarcomas, carcinomas, and metastasized colorectal cancers (2). Cisplatin reacts with DNA bases and cross-links the N7 atoms of adjacent purines, preferably guanines, either within one strand (intrastrand) or between strands (interstrand) (3). The main intrastrand cross-linked products are cis-Pt-1,2-d(GpG) (Pt-GG, 65%), 1,2-d(ApG) (Pt-AG, 25%), and 1,3-d(GpNpG) (Pt-GNG, 10%). The antitumor activity of cisplatin-DNA adducts is believed to stem from blocking DNA replication and, thus, activating apoptosis in rapidly dividing cells (3, 4). Resistance to cisplatin treatment arises from (i) reduced cisplatin accumulation inside cells because of deactivation of influx transporters (5), (ii) removal of cisplatin DNA adducts by nucleotide excision repair (NER) (6, 7), and (iii) bypass of cisplatin adducts during replication by specialized DNA polymerases in a process called translesion DNA synthesis (TLS) (8–11).
Among many polymerases tested in vitro, the Y-family DNA polymerase η (Pol η) is the most efficient and accurate at bypassing Pt-GG lesions (12–14). Mutations of the human POLH gene, which encodes polymerase η, are directly linked to a variant form of the cancer predisposition syndrome xeroderma pigmentosum (XP-V) (15, 16). XP-V patients have normal NER but are deficient in TLS of UV-induced cyclobutane pyrimidine dimers (CPD) and are hypersensitive to sunlight (17). XP-V cell lines are also hypersensitive to cisplatin (18, 19). The expression level of Pol η is induced by cisplatin or oxaliplatin treatment, and the increased Pol η level reduces the effectiveness of chemotherapy and the survival time of patients with non–small-cell lung cancer or metastatic gastric adenocarcinoma (20, 21).
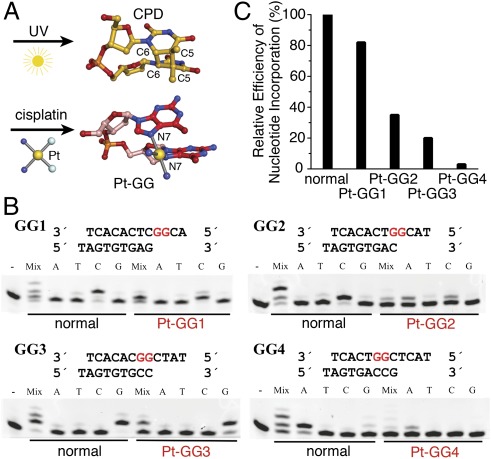
Cross-linked base dimers pose a major roadblock to all replicative polymerases and many specialized TLS polymerases, which typically accommodate a single template base in the active site (22). When the archaeal Y-family polymerase Dpo4 was cocrystallized with Pt-GG, the 3′G that is supposed to base pair with an incoming nucleotide is displaced from the normal templating position by 5 Å, and the lesion is extruded from the active site (23) (SI Appendix, Fig. S1 A and B). Dpo4 is very inefficient in dCTP incorporation opposite Pt-GG (24). Pol η, however, can accommodate two pyrimidine bases in its active site, whether UV–cross-linked (25, 26), and catalyze efficient and accurate primer extension opposite and beyond typical CPD cis-syn thymine dimers (27–29). Structurally, the major species of cisplatin–cross-linked guanines (Pt-GG) is related to CPD (Fig. 1A). A DNA duplex containing either CPD or Pt-GG is distorted and bent at the lesion (30–32). However, the Pt-GG lesion is much larger and more flexible than CPD, owing to the single Pt-bridged cross-link. Yeast Pol η was crystallized with Pt-GG (33), but as shown by the Prakash and Aggarwal team (26), the lattice contacts distorted the essential protein–DNA interactions such that DNA is displaced by >3 Å and the templating base, the 3′ primer end, and the A-site metal ion are dislocated and misaligned relative to the catalytic center (SI Appendix, Fig. S1 C and D). The mechanism for Pt-GG bypass and chemoresistance in humans remained unknown.
Fig. 1.
Efficiency of Pt-GG bypass by hPol η. (A) Structures of a thymine dimer (CPD) and a Pt-GG lesion. (B) Base selection and TLS efficiency of hPol η in four consecutive steps of bypassing a Pt-GG. DNA oligos used in the assays are shown above each gel. The results with normal DNA of the same sequences are shown for comparison. Reactions were carried out with a single type or a mixture of all four dNTPs as indicated at a total concentration of 0.1 mM. (C) A plot of relative polymerase efficiency (kcat/KM) of single nucleotide incorporation in each step of Pt-GG bypass normalized to the average efficiency with normal DNA as summarized in SI Appendix, Table S2.
To elucidate how human Pol η, which is a cancer suppressor guarding against UV damage in DNA, becomes a “double agent” that counteracts the effect of cisplatin in anticancer treatment, we have determined four structures of the polymerase domain of human Pol η (1–432 aa, hPol η for short) complexed with four Pt-GG DNAs representing four consecutive steps of primer extension opposite and beyond the lesion. For comparison, we have also determined two structures of hPol η complexed with normal guanines (SI Appendix, Table S1).
Results
Kinetic Analysis of Pt-GG Bypass by hPol η.
We first determined the catalytic efficiency and nucleotide preferences of hPol η (1–432 aa) when it extends a primer opposite and beyond a Pt-GG in four consecutive steps, labeled as Pt-GG1 to Pt-GG4 (Fig. 1 B and C). The KM and kcat measurements were made with Pt-GG–containing and normal DNA of the same sequence (SI Appendix, Table S2). Our results for normal DNA are comparable to published values obtained with the full-length human Pol η (25, 34), indicating that the N-terminal 432 residues of human Pol η are necessary and sufficient for catalysis. hPol η accurately inserts dC opposite the 3′G of Pt-GG (the first step of bypass) at 82% catalytic efficiency (kcat/KM) of nucleotide incorporation on undamaged DNA, whereas the second dC insertion opposite the 5′G has a lower but respectable efficiency of 35%. Although hPol η can misincorporate dATP opposite the 5′-G of Pt-GG, the KM for dATP is much higher than for dCTP, and the relative efficiency of dA incorporation is <1% (SI Appendix, Table S2) (12). Moreover, human Pol η is unable to extend after dA misincorporation (13). As indicated by cell-based studies (35), the catalytic efficiency of primer extension by hPol η drops sharply and is <3% at the second nucleotide after the lesion (Pt-GG4) (Fig. 1C).
Because a thymine dimer fills the active site of hPol η (25), we thought that the catalytic efficiency might differ when a purine rather than a pyrimidine is downstream of the templating base. However, kcat and KM are similar regardless of whether the template has GG, GC, or CT (in the 3′ to 5′ direction) in the active site (Fig. 1B and SI Appendix, Table S2).
Structural Features of hPol η–Pt-GG Complexes.
The crystal structures of hPol η (1–432 aa) complexed with Pt-GG DNA and a correct nonhydrolyzable incoming dNMPNPP opposite the 3′G (Pt-GG1), the 5′G (Pt-GG2) of the lesion, or one or two nucleotides after the lesion (Pt-GG3 and Pt-GG4, respectively) were determined (SI Appendix, Tables S1 and S3). For comparison, structures of hPol η complexed with a normal G at the templating position followed by a downstream G (GG0a) or C (GG0b) were also determined. All six hPol η complexes crystallized in the P61 space group with one complex per asymmetric unit, and the structures were refined to resolutions between 2.0 and 2.9 Å (Materials and Methods). In five structures, hPol η forms a ternary complex, but the Pt-GG4 structure is a binary complex of the polymerase and DNA without a dNTP (see details below).
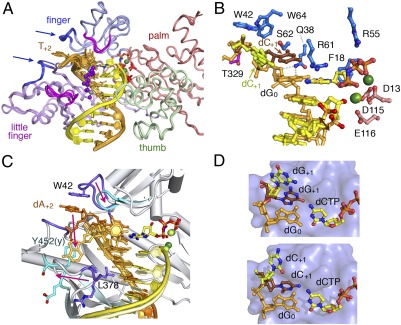
The Pt-GG structures are remarkably similar to the normal hPol η ternary complexes (Fig. 2A). Of the four structural domains of hPol η, the thumb (240-306 aa) and LF (little finger, 307–432 aa) domains hold the template-primer duplex, and the palm (1–15 and 88–239 aa) and finger domains (16–87 aa) surround the replicating base pair (25). Contrary to the general impression that Y-family polymerases are flexible, the hPol η structures are superimposable, regardless of the presence or position of a lesion (Fig. 2A). The pairwise rmsd (root mean square deviation) of hPol η among the six structures (including the binary Pt-GG4) ranges from 0.3 to 0.4 Å over 390–396 Cα atoms. From Pt-GG1 to Pt-GG3, the catalytic triad Asp13, Asp115, and Glu116 in the palm domain coordinates two Mg2+ ions for catalysis (Fig. 2B). The two uniquely conserved residues among Pol η homologs, Q38 and R61, hold the replicating base pair in place. Q38 donates H bonds to both the O4′ and the O2 (pyrimidine) or N3 (purine) of the templating nucleotide, and R61 interacts with the phosphate and base of the incoming nucleotide (Fig. 2B).
Fig. 2.
Structural features of hPol η ternary complexes. (A) Superposition of six hPol η–DNA complexes. The protein is shown as Cα traces, and the four domains are color-coded. The DNA template strand is drawn in orange and primer in yellow. Cisplatin is shown as purple and gray spheres. Mg2+ ions (green spheres), catalytic carboxylates (red sticks), and incoming nucleotides (yellow and multicolored sticks) are also shown. Two generally mobile loops are shown in blue and pointed out by arrows, and the two, whose movement is coupled with the presence of DNA lesions, are shown in magenta. (B) The active site of GG0b. The nucleotide downstream of the templating base (dC+1) has two conformations; so does the 3′ nucleotide of the primer strand. (C) Superposition of human (PDB ID code 3MR3) (25) and yeast Pol η (PDB ID code 3MFI) (26) complexed with CPD. The protein loops involved in template-strand binding, the DNA, and metal ions are shown in darker color in the human complex than in the yeast. Pink arrows indicate the changes from human to yeast. (D) Two conformations of the nucleotide downstream of the templating base. Upper is GG0a (dG+1), and Lower is GG0b (dC+1).
Four small protein loops show noticeable deviations (Fig. 2A), two of which contact the downstream single-stranded template and are flexible even in the complexes with normal DNA substrate. The third flexible loop (R61-M63 in the finger domain) contacts the replicating base pair; the fourth (Q373-S379 in the LF) contacts the major groove of the upstream duplex. Interestingly, two of these four loops also contain the largest structural deviations between human and yeast Pol η (Fig. 2C). The loop containing W42, which stacks with the +2 base of the template strand in the human case, is not conserved and much shorter in yeast. Moreover, the loop equivalent to human Q373-S379 in yeast Pol η has a different sequence and an entirely different conformation (Fig. 2C). Consequently, the major groove of the DNA duplex is exposed to solvent and the downstream template makes a U-turn in the yeast Pol η complex (26).
Although DNA with a cisplatin adduct is bent by ≈30° in the absence of any protein (31, 32, 36, 37), in the ternary complexes with hPol η, the DNA appears more-or-less straight (Fig. 2A, see details below). The backbone of the template DNA upstream from the active site is held tightly by hPol η (known as the molecular splint effect; ref. 25) and exhibits little difference among nearly a dozen ternary complex structures. However, the nucleotide downstream from the templating base (+1) is flexible. In the normal DNA–hPol η complexes, it can stack with the templating base because of the enlarged active site as reported (25) or flip out into the major groove (Fig. 2 B and D). The stacked-in conformation occurs whether the base is pyrimidine or purine. In GG0a, the downstream guanine adopts a syn conformation to fit in the pocket, which is too small to accommodate the anti conformation.
Pt-GG1: Insertion of dC Opposite the 3′G.
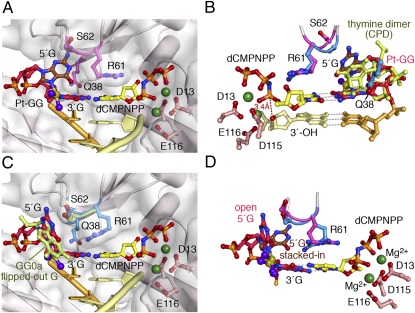
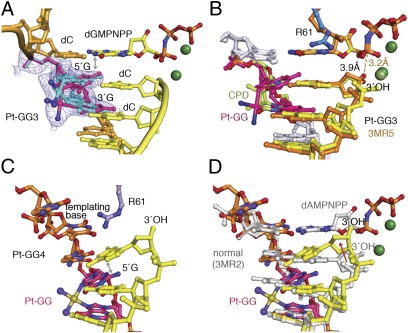
In Pt-GG1, the 3′G of the lesion serves as the template and forms a Watson–Crick pair with an incoming dCMPNPP. The active site is well-formed, and the 3′-OH of the primer strand is aligned with the α-phosphate for the chemical reaction (Fig. 3 A and B). The electron density at 2.0-Å resolution clearly shows that the 5′G of Pt-GG has two conformations (SI Appendix, Fig. S2A). In the stacked-in conformation, the cross-linked Gs are at an acute angle and are both accommodated in the active site like the CPD in the hPol η ternary complex TT1 (PDB ID code 3MR3) (25) (Fig. 3B). In the open conformation, the platinum and 5′ G can be overlaid with the flipped-out normal downstream guanine, and the R61-M63 loop is slightly shifted to accommodate the lesion (Fig. 3C). Neither of the Pt-GG conformations observed here can fit without clashes into the active site of a replicative polymerase or even other Y-family polymerases including Dpo4.
Fig. 3.
The Pt-GG1 structure and comparison with TT1. (A) When the Pt-GG is in the stacked-in conformation, the R61-M63 loop (in magenta) is lifted up. (B) Overlay of the stacked-in Pt-GG with CPD in TT1 [PDB ID code 3MR3 (25), blue R61-S62]. R61 and S62 in Pt-GG1 (magenta) are altered. Gray dashed lines represent hydrogen bonds. The red dashed line connects the 3′-OH and the α-phosphate. This view is roughly the back side of A. (C) With the open conformation of Pt-GG, the R61-M63 loop (blue) is slightly shifted compared with the normal structure (GG0a, shown in light green). (D) Superposition of the stacked-in (brown 5′G) and open structures of the Pt-GG (dark pink).
To accommodate the stacked-in Pt-GG, which is larger than a CPD, the protein loop containing R61, S62, and M63 (R61-M63) changes conformation locally to avoid clashes with the lesion bases (Figs. 2A and 3). Because the cross-link prevents the 5′G from adopting the syn conformation, the Cα of S62 moves ≈1.7 Å and the peptide bond between R61 and S62 flips 180° to enlarge the templating base-binding pocket (Fig. 3D and Movie S1). Interestingly, S62 is not conserved among Pol η homologs and varies among Gly, Ser, Asn, Met, and His (25). Replacing S62 with a more flexible Gly in human Pol η makes the mutant protein more efficient in TLS (38). In yeast Pol η, the S62-equivalent M74 and the surrounding residues I71 to T76 undergo a reverse conformational change from open in the apo-protein to closed in the ternary complex structures (26, 33). In the “open” conformation yeast, Pol η cannot bind an incoming dNTP. In contrast, in hPol η, opening of the R61-M63 loop allows accommodation of Pt-GG in the active site without altering dNTP binding, thus leading to accurate incorporation of dC opposite the 3′G.
Pt-GG2: Insertion of dC Opposite the 5′G.
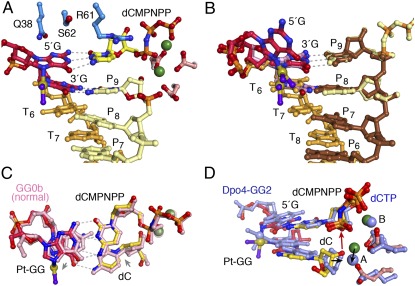
In the second insertion step opposite a Pt-GG, the ≈30° roll angle imposed by the cisplatin cross-link (32) is absorbed mainly on the 3′G, thus allowing the 5′G to serve as the templating base with little deviation from the undamaged DNA (Fig. 4). Both guanines of Pt-GG make three hydrogen bonds with their base-pairing partners, the incoming dCMPNPP and dC at the primer 3′ end. The roll of the 3′G is dissipated to the surrounding primer strand and dCMPNPP (Fig. 4 A and C). The distance between the reactants (3′-OH and α-phosphate) is slightly increased (from 3.3 Å to 3.7Å), and kcat decreases twofold (Fig. 1C and SI Appendix, Table S2). The Pt-GG2 ternary complex is similar to one of the two DNA alternate conformations observed in the equivalent step of Pt-GG bypass by Dpo4 (23) (Fig. 4D), suggesting that, unlike Pt-GG1, template-dependent nucleotide incorporation can be achieved by other Y-family polymerases in the Pt-GG2 step. However, Dpo4 is extremely inefficient (24), which may be explained by the poor alignment of the 3′-OH and α-phosphate compared with the hPol η complex (Fig. 4D).
Fig. 4.
The Pt-GG2 structure and comparison with GG0b and Dpo4-GG2. (A) The active site of the ternary complex is shown in ball-and-stick model. The Pt-GG makes all of the hydrogen bonds for base pairing. (B) Overlay of the two DNA populations. The primer (P6 to P9) of the binary complex is shown in brown, the template strand (T6 to T8) in orange, and Pt-GG in red and blue. The Pt-GG in the ternary complex is shown in pink, and its base-pairing partners are shown in light yellow with the phosphorus atoms of the incoming nucleotide highlighted in orange. (C) Comparison of Pt-GG2 ternary complex and GG0b. The Pt-GG base pairs are colored as in A, and those in GG0b are in pink. Gray arrows indicate the slight shifts of template and primer bases in Pt-GG2. (D) Comparison of the Pt-GG2 ternary complexes of hPol η and Dpo4 (PDB ID code 3M9N, blue color) (23). The displacement of the metal ion (2.7 Å) and the primer ends (1.5 Å) in Dpo4 are indicated by black arrows.
The reduced efficiency in nucleotide incorporation at this step may stem from a second cause. Close inspection of the hPol η electron density maps reveals two DNA populations (SI Appendix, Fig. S2B). In addition to the ternary complexes, the second DNA population forms a binary complex with hPol η, mimicking the reaction product of the Pt-GG1 step before translocation. A 5′-overhang nucleotide on the primer strand, which is flipped out and disordered in the ternary complex, is stacked in to form lattice contacts in the binary complex. The two DNA populations overlap in the phosphosugar backbones, but the bases are out of register by one. For instance, the primer 3′-end in the binary complex completely overlaps with the incoming nucleotide of the ternary complex (Fig. 4B). The existence of the binary complex illustrates the instability of the ternary complex. It also provides a glimpse of hPol η with the DNA product still in the active site (Movie S2).
Pt-GG3: Primer Extension Immediately After the Lesion.
To find why hPol η has reduced efficiency in primer extension after Pt-GG (Fig. 1C), the structure of a ternary complex with the base downstream of the Pt-GG in the active site was determined (Fig. 5A and SI Appendix, Table S3). Compared with the CPD structure, the cisplatin protrudes into the major groove (Fig. 5B). To accommodate it, the loop Q373-S379 unique to hPol η shifts away (Fig. 2A), and the entire LF is rotated slightly outwards (SI Appendix, Fig. S3A). The electron density at 2.5-Å resolution clearly shows two conformations of the Pt-GG lesion (Fig. 5A and SI Appendix, Fig. S2C). In each conformation, one of the cross-linked G’s (3′ or 5′) forms a planar base pair with the primer strand and the other G rolls by 30°. With the template backbone fixed by the molecular splint, a composite of the two conformations gives the appearance of normal straight DNA (Fig. 5A). However, when the 5′G has a roll angle, it clashes with the incoming dGMPNPP (Fig. 5B), and the base of the dGMPNPP has B values near 50 Å2 instead of ≈25 Å2 as in the normal DNA and Pt-GG1 complexes. This nonideal conformation may explain the fivefold increase in KM (SI Appendix, Table S2) and a larger separation between the 3′-OH of the primer and the α-phosphate (3.9 Å) than the normal 3.3 Å. Interestingly, at the same stage of Pt-GG bypass by Dpo4, the lesion adopts a single conformation with the 5′G clashing with the incoming dATP (PDB ID code 3M9O) (23) (SI Appendix, Fig. S1B).
Fig. 5.
The Pt-GG3 and Pt-GG4 structures. (A) The two conformations of Pt-GG (cyan and magenta) are shown with the refined 2Fo-Fc map contoured at 1σ. A double arrowhead marks close contacts between the magenta 5′G and dGMPNPP. (B) Superposition of Pt-GG3 and TT3 structures [PDB ID code 3MR5 (25); orange colored primer and incoming nucleotide and dark green Mg2+]. In Pt-GG3 (yellow, silver, and magenta), the cisplatin protrudes into the major groove and the 3′-primer end is shifted away from the α-phosphate of dNMPNPP (yellow dash). (C) The DNA in the Pt-GG4 structure. The template strand is shown in orange with Pt-GG highlighted in magenta. Notice the bent 3′-primer end and its stacking with the 5′G of Pt-GG (double arrowhead). (D) Superposition of the DNA in Pt-GG4 (colored as in C) and that in the normal ternary complex (PDB ID code 3MR2, gray) (25). The displacement of the primer end in Pt-GG4 is indicated by the red arrow.
Pt-GG4: A Binary Complex.
In the Pt-GG4 crystal structure, which was designed to capture the primer extension two nucleotides after the lesion, no incoming nucleotide is observed. Neither increasing the concentration of dAMPNPP in the crystallization buffer nor soaking crystals in a dAMPNPP solution led to the intended ternary complex. At 2.0-Å resolution, both the Pt-GG and DNA are well-defined (Fig. 5C). The Pt-GG assumes a conformation similar to the protein-free structure reported previously, including the local A-form (3′-endo) conformation of the Pt-GG (31, 36, 37). The cross-linked Gs are at a 30° angle. Because of the tight interaction between hPol η and the template backbone, the distortion is transmitted to the free 3′-primer end, which rolls toward the major groove. The 3′-OH is >4 Å from the normal reactive position, and the bent DNA precludes binding of an incoming nucleotide (Fig. 5D). Without an incoming nucleotide, no magnesium ion is in the active site and the three catalytic carboxylates, D13, D115, and E116, assume the most favorable rotamer conformations (SI Appendix, Fig. S3B).
Hydrophobic Pocket for Inhibitor Binding.
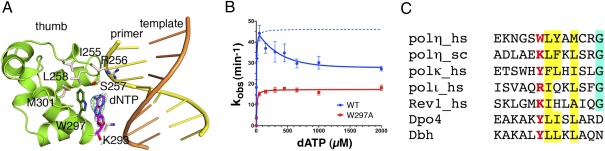
In all 11 crystal structures of hPol η–DNA complexes (three normal and eight with a lesion DNA), a hydrophobic pocket between the upstream minor groove and W297 in the thumb domain contains extra electron density, which is interpreted as the base of a dNMPNPP present at 1 mM concentration in all crystallization buffers (Fig. 6A) (25). The sugar and phosphate moieties of this dNMPNPP are disordered. After soaking crystals in a stabilization buffer without dNMPNPP, the density near W297 disappeared. Consistent with this crystallographic observation, concentrations of dNTP >0.5 mM have an inhibitory effect on the catalytic rate of hPol η (Fig. 6B). Inhibition by dNTP has not been observed with other DNA polymerases. Structure-based sequence comparison of the Y-family polymerases reveals that W297 is unique in human Pol η (Fig. 6C). When W297 is replaced by Ala, which likely destabilizes the thumb domain and DNA binding (Fig. 6A), the mutant hPol η has a reduced kcat (SI Appendix, Fig. S5) but is no longer sensitive to inhibition by high concentrations of dNTP (Fig. 6B).
Fig. 6.
A unique hydrophobic pocket potentially binds hPol η inhibitors. (A) A nucleotide (dATP) is bound between a tryptophan (W297) in the thumb domain (green ribbon diagram) and the minor groove of DNA. An omit electron density map contoured at 2σ is superimposed on the dATP. Residues surrounding W297, shown as stick models, form a hydrophobic core or interact with the DNA. (B) At dATP concentrations >0.5 mM, WT hPol η is inhibited (blue curve) and the W297A mutant protein is unaffected (red curve). The blue dotted line indicates the expected values if WT is not inhibited. (C) Sequence alignment of Y-family polymerases around the α-helix containing W297 (colored red). Human and yeast proteins are denoted by hs or sc. Conserved hydrophobic residues are highlighted in yellow and others in cyan.
Discussion
Requirement for Nucleotide Incorporation Opposite Pt-GG.
Pol η is the only DNA polymerase that has an enlarged active site to accommodate the entire CPD or Pt-GG for the 3′ base to instruct correct nucleotide incorporation. The active site of hPol η is optimized for CPD bypass. To accommodate the larger Pt-GG and enable its 3′G to template dCTP incorporation (Pt-GG1), the R61-M63 loop of hPol η is unexpectedly shifted, thus opening the active site to admit the cisplatin lesion in the stacked-in conformation while maintaining dCTP binding (Fig. 3). Although the 5′G of Pt-GG can flip out and allow Pol η to catalyze nucleotide incorporation without altering the R61-M63 loop, the flipped-out Pt-GG would prevent or slow down DNA translocation for the second dC incorporation as shown in the Pt-GG2 structure (Fig. 4). The plasticity of R61-M63 is probably also necessary for Pol η to bypass another anticancer agent, oxaliplatin [(1R,2R)-diaminocyclohexane)platinum(II)] (SI Appendix, Fig. S4) (10). In addition to the lesion accommodation, human Pol η keeps the upstream DNA duplex in place with the molecular splint and holds the downstream template strand between the finger (W42) and LF domain, thus enabling a templating base to pair with an incoming nucleotide even in the absence of base stacking. Unlike many Y-family polymerases, which allow dNTP to sample different conformations (39–41), Q38 and R61, which are conserved among Pol η homologs, keep the replicating base pair in the single conformation optimal for the reaction (Figs. 3–5) (25).
DNA Backbone Flexibility in Primer Extension.
The structures reported here provide a molecular basis for why two TLS polymerases are needed for efficient bypass of Pt-GG, one for nucleotide insertion opposite the lesion and another for primer extension (42, 43). The molecular splint, which keeps CPD-containing DNA straight and allows hPol η to complete bypass synthesis in four consecutive steps (25, 29), fails to support complete bypass of Pt-GG. This distinction is likely due to the size difference between Pt-GG and CPD. Because the DNA backbone is rigidified by Pol η, the roll of a cross-linked G in the template strand often perturbs the primer strand or the incoming dNTP (Fig. 5) and inhibits the catalysis.
Pol ζ, a member of the B-family polymerases, has been shown to be a general extender in TLS (35). Although little is known about Pol ζ, its homolog Escherichia coli DNA polymerase II has a well-characterized ability to extend primers after lesions. DNA pol II takes advantage of the reduced base stacking caused by lesions (44) to loop out damaged bases and uses normal template bases downstream to direct primer extension (45). DNA backbone flexibility is likely exploited by Pol ζ to accommodate DNA lesions for primer extension.
Concluding Remarks.
Our biochemical and structural studies suggest an alternative strategy for cancer therapy. Although several other DNA polymerases in humans may carry out TLS, they do so with low efficiency and high error rates (42, 43). Accurate bypass of Pt-GG appears to depend on Pol η to first incorporate dCTP opposite the cross-linked bases. Inhibition of Pol η may thus reduce chemoresistance to platinum-based drugs caused by translesion DNA synthesis. Our finding of the unique hydrophobic pocket lined by W297 provides a specific target for developing inhibitors of human Pol η. A combination of cisplatin and Pol η inhibitors may allow reduced cisplatin dosage and increase the efficacy of anticancer treatment.
Materials and Methods
Each oligonucleotide containing a single pair of adjacent guanines was cross-linked by activated cisplatin and purified by using HPLC or PAGE. Protein preparation, crystallization, structure determination, and mutagenesis were carried out as described (25). The primer extension and steady-state Km and kcat were measured in 2.5–17 nM Pol η, 5 μM primer-template substrate and 0–200 μM dNTP or with 0–2,000 μM dATP for the inhibition assay. Reaction products were analyzed as described (45). Detailed materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. R. Craigie, D. Leahy, and M. Gellert for critical reading of the manuscript. This work was supported by the Intramural Research Program of National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (Y.Z., C.B., M.T.G., and W.Y.), National Natural Science Foundation of China Grant 30830006 (to Y.-J.H.), and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 22131008 and 22249005 (to F.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4DL2, 4DL3, 4DL4, 4DL5, 4DL6, and 4DL7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202681109/-/DCSupplemental.
References
- 1.Rosenberg B, VanCamp L, Trosko JE, Mansour VH. Platinum compounds: A new class of potent antitumour agents. Nature. 1969;222:385–386. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 2.Wheate NJ, Walker S, Craig GE, Oun R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010;39:8113–8127. doi: 10.1039/c0dt00292e. [DOI] [PubMed] [Google Scholar]
- 3.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 4.Cepeda V, et al. Biochemical mechanisms of cisplatin cytotoxicity. Anticancer Agents Med Chem. 2007;7:3–18. doi: 10.2174/187152007779314044. [DOI] [PubMed] [Google Scholar]
- 5.Howell SB, Safaei R, Larson CA, Sailor MJ. Copper transporters and the cellular pharmacology of the platinum-containing cancer drugs. Mol Pharmacol. 2010;77:887–894. doi: 10.1124/mol.109.063172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 7.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 8.Strauss BS. Translesion DNA synthesis: Polymerase response to altered nucleotides. Cancer Surv. 1985;4:493–516. [PubMed] [Google Scholar]
- 9.Mamenta EL, et al. Enhanced replicative bypass of platinum-DNA adducts in cisplatin-resistant human ovarian carcinoma cell lines. Cancer Res. 1994;54:3500–3505. [PubMed] [Google Scholar]
- 10.Chaney SG, Campbell SL, Bassett E, Wu Y. Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit Rev Oncol Hematol. 2005;53:3–11. doi: 10.1016/j.critrevonc.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci USA. 2010;107:20792–20797. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry. 2000;39:4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 13.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett E, et al. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res. 2004;64:6469–6475. doi: 10.1158/0008-5472.CAN-04-1328. [DOI] [PubMed] [Google Scholar]
- 15.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann AR. Replication of damaged DNA in mammalian cells: New solutions to an old problem. Mutat Res. 2002;509:23–34. doi: 10.1016/s0027-5107(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 18.Albertella MR, Green CM, Lehmann AR, O’Connor MJ. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005;65:9799–9806. doi: 10.1158/0008-5472.CAN-05-1095. [DOI] [PubMed] [Google Scholar]
- 19.Chen YW, Cleaver JE, Hanaoka F, Chang CF, Chou KM. A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Mol Cancer Res. 2006;4:257–265. doi: 10.1158/1541-7786.MCR-05-0118. [DOI] [PubMed] [Google Scholar]
- 20.Ceppi P, et al. Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2009;15:1039–1045. doi: 10.1158/1078-0432.CCR-08-1227. [DOI] [PubMed] [Google Scholar]
- 21.Teng KY, et al. DNA polymerase η protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy. J Transl Med. 2010;8:126. doi: 10.1186/1479-5876-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suo Z, Lippard SJ, Johnson KA. Single d(GpG)/cis-diammineplatinum(II) adduct-induced inhibition of DNA polymerization. Biochemistry. 1999;38:715–726. doi: 10.1021/bi981854n. [DOI] [PubMed] [Google Scholar]
- 23.Wong JH, et al. Structural insight into dynamic bypass of the major cisplatin-DNA adduct by Y-family polymerase Dpo4. EMBO J. 2010;29:2059–2069. doi: 10.1038/emboj.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown JA, Newmister SA, Fiala KA, Suo Z. Mechanism of double-base lesion bypass catalyzed by a Y-family DNA polymerase. Nucleic Acids Res. 2008;36:3867–3878. doi: 10.1093/nar/gkn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biertümpfel C, et al. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silverstein TD, et al. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masutani C, et al. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 29.McCulloch SD, et al. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 30.Park H, et al. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc Natl Acad Sci USA. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, et al. Solution structures of a DNA dodecamer duplex with and without a cisplatin 1,2-d(GG) intrastrand cross-link: Comparison with the same DNA duplex containing an oxaliplatin 1,2-d(GG) intrastrand cross-link. Biochemistry. 2007;46:6477–6487. doi: 10.1021/bi062291f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 33.Alt A, et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 34.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 35.Shachar S, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spingler B, Whittington DA, Lippard SJ. 2.4 A crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex. Inorg Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- 37.Todd RC, Lippard SJ. Structure of duplex DNA containing the cisplatin 1,2-Pt(NH3)22+-d(GpG) cross-link at 1.77 A resolution. J Inorg Biochem. 2010;104:902–908. doi: 10.1016/j.jinorgbio.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glick E, Vigna KL, Loeb LA. Mutations in human DNA polymerase eta motif II alter bypass of DNA lesions. EMBO J. 2001;20:7303–7312. doi: 10.1093/emboj/20.24.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol Cell. 2008;29:767–779. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Kirouac KN, Ling H. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase iota. EMBO J. 2009;28:1644–1654. doi: 10.1038/emboj.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 43.Hirota K, et al. Simultaneous disruption of two DNA polymerases, Polη and Polζ, in Avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet. 2010;6:e1001151. doi: 10.1371/journal.pgen.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair. 2006;5:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.