Abstract
In the preimplantation mouse embryo, TEAD4 is critical to establishing the trophectoderm (TE)-specific transcriptional program and segregating TE from the inner cell mass (ICM). However, TEAD4 is expressed in the TE and the ICM. Thus, differential function of TEAD4 rather than expression itself regulates specification of the first two cell lineages. We used ChIP sequencing to define genomewide TEAD4 target genes and asked how transcription of TEAD4 target genes is specifically maintained in the TE. Our analyses revealed an evolutionarily conserved mechanism, in which lack of nuclear localization of TEAD4 impairs the TE-specific transcriptional program in inner blastomeres, thereby allowing their maturation toward the ICM lineage. Restoration of TEAD4 nuclear localization maintains the TE-specific transcriptional program in the inner blastomeres and prevents segregation of the TE and ICM lineages and blastocyst formation. We propose that altered subcellular localization of TEAD4 in blastomeres dictates first mammalian cell fate specification.
Allocation of blastomeres to outside and inside positions during preimplantation mammalian development initiates specification of the first two cell lineages, the trophectoderm (TE) and the inner cell mass (ICM) (1, 2). Successful progression of TE and ICM fate specification and proper development of the preimplantation embryo depends on differential transcriptional programs that are instigated and maintained within the outer and inner cells. Gene-KO studies in mice showed TEAD4 as the master orchestrator of the TE-specific transcriptional program (3–5). TEAD4-null embryos do not mature to the blastocyst stage and TEAD4-null blastomeres lack expression of TE-specific master regulators like CDX2, GATA3, and EOMES (3, 4). However, they maintain expression of ICM-specific factors like OCT4 and NANOG.
Interestingly, TEAD4 expression is maintained both in cells of TE and ICM lineages, as well as in the TE-derived trophoblast stem cells (TSCs) and ICM-derived ES cells (ESCs) (5, 6). Thus, questions are raised as to how TEAD4 selectively orchestrates the TE/TSC-specific transcriptional program but not the ICM/ESC-specific transcriptional program. The current model predicts that the presence vs. the absence of a TEAD4 cofactor, yes-associated protein (YAP), modulates TEAD4 function at its target genes in outer vs. inner blastomeres (6), leading to the segregation of the TE and ICM lineages. However, YAP-null mouse embryos do not show preimplantation developmental defects (7), indicating that, unlike TEAD4, YAP function is dispensable during TE and ICM fate determination. It is proposed that another YAP-related cofactor, WWTR1 (i.e., TAZ), could compensate for the absence of YAP during early development (6). However, the mode of TAZ function during TE and ICM specification is unknown. Furthermore, direct targets of TEAD4 have not been identified in the TE or in trophoblast cells. Thus, definitive experiments have not been performed to conclude that loss of cofactor function/recruitment is the crucial mechanism to impair transcription of TEAD4 target genes in the ICM. Therefore, in this study, we used a ChIP sequencing (ChIP-seq) analysis to determine TEAD4 target genes in mouse TSCs (mTSCs), validated those targets in the early mouse embryos, and asked how TEAD4-target genes are differentially regulated in inner vs. outer blastomeres during preimplantation development. Our analyses revealed an evolutionarily conserved mechanism, in which altered subcellular localization of TEAD4 orchestrates differential transcriptional program in outer vs. inner blastomeres and determines the first cell fate decision during preimplantation mammalian development.
Results
Identifying Direct Targets of TEAD4 in mTSCs.
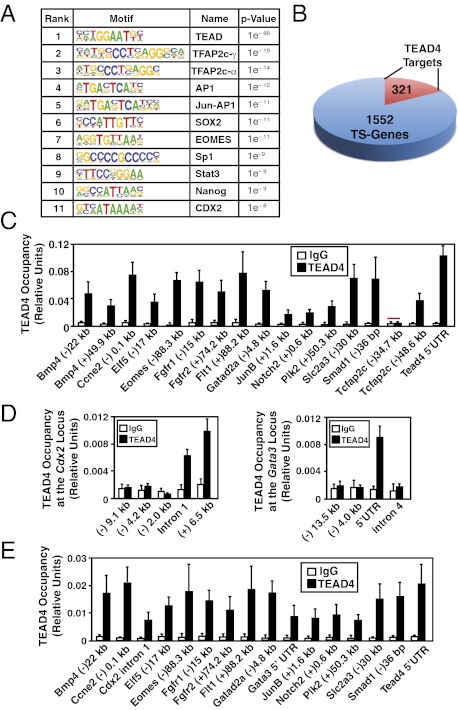
We conducted ChIP-seq in mTSCs to identify genomewide targets for TEAD4. Immunoprecipitated chromatin fragments were used to prepare libraries for deep sequencing, and sequences were mapped to the University of California, Santa Cruz, mouse genome assembly. Two control libraries were also generated and sequenced: one from total chromatin fragments before immunoprecipitation and the other from immunoprecipitated chromatin fragments using a nonspecific antibody (mouse IgG). These sequenced libraries were analyzed to detect TEAD4 ChIP peaks (SI Appendix, Fig. S1), and specific targets of TEAD4 were identified by using a model-based analysis for ChIP-seq (Model-based Analysis of ChIP-Seq) algorithm (8). By using a very high significance value (1 × 10−5) for binding sites, we identified 4,949 TEAD4 binding regions (Dataset S1). We used the overrepresented motif detection algorithm Hypergeometric Optimization of Motif Enrichment (HOMER), which ranks motifs on the basis of their statistical enrichment (9), to identify motifs localized within TEAD4 binding regions. HOMER analysis showed that the TEAD motif is the most prevalent motif at the identified binding regions, with a P value of 1 × 10−89 (Fig. 1A). Interestingly, TEAD4 binding regions are also enriched with binding motifs of other TSC-specific transcription factors (Fig. 1A), like TCFAP2c, SOX2, JUNB, EOMES, and CDX2 (10–14).
Fig. 1.
Direct regulation of core TSC genes by TEAD4. (A) HOMER analysis shows most abundant transcription factor-binding motifs at the ChIP-seq–identified TEAD4 binding regions along with log P values. (B) Pie chart shows numbers of core mTSC-specific genes with identified TEAD4 binding regions within ±50 kb of the transcription start site. (C) Quantitative ChIP analyses (mean ± SE; n = 3) shows TEAD4 chromatin occupancy at 18 regions that were identified by ChIP-seq analysis. The red bar shows a region in which TEAD4 occupancy was not detected. (D) ChIP analysis shows TEAD4 occupancy (mean ± SE; n = 3) at Gata3 and Cdx2 loci in mTSCs. (E) ChIP analyses (mean ± SE; n = 3) in mouse blastocysts show TEAD4 occupancy at the chromatin domains of mTSC-specific genes, including Gata3 and Cdx2.
Through comparative gene expression analyses, the Ralston et al. (4) defined 1,928 core mTSC-specific genes, of which 1,552 corresponded to mouse RefSeq transcripts. Among those 1,552 genes, ChIP-seq detected TEAD4 binding peaks within ±50 kb of 321 genes (Fig. 1B and Dataset S2), indicating that these might be the direct targets of TEAD4 in mTSCs. To validate genomewide analysis, we tested TEAD4 occupancy by conventional, real-time PCR-based quantitative ChIP (15) analysis at a subset of binding regions that are detected by ChIP-seq analysis. We selected 17 binding regions (Fig. 1C) near or within genes, which have mTSC-specific expression patterns (4) and have been implicated in trophoblast biology or in normal development (4, 16). The conventional quantitative ChIP analyses detected TEAD4 occupancy in 16 of those 17 regions, including a site at the 5′UTR of its own locus (Fig. 1C). We also tested functional importance of TEAD4 in maintaining expression of these putative target genes in mTSCs. We specifically depleted TEAD4 expression in mTSCs by RNAi (SI Appendix, Fig. S2 A and B), and loss of TEAD4 resulted in strong transcriptional repression of TEAD4 target genes (SI Appendix, Fig. S2C).
In preimplantation mouse embryos, TEAD4 is essential for the expression of GATA3 and CDX2 (3, 4). RNAi analysis showed that maintenance of GATA3 and CDX2 expression in mTSCs is also TEAD4-dependent (SI Appendix, Fig. S2 D and E). However, our initial analysis of ChIP-seq data did not identify any TEAD4 binding region within the Cdx2 locus, and a binding site was detected at a nonconserved (+)36.3 kb region of the mouse Gata3 locus. Therefore, we hypothesized that we might have obtained false-negative results for some TEAD4-binding regions at the Gata3 and Cdx2 loci as a result of the stringency of our ChIP-seq analysis. Therefore, we analyzed Gata3 and Cdx2 loci for the presence of conserved TEAD (GGAATG) motifs. Our analyses revealed the presence of multiple conserved TEAD motifs within ±15 kb of the transcription start sites of mouse Cdx2 and Gata3 loci (SI Appendix, Fig. S3A). Interestingly, when we compared TEAD4 ChIP-seq data with the Model-based Analysis of ChIP-Seq algorithm using only IgG ChIP-seq data set as control, we detected TEAD4-binding peaks at the Cdx2 intron 1, Cdx2 (+)6.5 kb, and Gata3 5′UTR regions (SI Appendix, Fig. S3B). To definitively conclude that TEAD4 occupies those regions of Cdx2 and Gata3 loci in mTSCs, we performed quantitative ChIP with mTSCs and detected TEAD4 occupancy at the conserved TEAD motifs within the intron 1 and 3′UTR regions of the Cdx2 locus and at the 5′UTR region of the Gata3 locus (Fig. 1D).
Next, we tested TEAD4 occupancy at the chromatin domains of mTSC-specific genes in mouse blastocysts. To test TEAD4 chromatin occupancy in blastocysts, we combined ChIP analyses with whole-genome amplification (15) and detected TEAD4 occupancy at all mTSC-specific gene loci, including Cdx2 and Gata3 (Fig. 1E). Collectively, the ChIP-seq and quantitative ChIP analyses showed that TEAD4 directly regulates transcription of key TE-specific regulators including Gata3 and Cdx2 in both mTSCs and preimplantation mouse embryos.
Lack of TEAD4 Nuclear Localization Impairs Its Target Gene Activation in ESCs.
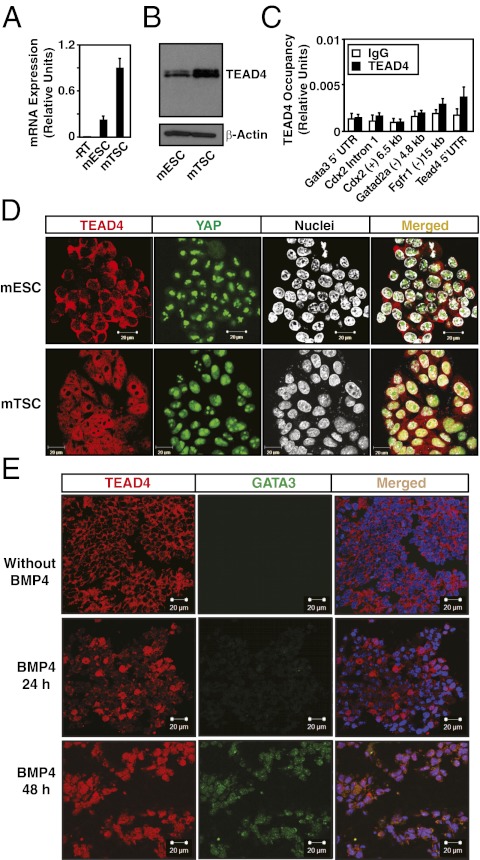
Although TEAD4 is expressed in the TE and the ICM, TEAD4 target genes such as Cdx2 and Gata3 are not expressed in the mouse ICM or ICM-derived mESCs, indicating that TEAD4-dependent transcriptional activation of these genes is impaired in those contexts. However, it has not been tested whether TEAD4 actually occupies the target chromatins in mESCs. We therefore tested TEAD4 expression and chromatin occupancy at its target genes in mESCs. Expression analyses revealed that, although in low quantity compared with mTSCs, TEAD4 mRNA and proteins are expressed in mESCs (Fig. 2 A and B). However, ChIP analyses revealed that TEAD4 chromatin occupancy at its target loci is severely impaired in mESCs (Fig. 2C). We asked whether the lack of TEAD4 chromatin occupancy is a result of the failure of TEAD4 to accumulate in mESCs nuclei. We performed immunofluorescence analysis to define the subcellular localization of TEAD4 and found that TEAD4 is localized only in the cytoplasm of mESCs (Fig. 2D, Upper). However, in mTSCs, TEAD4 is present in the cytoplasm and in nuclei, with more abundance in nuclei (Fig. 2D, Lower). We also looked at the localization of YAP, and found that, unlike TEAD4, YAP is mostly concentrated in the nuclei of both mESCs and mTSCs. These results strongly indicate that, although TEAD4 is expressed in mESCs and mTSCs, nuclear localization of TEAD4 and therefore activation of its target genes is impaired in mESCs.
Fig. 2.
TEAD4 nuclear localization is impaired in ESCs. Quantitative RT-PCR (A) and Western blot (B) analyses of TEAD4 expression in undifferentiated mESCs and mTSCs. (C) Quantitative ChIP analyses for TEAD4 occupancy at its target loci in undifferentiated mESCs (mean ± SE; n = 3). (D) Confocal images of undifferentiated mESCs and mTSCs show cellular localization of TEAD4 with respect to nuclei. (E) H9 hESCs were treated with BMP4 for different time intervals, and confocal images were taken to show that TEAD4 protein (red) localization in the nuclei (blue) precedes GATA3 (green) expression.
Next, we tested whether TEAD4 nuclear localization is altered in human ESCs (hESCs). As BMP4 treatment robustly induces trophoblast fate in hESCs (17, 18), we treated H9 hESCs with BMP4 and assessed TEAD4 nuclear localization and its target gene, GATA3, expression. In hESCs, GATA3 is strongly induced upon BMP4 treatment, and TEAD4 mRNA and protein are abundantly expressed in undifferentiated and BMP4-treated hESCs (SI Appendix, Fig. S4A). However, immunofluorescence studies revealed that undifferentiated hESCs lack TEAD4 protein in their nuclei (SI Appendix, Fig. S4B), and, upon BMP4 treatement, TEAD4 nuclear localization precedes GATA3 expression (Fig. 2E). Interestingly, unlike TEAD4, YAP nuclear localization is detected in BMP4-treated and untreated hESCs (SI Appendix, Fig. S4B). We also performed quantitative ChIP analysis to determine TEAD4 chromatin occupancy at the GATA3 chromatin domain in hESCs. In correlation with the subcellular localization pattern, TEAD4 occupancy at the GATA3 chromatin domain was detected only in the BMP4-treated cells (SI Appendix, Fig. S4C). Collectively, these results showed that BMP4 treatment induces TEAD4 nuclear localization in hESCs, activates its target genes like GATA3, and could contribute toward induction of trophoblast fate.
Impaired Nuclear Localization of TEAD4 Represses TE-Specific Genes in Nascent ICM.
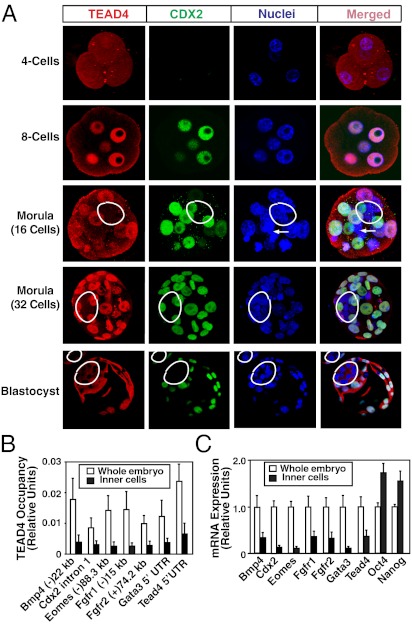
As TEAD4 protein was absent in nuclei of ICM-derived mESCs, we tested subcellular localization pattern of TEAD4 in preimplantation mouse embryos. Immunofluorescence analyses of preimplantation mouse embryos showed strong TEAD4 nuclear localization in all the blastomeres of four- and eight-cell embryos (Fig. 3A and SI Appendix, Fig. S5). However, in compacted morula-stage embryos, TEAD4 nuclear localization was observed only in the outer cells (Fig. 3A and SI Appendix, Fig. S5). Similarly, at the blastocyst stage, TEAD4 protein was detected only in the cytosol of the ICM lineage cells (Fig. 3A and SI Appendix, Fig. S5). By using two widely used, specific antibodies (SI Appendix, SI ExperimentalProcedures), we also tested YAP localization in mouse morula/blastocysts. Surprisingly, in contrast to earlier reports (6, 19), we detected YAP proteins only within the nuclei of ICM lineage cells (SI Appendix, Fig. S6A). Analyses of Yap−/− embryos confirmed specificity of anti-YAP antibodies (SI Appendix, Fig. S6B).
Fig. 3.
Impaired nuclear localization of TEAD4 represses target genes in the inner blastomeres of a preimplantation mouse embryo. (A) Preimplantation mouse embryos at different developmental stages were analyzed by confocal microscopy for TEAD4 (red) and CDX2 (green) expression with respect to nuclei (blue). Three-dimensional projections of confocal images at morula-stage embryos demonstrate lack of TEAD4 nuclear localization and CDX2 expression in the inner cells (white border). The white arrows show a dividing cell. Bottom: Micrographs show confocal images of blastocysts with near-complete exclusion of TEAD4 and loss of CDX2 expression in the nuclei of the ICM (white borders). Quantitative ChIP (B) and RT-PCR (C) analyses show decreased TEAD4 chromatin occupancy at its target loci, and repression of TEAD4 target genes in the inner cells of late morula/early blastocyst-stage mouse embryos compared with whole embryos (mean ± SE; n = 3). Note high levels of Oct4 and Nanog mRNA expression in the inner cells.
Next, we tested whether TEAD4 chromatin occupancy at its target loci is impaired within the nascent ICM lineage of developing mouse embryos. For this study, we took embryos at the early blastocyst stage and isolated inner ICM lineage cells by immunosurgery (15). With isolated inner cells, we performed ChIP/whole-genome amplification to test TEAD4 chromatin occupancy. We detected significant loss of TEAD4 chromatin occupancy at its target genes within the inner cells compared with that in the whole embryos (Fig. 3B), and this loss of chromatin occupancy was associated with significant loss in mRNA expression of TEAD4 target genes, including Tead4 itself (Fig. 3C). These results strongly indicate that the absence of TEAD4 protein in the nuclei of inner cells impairs TEAD4 chromatin occupancy resulting in decreased transcription of Tead4 itself and other target genes.
Restricted TEAD4 Nuclear Localization in TE Lineage Is a Conserved Phenomenon.
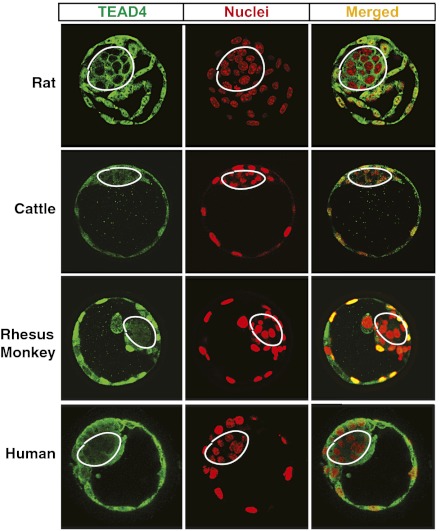
We tested whether restricted nuclear localization of TEAD4 to TE lineages is a conserved phenomenon in other mammalian species. Therefore, we investigated TEAD4 localization in blastocysts from rats, cattle, rhesus monkeys, and humans. Immunofluorescence studies revealed that, similar to the mouse, ICM nuclei of these species also lack TEAD4 protein (Fig. 4). Thus, restricted nuclear localization of TEAD4 to the TE lineages is a conserved phenomenon during preimplantation development in other mammalian species, including human.
Fig. 4.
Impaired TEAD4 nuclear localization in the ICM lineages is a conserved phenomenon during mammalian preimplantation development. Immunofluorescence analyses show localization of TEAD4 protein (green) with respect to nuclei (red) in rat, cattle, monkey, and human blastocysts as observed by confocal microscopy.
Induced TEAD4 Nuclear Localization in Inner Cells of Preimplantation Embryos Prevents Segregation of TE and ICM Lineages.
Analyses of mRNA expression revealed that TEAD4 expression is repressed in mESCs (Fig. 2A) as well as in the ICM lineage cells (Fig. 3C). Interestingly, in mESCs and in ICM lineage, loss of TEAD4 transcription is associated with loss of TEAD4 occupancy at the 5′UTR region of its own locus (Figs. 2C and 3B). Based on these observations, we hypothesized that TEAD4 positively autoregulates its own transcription in mTSCs and in the cells of TE lineage. However, impaired nuclear localization of TEAD4 disrupts the positive autoregulation in the ICM lineage and in mESCs, resulting in reduced TEAD4 protein levels compared with that in mTSCs (Fig. 2B). We also hypothesized that reduced protein level facilitates cytosolic retention of TEAD4 in the mESCs and in the mouse ICM lineage. This hypothesis is supported by an earlier observation that overexpression of a TEAD4 construct induces trophoblast fate in mESCs (6). Therefore, in the next set of experiments, we assessed positive autoregulation of TEAD4 and tested whether ectopic induction of TEAD4 induces its nuclear localization in mESCs and in the ICM lineage.
To test positive autoregulation, we assessed functional importance of conserved TEAD motifs at the TEAD4 5′UTR in transient transfection assays. We fused mouse TEAD4 promoter along with the 5′UTR region in front of a luciferase reporter gene (SI Appendix, Fig. S7A). The TEAD4 5′UTR region contains two adjacent putative TEAD motifs (SI Appendix, Fig. S7A). We found that deletion of those adjacent TEAD motifs significantly reduced the expression of luciferase gene from the reporter construct in mTSCs (SI Appendix, Fig. S7A, Right). These data strongly implicate those conserved TEAD motifs in enhancing transcription through TEAD4 promoter region in mTSCs, supporting a positive autoregulation.
To determine whether increased TEAD4 protein levels induce its nuclear localization, we generated a TEAD4-inducible mESC line (iTEAD4 cell), in which TEAD4 can be conditionally overexpressed in a tetracycline-inducible fashion. We efficiently induced TEAD4 protein expression in multiple clones of iTEAD4 cells (SI Appendix, Fig. S7B) with doxycycline and evaluated its nuclear localization and chromatin occupancy. We found that ectopic overexpression resulted in TEAD4 nuclear localization (SI Appendix, Fig. S7C) and chromatin occupancy at the target genes (SI Appendix, Fig. S7D).
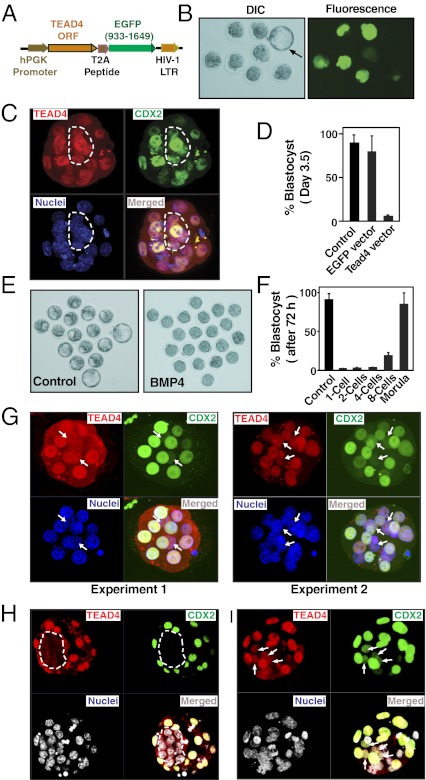
We next determined whether ectopic expression of TEAD4 induces its nuclear localization, thereby inducing expression of its target TE-specific genes in the inner blastomeres of a developing mouse embryo. We ectopically expressed TEAD4 in blastomeres by using a lentiviral construct. The TEAD4-expressing construct also expressed an EGFP protein that was linked with TEAD4 gene with a T2A linker (Fig. 5A). Thus, ectopic expression of TEAD4 in developing embryos was monitored by EGFP expression (Fig. 5B). We injected lentiviral particles at the one-cell–stage embryos to induce TEAD4 expression and tested their development ex vivo. We found that ectopic expression of TEAD4 induced its nuclear localization and activated CDX2 expression in the inner blastomeres (Fig. 5C) and strongly inhibited morula to blastocyst transition of developing embryos (Fig. 5D).
Fig. 5.
Induced TEAD4 nuclear localization in the inner blastomeres impairs preimplantation mouse development. (A) Schematic diagram of the Tead4-T2A-EGFP lentiviral construct. (B) One-cell mouse embryos were infected with the Tead4-T2A-EGFP lentiviral particles and were cultured for 96 h. Differential interference contrast (Left) and fluorescence (Right) micrographs show that development of embryos with ectopic TEAD4 expression (detected by EGFP expression) was arrested at the morula stage. A control embryo without lentiviral infection (black arrow, differential interference contrast micrograph) developed to the late blastocyst stage when cultured for the same period. (C) Three-dimension projection of confocal images of a TEAD4-overexpressing embryo. Presence of TEAD4 and CDX2 in all the nuclei is indicated with white borders. (D) Graph shows percentage of embryos maturing to the blastocyst stage after 96 h of culture. (E) One-cell mouse embryos were treated with or without BMP4, and preimplantation embryonic development was monitored. Micrographs show that, after 72 h, most embryos without BMP4 treatment are maturing to the blastocyst stage; BMP4 treatment arrested embryos at the morula stage. (F) One-cell mouse embryos were cultured for 72 h and treated with BMP4 at different developmental stages. The graph shows the percentage of embryos maturing to the blastocyst stage. (G) Three-dimensional projections of confocal images of morula-stage embryos treated with BMP4 from the four-cell stage. Two different embryos from two independent experiments are shown. The images show presence of TEAD4 and CDX2 in inner cell nuclei (white arrows). (H) Confocal images of a vehicle-injected blastocyst shows a lack of TEAD4 nuclear localization and CDX2 expression in the ICM. (I) Three-dimensional projection of confocal images of a BMP4-injected embryo shows nuclear TEAD4 and CDX2 expression in the ICM cells (white arrows).
Next, we sought to examine whether induction of TEAD4 nuclear localization at physiological concentration affects transcriptional program in the inner blastomeres of a developing embryo. To that end, we focused on BMP4 signaling. BMP4 treatment could induce trophoblast fate in ESCs from different mammalian species like mouse, rabbit, and human (17, 20, 21). Furthermore, we showed that BMP4 treatment induces TEAD4 nuclear localization in hESCs (Fig. 2E). Thus, we asked whether enhanced BMP4 signaling induces TEAD4 nuclear localization in the inner cells of a developing mouse embryo. We exposed mouse embryos to BMP4 at different developmental stages. We found a strong delay in blastocyst formation when BMP4 treatment was initiated at the one- to eight-cell stage of a mouse embryo (Fig. 5 E and F). Furthermore, immunofluorescence analyses in BMP4-treated embryos revealed that the inhibition in development is associated with induced TEAD4 localization and CDX2 expression in the nuclei of inner cells (Fig. 5G and SI Appendix, Figs. S8 and S9). Thus, we concluded that induction of TEAD4 nuclear localization in all blastomeres of an early preimplantation embryo inhibits its development to the blastocyst stage.
Interestingly, when BMP4 treatment was initiated at the late morula stage, no significant inhibitory effect was observed on blastocyst formation (Fig. 5F). We hypothesized that lack of accessibility of BMP4 to the inner cells of a morula-stage embryo could be the reason behind this observation. Therefore, we injected BMP4 into the blastocoel cavities of early-stage mouse blastocysts (SI Appendix, Fig. S10). We found that exposure to BMP4 at that stage did not inhibit blastocyst expansion. However, TEAD4 nuclear localization and CDX2 expression was induced in the ICM lineage cells (Fig. 5 H and I). These results further confirm that absence of nuclear TEAD4 protein is important to suppress TE-specific transcriptional program in the ICM-lineage cells.
Discussion
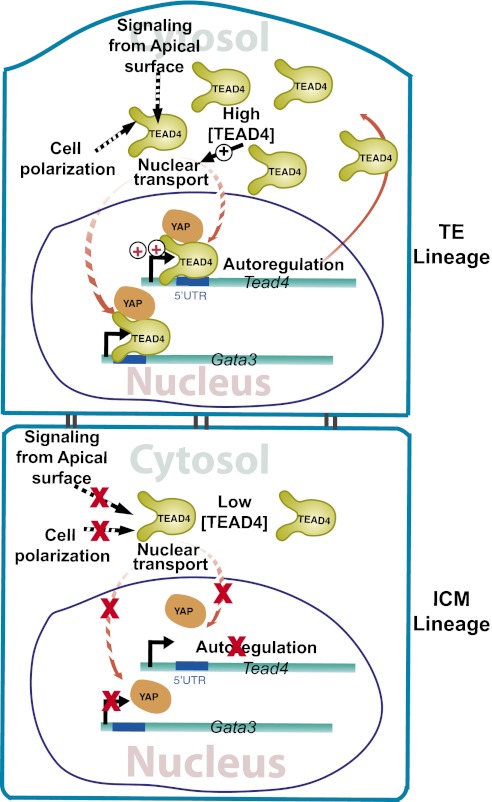
The inside-outside model (22) and the polarity model (23) propose that, in preimplantation embryos, inner blastomeres are functionally different from outer blastomeres. However, molecular mechanisms that specify and maintain differential transcriptional programs in outer vs. inner cells remain largely unknown. In this study, we demonstrated that transcription of several key TE-specific genes, as well as Tead4 itself, is directly regulated by TEAD4. However, transcription of those genes is impaired in the nascent ICM lineage because of the absence of nuclear TEAD4. We showed that differential subcellular localization of TEAD4 between TE and ICM lineages is a conserved event in several mammalian species, including human. We also showed that ectopic expression of TEAD4 induces its nuclear localization in the inner blastomeres, induces TE-specific genes, and prevents proper preimplantation development. Taken together, our results suggest a model (Fig. 6) in which reduced TEAD4 expression caused by disruption of a positive autoregulatory loop contributes to impaired TEAD4 nuclear localization in the inner blastomeres, thereby suppressing a TE-specific transcriptional program. We also propose that altered nuclear localization of TEAD4 is a conserved mechanism responsible for regulating distinct transcriptional programs in the nascent TE vs. ICM lineages, thereby allowing their specification and maturation of a preimplantation mammalian embryo to the blastocyst stage.
Fig. 6.
TEAD4 mediated specification of TE and ICM lineages in a preimplantation mammalian embryo. The model illustrates that nuclear localization of TEAD4 in the outer TE lineage induces TE-specific genes like Gata3. TEAD4 nuclear localization positively autoregulates its own transcription and increases TEAD4 protein levels in the TE lineage. The high TEAD4 concentration facilitates its nuclear localization. The dotted line denotes other putative mechanisms, not yet supported by experimental evidence, which could regulate TEAD4 nuclear localization in polar outer cells. The model also predicts that low TEAD4, as well as the absence of other putative mechanisms, impairs TEAD4 nuclear localization in the ICM lineage, thereby limiting TEAD4 transcription and abrogating expression of other TE-specific genes, such as Gata3.
A recent study in the context of endothelial cells showed that altered nuclear localization regulates TEAD4 function (24). However, our findings regarding the exclusion of TEAD4 from the nuclei of the ICM lineage cells is different from an earlier report by Nishioka et al. (5). That report indicated that, at the blastocyst stage of a mouse embryo, TEAD4 protein is detectable in all nuclei of TE and ICM. We propose that different reagents and experimental procedures are probable reasons for the discrepancies in our findings and those of Nishioka et al. (5). As we were unable to analyze TEAD4-null mice or TEAD4-null mESCs, we validated the specificity of the TEAD4 antibody that is used in our study by three different tests: (i) by showing that TEAD motif is the most predominant motif in genomewide TEAD4-binding regions identified by ChIP-seq analysis; (ii) by specifically knocking down TEAD4 in mTSCs; and (iii) by overexpressing the mouse Tead4 gene in iTEAD4 cells and in preimplantation mouse embryos.
Our mechanistic analyses showed evidence that a positive autoregulatory mechanism of TEAD4 expression could be important for altered protein levels in TE vs. ICM lineage cells (Fig. 6), which in turn could contribute to altered subcellular localization of TEAD4 in these two cell lineages. However, the molecular mechanism that initiates impairment of TEAD4 nuclear localization in inner blastomeres remains to be identified. Although we have shown that exposure of developing embryos to BMP4 can induce TEAD4 localization in inner cells, the lack of preimplantation developmental defect in BMP4-null mice (25) indicates that BMP4-dependent mechanisms are not the sole regulator in altering subcellular localization of TEAD4 in developing embryos. We predict that lack of cell polarization and absence of signaling from apical surfaces in the inner cells also contributes toward loss of TEAD4 nuclear localization. Thus, defining signaling mechanisms that prevent nuclear localization of TEAD4 in inner cells is an important subject of further study. Interestingly, as the appropriate TEAD4 nuclear localization patterns are maintained in mESCs and mTSCs, these stem cell populations can be exploited for further mechanistic studies.
Finally, the present model predicts that, in the ICM lineage cells or in ICM-derived ESCs, TEAD4 function is impaired, as the coactivator YAP is excluded from the nuclei of those cells (6). It has been indicated that phosphorylation of YAP by hippo signaling component, LATS2 kinase, restricts its nuclear localization in the ICM lineage cells as well as in the nuclei of ICM-derived mESCs (6). However, we found that YAP is mainly present in the nuclei of undifferentiated mESCs (Fig. 2D). Our observation in mESCs supports two recent studies (26, 27). These studies indicated that YAP is predominantly found in the nuclei of undifferentiated mESCs and is important for inducing expression of pluripotency regulators OCT3/4 and NANOG to maintain mESC self-renewal. We also analyzed numerous mouse blastocysts and detected YAP in the ICM nuclei in all of them (SI Appendix, Fig. S6). As mentioned earlier, we confirmed specificity of YAP antibodies by using Yap−/− mouse embryos (SI Appendix, Fig. S6). These observations and our findings of impaired TEAD4 nuclear localization in the ICM/mESCs indicate that further study is important to definitively conclude involvement of a Hippo–LATS2–YAP axis in limiting TEAD4 function in those contexts.
Materials and Methods
ChIP and ChIP-Seq Analyses.
Quantitative ChIP analyses with cultured cells and preimplantation embryos were performed following published protocols (15, 28) and is described in SI Appendix, SI Experimental Procedures. For TEAD4 ChIP-seq in mTSCs, immunoprecipitated chromatin fragments from three independent experiments were pooled, and genomic libraries were sequenced in Illumina Genome Analyzer II and in Illumina HiSeq platforms to generate 35-bp single end reads. Detailed bioinformatic analysis is described in SI Appendix, SI Experimental Procedures.
Analyses of Mammalian Preimplantation Embryos.
Procedures for collecting and culturing embryos from mice, rats, cattle, and rhesus monkeys are described in SI Appendix, SI Experimental Procedures. The human preimplantation embryos were collected at Stanford University according to all institutional rules, a two-stage consent process (29), and approval from institutional committees at Stanford University and the University of Kansas Medical Center. Fixed, discarded, and deidentified human blastocysts were analyzed for TEAD4 expression pattern by immunofluorescence study following protocols described in SI Appendix, SI Experimental Procedures. Additional experimental procedures are detailed in SI Appendix, SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Drs. M. J. Soares and D. F. Albertini for important suggestions, Dr. M. W. Wolfe for experiments with human embryonic stem cells, and Dr. Udayan Apte for Yap−/− embryos. This research was supported by National Institutes of Health Grants HD062546, HL106311, HL094892, HL094892, RR21876, HD53925, and RR000167.
Footnotes
The authors declare no conflict of interest.
Data deposition: The ChIP-seq data sets are available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37350).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201595109/-/DCSupplemental.
References
- 1.Zernicka-Goetz M. Cleavage pattern and emerging asymmetry of the mouse embryo. Nat Rev Mol Cell Biol. 2005;6:919–928. doi: 10.1038/nrm1782. [DOI] [PubMed] [Google Scholar]
- 2.Cockburn K, Rossant J. Making the blastocyst: Lessons from the mouse. J Clin Invest. 2010;120:995–1003. doi: 10.1172/JCI41229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yagi R, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 4.Ralston A, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- 5.Nishioka N, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Nishioka N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Morin-Kensicki EM, et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russ AP, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 11.Keramari M, et al. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS ONE. 2010;5:e13952. doi: 10.1371/journal.pone.0013952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidder BL, Palmer S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 2010;20:458–472. doi: 10.1101/gr.101469.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuckenberg P, et al. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol. 2010;30:3310–3320. doi: 10.1128/MCB.01215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schorpp-Kistner M, Wang ZQ, Angel P, Wagner EF. JunB is essential for mammalian placentation. EMBO J. 1999;18:934–948. doi: 10.1093/emboj/18.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Home P, et al. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284:28729–28737. doi: 10.1074/jbc.M109.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts RM, Fisher SJ. Trophoblast stem cells. Biol Reprod. 2011;84:412–421. doi: 10.1095/biolreprod.110.088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu RH, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 18.Schulz LC, et al. Human embryonic stem cells as models for trophoblast differentiation. Placenta. 2008;29(suppl A):S10–S16. doi: 10.1016/j.placenta.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varelas X, et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Tan T, et al. Generation of trophoblast stem cells from rabbit embryonic stem cells with BMP4. PLoS ONE. 2011;6:e17124. doi: 10.1371/journal.pone.0017124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi Y, et al. BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell Dev Biol Anim. 2010;46:416–430. doi: 10.1007/s11626-009-9266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarkowski AK, Wróblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. 1967;18:155–180. [PubMed] [Google Scholar]
- 23.Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24:71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Zhao D, James L, Li J, Zeng H. Requirement of the nuclear localization of transcription enhancer factor 3 for proliferation, migration, tube formation, and angiogenesis induced by vascular endothelial growth factor. FASEB J. 2011;25:1188–1197. doi: 10.1096/fj.10-167619. [DOI] [PubMed] [Google Scholar]
- 25.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 26.Lian I, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamm C, Böwer N, Annerén C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci. 2011;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- 28.Ray S, et al. Context-dependent function of regulatory elements and a switch in chromatin occupancy between GATA3 and GATA2 regulate Gata2 transcription during trophoblast differentiation. J Biol Chem. 2009;284:4978–4988. doi: 10.1074/jbc.M807329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalista T, Freeman HA, Behr B, Pera RR, Scott CT. Donation of embryos for human development and stem cell research. Cell Stem Cell. 2011;8:360–362. doi: 10.1016/j.stem.2011.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.