Abstract
Lipopolysaccharide (LPS), also known as endotoxin, activates the innate immune response through toll-like receptor 4 (TLR4) and its coreceptor, MD-2. MD-2 has a unique hydrophobic cavity that directly binds to lipid A, the active center of LPS. Tetraacylated lipid IVa, a synthetic lipid A precursor, acts as a weak agonist to mouse TLR4/MD-2, but as an antagonist to human TLR4/MD-2. However, it remains unclear as to how LPS and lipid IVa show agonistic or antagonistic activities in a species-specific manner. The present study reports the crystal structures of mouse TLR4/MD-2/LPS and TLR4/MD-2/lipid IVa complexes at 2.5 and 2.7 Å resolutions, respectively. Mouse TLR4/MD-2/LPS exhibited an agonistic “m”-shaped 2:2:2 complex similar to the human TLR4/MD-2/LPS complex. Mouse TLR4/MD-2/lipid IVa complex also showed an agonistic structural feature, exhibiting architecture similar to the 2:2:2 complex. Remarkably, lipid IVa in the mouse TLR4/MD-2 complex occupied nearly the same space as LPS, although lipid IVa lacked the two acyl chains. Human MD-2 binds lipid IVa in an antagonistic manner completely differently from the way mouse MD-2 does. Together, the results provide structural evidence of the agonistic property of lipid IVa on mouse TLR4/MD-2 and deepen understanding of the ligand binding and dimerization mechanism by the structurally diverse LPS variants.
Toll-like receptors (TLRs) recognize and respond to diverse pathogenic components of microorganisms and provide the first line of defense against microbial infection (1, 2). Among the microbial components, endotoxic lipopolysaccharide (LPS) from a membrane component of Gram-negative bacteria elicits the potent innate immune response through the receptor complex of TLR4 and MD-2 (3, 4). Excessive exposure to LPS often causes exaggerated signaling via TLR4 and fatal septic shock (5, 6), which is associated with a high mortality (20–30%) and is the most common cause of death in intensive care units (5, 6).
The lipid A moiety of LPS, which anchors LPS to the outer membrane of Gram-negative bacteria, is responsible for the immunostimulatory activity of LPS (7, 8). Lipid A consists of a 1,4′-bis-phosphorylated diglucosamine backbone to which variable lengths and numbers of acyl chains are covalently linked (8). The two phosphate groups are also important for the agonistic activity of lipid A because deletion of either phosphate group reduces the endotoxic activity (9, 10).
TLR4 is a type I transmembrane protein composed of 22 extracellular leucine-rich repeats (LRRs), a transmembrane domain, and the Toll/IL-1 receptor domain (TIR domain) that is essential for TLR signaling and conserved among members of the Toll receptor family (1). TLR4 alone does not directly bind LPS and requires the coreceptor MD-2 (11). MD-2 is associated with the extracellular domain of TLR4 and is indispensable for LPS recognition (4). A member of the MD-2–related lipid-recognition protein family (12), MD-2 directly binds to LPS in its hydrophobic cavity with high affinity (13).
Recently, the crystal structure of human TLR4/MD-2/Ra-LPS (Ra chemotype of Escherichia coli LPS) complex (14) was solved, which revealed that five of the six acyl chains of LPS are buried inside the MD-2 cavity. The sixth acyl chain lies on the surface of MD-2, partially exposed to the solvent. Together with the hydrophobic residues of MD-2, the partially exposed acyl chain constitutes the secondary binding site for the hydrophobic patch on the C-terminal convex face of the horseshoe structure of TLR4, leading to the formation of the “m”-shaped 2:2:2 hTLR4/MD-2/LPS complex. The close proximity of the C terminus of the extracellular domain in the complex induced by binding to LPS may allow for dimerization and signaling by the intracellular TIR domains (15, 16).
The number and length of the acyl chains determine the agonistic property of lipid A (17–19). E. coli lipid A is usually hexaacylated and acts as a potent agonist for all mammalian cells. In contrast, tetraacylated lipid IVa, the precursor of E. coli LPS, acts as an agonist only for some mammalian species. In particular, it acts as a weak agonist on mouse and as an antagonist on human cells (20, 21). Although several studies have investigated the species-specific activity of lipid IVa (22–28), these studies primarily used mutational and computational simulation methods. Structural information on the agonistic form of TLR4/MD-2 is limited to the hTLR4/MD-2/LPS complex; no structures of mTLR4/MD-2 complexed with LPS or lipid IVa are currently available. Structural knowledge may provide critical clues regarding the agonistic and antagonistic mechanisms by LPS and lipid IVa ligands that underlie species specificity.
Here, we present the two agonistic structures of mouse TLR4/MD-2/Re-LPS (Re chemotype of E. coli LPS) and TLR4/MD-2/lipid IVa complexes at 2.5 and 2.7 Å resolutions, respectively. This structural study will provide better understanding of the LPS recognition and signaling mechanism and will contribute to the development of therapeutic antiseptic shock drugs targeting TLR4/MD-2.
Results and Discussion
Oligomerization State Induced by a Lipid in Solution.
To address the species-specific response of TLR4/MD-2 induced by LPS and lipid IVa, we prepared mouse TLR4/MD-2 (mTLR4/MD-2) in complex with hexaacylated Re-LPS and tetraacylated lipid IVa. The mTLR4/MD-2/Re-LPS complex was dimeric in solution, as observed by gel filtration chromatography (Fig. 1A), consistent with the previous observation that LPS binding induces the dimerization of hTLR4/MD-2 (29). In contrast, the observed mTLR4/MD-2/lipid IVa complex was monomeric (Fig. 1A). Similarly, native gel electrophoresis analyses of mTLR4/MD-2 with hexaacylated LPS ligands (such as lipid A or various chemotypes of E. coli. LPS) showed an upward band shift (Fig. 1B, lane 3–6), whereas mTLR4/MD-2 with lipid IVa showed no upward band shift (Fig. 1B, lane 2). The upward band shift can be interpreted to be mTLR4/MD-2 dimerization following binding to LPS. We also confirmed the oligomerization state by the small-angle X-ray scattering (SAXS) analysis. The Rg calculated from SAXS Guinier analysis are 32.6 Å and 35.9 Å for mTLR4/MD-2 and mTLR4/MD-2/lipid IVa complexes, respectively (Table S1). The estimated molecular weights of these complexes from SAXS data (119 kDa for mTLR4/MD-2 and 129 kDa for mTLR4/MD-2/lipid IVa) agree with the calculated molecular mass of the monomeric mTLR4/MD-2 complex (119 kDa, N-glycan was estimated to be 2 kDa per site), suggesting that both complexes are monomeric in solution. The ligand-free form of hTLR4/MD-2 and hTLR4/MD-2/lipid IVa are also monomeric, whereas lipid A induces a dimerization for hTLR4/MD-2 (Table S1). The differences between hexaacylated LPS ligands and tetraacylated lipid IVa, although both act as agonists on mTLR4/MD-2, probably resulted from the low agonistic activity of lipid IVa compared with hexaacylated LPS.
Fig. 1.
(A) Gel filtration chromatography of mTLR4/MD-2. The mTLR4/MD-2 (black), mTLR4/MD-2/Re-LPS (blue), and mTLR4/MD-2/lipid IVa (green) complexes were analyzed by Superdex 200 gel filtration chromatography. Columns were calibrated with molecular weight standards (apoferritin and BSA), as indicated by arrows. Reported apparent molecular weight of the proteins was calculated from the elution volumes. (B) Ligand binding assay on native PAGE. Purified mTLR4/MD-2 (4 μg) was incubated with lipid IVa (0.5 μg), lipid A (0.5 μg), Ra-LPS (1.0 μg), Rd-LPS (1.0 μg), and Re-LPS (1.0 μg) at 37 °C for 2 h and then subjected to PAGE analyses.
LPS and Lipid IVa Induce Agonistic Dimeric Structures of Mouse TLR4/MD-2.
To gain structural insight into the species-specific activation mechanism, we determined the crystal structures of mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa at 2.5 and 2.7 Å resolutions, respectively. To facilitate crystallization, partial deglycosylation of mTLR4/MD-2 was used to prepare the mTLR4/MD-2/Re-LPS complex. Structures were determined by the molecular replacement method with the structure of the 1:1 mTLR4/MD-2 complex (30) [Protein Data Bank (PDB) code: 2Z64]. The ligand molecules, the lipid A moiety of Re-LPS and lipid IVa, were unambiguously modeled into the continuous electron densities observed in the hydrophobic cavity of MD-2 (Fig. 2 A and B). The outer two saccharides of Re-LPS were not modeled because of the poor electron density. In both complexes, the crystallographic asymmetric unit contained two copies of the 1:1:1 mTLR4/MD-2/Re-LPS or mTLR4/MD-2/lipid IVa complex related by noncrystallographic twofold symmetry, generating a dimeric form of the mTLR4/MD-2/Re-LPS or mTLR4/MD-2/lipid IVa complex (Fig. 2 C and D).
Fig. 2.
Dimeric structure of mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa. (A and B) Chemical structures and electron density maps for the bound ligands. Electron densities contoured with black at the 3.0-σ level in the Fo-Fc map, superimposed onto the refined model of Re-LPS (A) and lipid IVa (B). C, N, O, and P atoms are shown in yellow, blue, red, and orange, respectively. (C and D) Dimeric structure of the mTLR4/MD-2/Re-LPS (C) and mTLR4/MD-2/lipid IVa (D). TLR4 and MD-2 are shown in green and gray, respectively, and their dimerization partners are shown in blue and gray, respectively. Re-LPS and lipid IVa molecules bound to the hydrophobic cavity of MD-2 are shown in red as stick representations. Primary interfaces of mTLR4/MD-2/lipid IVa are encircled by orange dashes. Overall interfaces between the two 1:1:1 TLR4/MD-2/ligand complexes in the 2:2:2 TLR4/MD-2/ligand complex are indicated with black dashed lines. Dimerization interface of mTLR4/MD-2/lipid IVa is encircled by red dashes. Dimerization partners of the 1:1:1 mTLR4/MD-2/lipid IVa complex are indicated by asterisks. (E) Superposition of mTLR4/MD-2/lipid IVa (green), mTLR4/MD-2/Re-LPS (cyan), and hTLR4/MD-2/Ra-LPS (gray, PDB code 3FXI) with the same orientation as in Fig. 2B, Upper.
As expected from the crystal structure of the hTLR4/MD-2/Ra-LPS complex (14), the mTLR4/MD-2/LPS complex showed an m-shaped architecture, in which the N termini of TLR4 extended to opposite ends and the C termini were close to the middle. MD-2 and bound LPS molecules mediated dimerization of the two TLR4 molecules. Unexpectedly, the mTLR4/MD-2/lipid IVa complex, despite being monomeric in solution, showed a similar architecture to the 2:2:2 complex (Fig. 2 D and E). The difference between the oligomerization states of mTLR4/MD-2/lipid IVa in solution and in crystal may reflect the weak affinity for the association between the 1:1:1 mTLR4/MD-2/lipid IVa protomers. Throughout this paper, we indicate the second TLR4, MD-2, and their residues in the 2:2:2 TLR4/MD-2/ligand complex with asterisks to distinguish them from those of the 1:1 TLR4/MD-2 complex.
The surface area of the overall interfaces, indicated with black dashed lines in Fig. 2 C and D, are 1,799 and 1,764 Å2 for mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa, respectively. These values are comparable to that of hTLR4/MD-2/Ra-LPS (1,900 Å2) and sufficient for dimerization.
We only observed the 2:2:2 mTLR4/MD-2/lipid IVa complex in the crystal form, suggesting that the dimerization interaction of two 1:1:1 mTLR4/MD-2/lipid IVa complexes is weak in solution. Nevertheless, the dimerization would be enhanced on the cell surface in which the effective concentration of 1:1:1 mTLR4/MD-2/lipid IVa would be increased by the restriction of 2D movement in the membrane. Moreover, the transmembrane and cytoplasmic TIR domains of TLR4 may facilitate dimerization (23). In fact, the TIR domains of TLR1 or TLR2 are self-associated (31). The proposed dimeric structure of the TIR domains of TLR10 suggests that the TIR domains of the TLR family have the inherent property of self-association (32). Recently, the ectodomain of TLR5 in complex with its ligand has been shown to be monomeric in solution but dimeric in the crystal (33). The undistinguishable dimeric forms of human and mouse TLR4/MD-2 in complex with agonistic ligands suggest that Re-LPS and lipid IVa, which are agonistic ligands for mouse, induce agonistic dimerization in mTLR4/MD-2 as observed in hTLR4/MD-2/Ra-LPS.
LPS and Lipid IVa Induce Similar Structural Rearrangements in Mouse TLR4/MD-2.
The extracellular domain of TLR4 is divided into the N-terminal domain (LRRNT and LRR1–6), central domain (LRR7–LRR12), and C-terminal domain (LRR13–22 and LRRCT) (30). Compared with the unliganded 1:1 mTLR4/MD-2 structure (PDB ID: 2Z64), the C-terminal domain of TLR4 in mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa was bent by about 10° relative to the horseshoe plane (7–10 Å shifts in distances), to bring the two C-terminal domains of TLR4 into close proximity (10–15 Å distances) (Fig. S1A). These structural changes were also observed in the hTLR4/MD-2/Ra-LPS complex (14).
In the hTLR4/MD-2/Ra-LPS complex (14), MD-2 undergoes local structural changes in the Phe126 loop between the βG and βH strands (resides 123–129) by binding to LPS and through dimerization of TLR4/MD-2. The Phe126 side chain was exposed to solvent in the absence of agonistic ligand, as shown in the structures of mTLR4/MD-2, hMD-2, hMD-2/lipid IVa, and hTLR4/MD-2 in complex with an antagonistic ligand, eritoran (30, 34). This side chain was flipped into the hydrophobic cavity of MD-2 when complexed with agonistic LPS, where it made extensive hydrophobic contacts with the surrounding MD-2 residues and with the R2 and R3 acyl chains of LPS.
In both mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa, mMD-2 underwent similar structural changes around the Phe126 loop as observed in the hTLR4/MD-2/LPS complex (Fig. S1B). The Cα atom positions of Phe126 in mTLR4/MD-2/LPS and mTLR4/MD-2/lipid IVa were shifted 4.7 and 4.3 Å, respectively, toward the MD-2 cavity. The side chain of Phe126 interacted with Ile124, Leu54, Tyr131, and the R2 and R3 acyl chains of Re-LPS or lipid IVa, and also with the tip end of R2′′ acyl chain of Re-LPS in the Re-LPS complex (Fig. S1B). The Phe126 loop and βE–βF loop (residues 82–85), together with the partially exposed R2 acyl chain of lipid IVa, formed the hydrophobic core of the dimerization interface for another TLR4/MD-2 molecule (Fig. 3). This hydrophobic surface of MD-2 was suitable for interaction with the TLR4 hydrophobic patch located on the convex surface of TLR4 spanning LRR15 to LRR17 (Fig. 3). Residues in this region are thought to play an important role in LPS binding and receptor dimerization, as reported in several mutational studies (30, 35, 36).
Fig. 3.
Details of the dimerization interface of mouse TLR4/MD-2/lipid IVa. Dimerization interfaces of mTLR4/MD-2/lipid IVa. (Upper) Overall dimerization interface. (Lower) Interface is split and rotated to show the residues involved in recognition (MD-2 and lipid IVa, Left and TLR4*, Right). Ligand molecules and side chains directly interacting with each other within a radius of 4.5 Å are shown as stick representations. TLR4, TLR4*, and MD-2 are shown in green, blue, and gray, respectively.
Interactions Between Re-LPS or Lipid IVa and Mouse TLR4/MD-2.
Re-LPS and lipid IVa made many hydrophilic and hydrophobic interactions with mTLR4/MD-2, in a similar manner as with the hTLR4/MD-2/Ra-LPS complex (Fig. S2 and S3). At the cavity entrance of mMD-2, the glucosamine phosphate backbones of the ligands were aligned with the βG strand of MD-2 (Fig. S2 A and B). A total of eight and seven hydrogen bonds were formed between Re-LPS and mTLR4/MD-2, and lipid IVa and mTLR4/MD-2, respectively. In mTLR4/MD-2/lipid IVa, 3-OH of the R3′ acyl chain made hydrogen bonds with the side chain of Tyr102 of MD-2 (Fig. S2A). This interaction was specific for lipid IVa because hexaacylated LPS had an additional acyl chain, R3′′, at this position.
Two phosphate groups are important for the activation of TLR4/MD-2, and deletion of either of these phosphate groups reduces the endotoxic activity of the complex (9, 10). In fact, the two phosphate groups of Re-LPS or lipid IVa, 1-PO4 and 4′-PO4, made direct hydrogen bonds with TLR4 or TLR4*: 1-PO4 with Lys360 and Ser413* and 4′-PO4 with Lys263 in the mTLR4/MD-2/Re-LPS complex. In contrast, no direct hydrogen bonds were observed at the 1-PO4 group in the hTLR4/MD-2/Ra-LPS complex (Fig. S2C). In the mTLR4/MD-2/lipid IVa complex, 1-PO4 was surrounded by several positively charged residues, Lys341, Lys360, Lys367*, and Arg434* of mTLR4 and Arg90 of mMD-2, and 4′-PO4 was surrounded by Lys263, Arg266, Lys319, and Arg337 of mTLR4 (Fig. 4). In the hydrophobic cavity of MD-2, each acyl chain of each ligand was surrounded by the hydrophobic side chains of MD-2 and nearby ligand acyl chains (Fig. S3).
Fig. 4.
Electrostatic surface potentials of TLR4/MD-2. Electrostatic surface potentials of mTLR4/MD-2/lipid IVa (A) and hTLR4/MD-2/Ra-LPS (B). Positive and negative electrostatic potentials are shown in blue and red, respectively. Residues of interest are shown with their labels. Counterparts of human (A) and mouse (B) TLR4/MD-2 are shown in parentheses. Nonconserved residues between human and mouse TLR4/MD-2 are highlighted with boxes. Views are nearly the same as those in Fig. 3, Lower Left. Dimerization partner TLR4*/MD-2*/ligand* is omitted for clarity.
Lipid A Moiety Is Crucial for the Binding to TLR4/MD-2.
LPS molecules are embedded in the hydrophobic cavity of MD-2. The lipid A moiety of the LPS molecule occupied essentially the same space in hTLR4/MD-2/Ra-LPS (14) and mTLR4/MD-2/Re-LPS (Fig. 5A), despite differences of the LPS chemotype in the crystallization material (i.e., Re-LPS has an inner core disaccharide, whereas Ra-LPS has an inner core decasaccharide). The 4′-PO4 groups were located near the primary interface of TLR4/MD-2, and the 1-PO4 groups were located near the dimerization interface of TLR4/MD-2 in both complexes. The displacements of the phosphate atoms were 0.7 Å and 0.9 Å in 1-PO4 and 4′-PO4 groups, respectively, when superimposed with MD-2 molecules.
Fig. 5.
Structural comparison of the LPS ligand in the MD-2 cavity. Superposition of (A) mTLR4/MD-2/Re-LPS (blue) and hTLR4/MD-2/Ra-LPS (gray), (B) hMD-2/lipid IVa (magenta) and mTLR4/MD-2/lipid IVa (green), and (C) mTLR4/MD-2/Re-LPS (blue) and mTLR4/MD-2/lipid IVa (green).
All six acyl chains of LPS aligned in the cavity with identical conformations. The R3′ acyl chain was most deeply buried inside the cavity with ∼12 Å from the cavity entrance to its tip end. The R2 acyl chain was located in the shallow side of the cavity and was partially exposed to the solvent, with its tip end extended to interact with the hydrophobic patch on TLR4* (Fig. 5A). These structural observations confirm that the inner oligosaccharide moiety is not crucial for recognition by TLR4/MD-2. Moreover, they indicate that the recognition mechanism of hexaacylated LPS is conserved between human and mouse TLR4/MD-2, both of which are potently activated by hexaacylated LPS.
Lipid IVa and LPS Display the Same Overall Orientation in the MD-2 Cavity.
The lipid IVa structure in the dimeric mTLR4/MD-2 complex was compared with lipid IVa in the antagonistic hMD-2/lipid IVa complex (34). As previously found between hTLR4/MD-2/Ra-LPS and hMD-2/lipid IVa (14), the glucosamine phosphate backbone of lipid IVa was shifted upward by about 4–7 Å and rotated by about 180°. As a result, the arrangement of the acyl chains was completely different in the dimeric mTLR4/MD-2 complex compared with the antagonistic hMD-2/lipid IVa complex (Fig. 5B). The difference in lipid IVa orientation was not due to the absence of TLR4 but reflected the agonistic and antagonistic structure of lipid IVa. This conclusion can be made because eritoran (or E5564), a synthetic lipid A-derived antagonist against human and mouse TLR4/MD-2, when complexed with hTLR4/MD-2, had a similar orientation to that of lipid IVa in the hMD-2/lipid IVa complex (30).
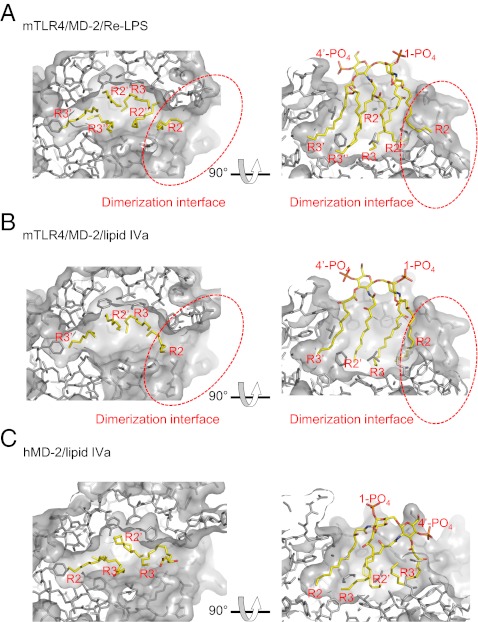
Surprisingly, although lipid IVa lacked two acyl chains compared with hexaacylated LPS, lipid IVa had the same overall orientation as LPS, with phosphate atom displacements of 1.3 and 2.6 Å in the 1-PO4 and 4′-PO4 groups, respectively, when superimposed with MD-2 molecules in mTLR4/MD-2/lipid IVa and mTLR4/MD-2/Re-LPS (Figs. 5C and 6 A and B). The R3, R2′, and R3′ acyl chains occupied essentially the same space in the cavity, with R3′ acyl chain most deeply buried inside the cavity with ∼11 Å from the cavity entrance to its tip end. The R2 acyl chain of lipid IVa folded back into the cavity, into the space where the R2′′ acyl chain of Re-LPS occupied. As a result, the tip ends of the R2 acyl chain of lipid IVa were shifted to the cavity bottom by 7.7 Å. The C3 to C7 carbon atoms of the R2 acyl chain constituted part of the dimerization interface, making van der Waals contacts with the side chain of Phe438* of TLR4*. The R2 acyl chain of Re-LPS in the mTLR4/MD-2/Re-LPS complex made a similar interaction, although all of the carbon atoms of the R2 acyl chain were involved in the interaction.
Fig. 6.
Arrangement of acyl chains in the hydrophobic cavity of MD-2. Surface cross-sections of mTLR4/MD-2/Re-LPS (A), mTLR4/MD-2/lipid IVa (B), and hMD-2/lipid IVa (C). Surfaces of MD-2 are represented with constituent residues in stick models. Surface cross-sections are in white. Ligand molecules are shown as stick models.
The interfaces between the two 1:1:1 TLR4/MD-2/ligand complexes in the 2:2:2 TLR4/MD-2/ligand complex comprised three distinct regions: MD-2/ligand vs. TLR4*, MD-2*/ligand* vs. TLR4, and TLR4 vs. TLR4*. The surface area of the dimerization interfaces (MD-2/ligand vs. TLR4*) were 500 Å2 (384 Å2 for MD-2/TLR4* and 116 Å2 for lipid IVa/TLR4*) and 519 Å2 (389 Å2 for MD-2/TLR4* and 130 Å2 for Re-LPS/TLR4*) for mTLR4/MD-2/lipid IVa and mTLR4/MD-2/Re-LPS, respectively. These values were comparable to the dimerization interface of hTLR4/MD-2/Ra-LPS (536 Å2). The space that R3′′ and R2′′ (C1 to C8) of Re-LPS occupied in the mTLR4/MD-2/Re-LPS complex was not occupied by lipid IVa (Fig. 6 A and B). Hence, interactions between acyl chains of lipid IVa and MD-2 were concentrated on one side of the cavity.
The arrangement of acyl chains of lipid IVa in the MD-2 cavity observed in the mTLR4/MD-2/lipid IVa complex is thought to be energetically unfavorable because one acyl chain (R2) is partially exposed to the protein surface, whereas the hydrophobic cavity is left unoccupied (Fig. 6B). We speculated that the unfavorable arrangement of acyl chains of lipid IVa may be partially compensated by the favorable interaction induced by dimerization of the 1:1:1 mTLR4/MD-2/lipid IVa complex, including the interaction of the exposed R2 chain with the hydrophobic patch of TLR4*.
Residues Involved in the Species Specificity for Lipid IVa.
The two phosphate groups of lipid IVa were surrounded by positively charged lysine and arginine residues of mTLR4/MD-2 (Fig. 4A). Of these residues, Arg266, Lys319, Arg337, Lys341, Lys367, and Arg434 of mTLR4 were not conserved between mTLR4 and hTLR4 (Fig. 4B). Notably, Lys367 and Arg434 were located near the dimerization interface. Meng and coworkers reported that when these residues were replaced with their human counterparts, Glu369 and Gln436, the responsiveness of mTLR4/MD-2 to lipid IVa was abolished (27). Hence, these two residues play an important role in species specificity for lipid IVa, possibly by augmenting the interaction with the 1-PO4* group of lipid IVa. These residues may modulate the electrostatic surface potentials to favor positioning of the negatively charged 1-PO4 group to be suitable for dimerization of mTLR4/MD-2/lipid IVa.
Meng and coworkers also reported that the mutation of Glu122 of mMD-2 to its human counterpart, Lys122, substantially reduced responsiveness to lipid IVa (26). Glu122 is located in the cavity entrance of MD-2. Mutation to the positively charged Lys residue would change the electrostatic potential of the cavity entrance and interaction with the charged phosphate group of lipid IVa, as shown in Fig. 4B. In the antagonistic structure of hMD-2/lipid IVa, lipid IVa was confined more deeply in the hydrophobic cavity of hMD-2 than it was in the mTLR4/MD-2/lipid IVa complex (34) (Fig. 6B). The side chain of Lys122 was oriented toward and partially covered the glucosamine backbone attached to the 4′-PO4 group. As a result, the side chain made hydrogen bonds with the O atom of the glucosamine residue and formed an electrostatic interaction with both of the phosphate groups of lipid IVa (Fig. S2D). Thus, Lys122 is likely to stabilize the antagonistic orientation of lipid IVa in hTLR4/MD-2.
The side chain of Glu122 of mMD-2 in the mTLR4/MD-2/lipid IVa complex was oriented away from lipid IVa (Fig. S2A). Owing to the repulsive effect of Glu122 against the phosphate groups of lipid IVa, mMD-2 would not form an antagonistic complex with lipid IVa as hMD-2. Hence, the repulsive effect of Glu122 against the phosphate groups of lipid IVa may be an important role in positioning lipid IVa at an appropriate orientation to allow the dimerization of TLR4.
In addition, mMD-2 had two hydrophobic residues (Leu125 and Pro127) in the Phe126 loop, corresponding to human Lys125 and Ser127, respectively (Fig. S4). Leu125 of mMD-2 in the mTLR4/MD-2/lipid IVa complex made van der Waals contacts with Asn415*, Met417*, Leu442*, and Ser443* of mTLR4*. Pro127 also made van der Waals contacts with Leu442*. These two residues reinforce the hydrophobic interaction in the peripheral region of the dimerization interface (Fig. 3) and may enable dimerization in the case of the lipid IVa complex. By mutagenesis, Leu125 of mMD-2 was previously shown to be important for the activation of mTLR4/MD-2 by lipid IVa (26).
These results suggest that, together with the different hydrophobicities of their dimerization interfaces, the different charge distributions of mouse and human TLR4/MD-2 affect the positions of the phosphate groups of lipid IVa, possibly by inducing the agonistic and antagonistic orientations of lipid IVa in mouse and human, respectively.
Conclusions
The agonistic structures of mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa provide structural evidence of how mTLR4/MD-2 is activated by tetraacylated lipid IVa and hexaacylated LPS. The unexpected orientation of lipid IVa in mTLR4/MD-2, which occupies the same conformational space as LPS, helps explain the agonistic effect by lipid IVa in mouse. The different charge distribution may be involved in proper positioning of the LPS ligand for the dimerization of TLR4/MD-2, especially for a relatively weak agonist such as lipid IVa. Our results may contribute to the development of therapeutic drugs that inhibit or modulate the responsiveness of TLR4/MD-2 on the basis of LPS variants.
Materials
Protein Expression, Purification, Crystallization, and Structure Determination.
The extracellular domain of mTLR4 (residues 22–627) and mMD-2 (residues 19–160) were inserted into the expression vector pMT/BiP/V5-His of the Drosophila Expression System. Protein was purified by IgG sepharose affinity chromatography, protein A tag cleavage by thrombin, saccharide trimming by endo Hf in the mTLR4/MD-2/LPS complex, and Superdex 200 gel filtration chromatography. Crystals were obtained using PEG3350 and the structural models of mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa were refined at 2.5 and 2.7 Å resolutions, respectively (Table S2). Details are described in SI Materials and Methods. Coordinates and structure factor data for the mTLR4/MD-2/Re-LPS and mTLR4/MD-2/lipid IVa complexes have been deposited to the PDB of the Research Collaboratory for Structural Bioinformatics as 3VQ1 and 3VQ2, respectively.
Supplementary Material
Acknowledgments
We thank the beamline staff at SPring-8 for their assistance with data collection. This work was supported by funding from Japanese Ministry of Education, Culture, Sports, Science and Technology Grant-in-Aid (to U.O., K.F., K.M., and T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (accession nos. 3VQ1 and 3VQ2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201193109/-/DCSupplemental.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 3.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 4.Nagai Y, et al. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 5.Angus DC, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Rangel-Frausto MS. Sepsis: Still going strong. Arch Med Res. 2005;36:672–681. doi: 10.1016/j.arcmed.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Schletter J, Heine H, Ulmer AJ, Rietschel ET. Molecular mechanisms of endotoxin activity. Arch Microbiol. 1995;164:383–389. doi: 10.1007/BF02529735. [DOI] [PubMed] [Google Scholar]
- 8.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rietschel ET, et al. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 10.Rietschel ET, et al. The chemical structure of bacterial endotoxin in relation to bioactivity. Immunobiology. 1993;187:169–190. doi: 10.1016/S0171-2985(11)80338-4. [DOI] [PubMed] [Google Scholar]
- 11.Shimazu R, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inohara N, Nuñez G. ML—a conserved domain involved in innate immunity and lipid metabolism. Trends Biochem Sci. 2002;27:219–221. doi: 10.1016/s0968-0004(02)02084-4. [DOI] [PubMed] [Google Scholar]
- 13.Viriyakosol S, Tobias PS, Kitchens RL, Kirkland TN. MD-2 binds to bacterial lipopolysaccharide. J Biol Chem. 2001;276:38044–38051. doi: 10.1074/jbc.M105228200. [DOI] [PubMed] [Google Scholar]
- 14.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 15.Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 17.Homma JY, Matsuura M, Kumazawa Y. Structure-activity relationship of chemically synthesized nonreducing parts of lipid A analogs. Adv Exp Med Biol. 1990;256:101–119. doi: 10.1007/978-1-4757-5140-6_6. [DOI] [PubMed] [Google Scholar]
- 18.Takada H, Kotani S. Structural requirements of lipid A for endotoxicity and other biological activities. Crit Rev Microbiol. 1989;16:477–523. doi: 10.3109/10408418909104475. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto Y, et al. Synthesis of lipid A and its analogues for investigation of the structural basis for their bioactivity. J Endotoxin Res. 2005;11:341–347. doi: 10.1179/096805105X76841. [DOI] [PubMed] [Google Scholar]
- 20.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golenbock DT, Hampton RY, Qureshi N, Takayama K, Raetz CR. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J Biol Chem. 1991;266:19490–19498. [PubMed] [Google Scholar]
- 22.Muroi M, Tanamoto K. Structural regions of MD-2 that determine the agonist-antagonist activity of lipid IVa. J Biol Chem. 2006;281:5484–5491. doi: 10.1074/jbc.M509193200. [DOI] [PubMed] [Google Scholar]
- 23.Saitoh S, et al. Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with Toll-like receptor 4 (TLR4)-MD-2 and ligand-induced TLR4 oligomerization. Int Immunol. 2004;16:961–969. doi: 10.1093/intimm/dxh097. [DOI] [PubMed] [Google Scholar]
- 24.Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3:354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- 25.Vasl J, Oblak A, Gioannini TL, Weiss JP, Jerala R. Novel roles of lysines 122, 125, and 58 in functional differences between human and murine MD-2. J Immunol. 2009;183:5138–5145. doi: 10.4049/jimmunol.0901544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng J, Drolet JR, Monks BG, Golenbock DT. MD-2 residues tyrosine 42, arginine 69, aspartic acid 122, and leucine 125 provide species specificity for lipid IVA. J Biol Chem. 2010;285:27935–27943. doi: 10.1074/jbc.M110.134668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng J, Lien E, Golenbock DT. MD-2-mediated ionic interactions between lipid A and TLR4 are essential for receptor activation. J Biol Chem. 2010;285:8695–8702. doi: 10.1074/jbc.M109.075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh C, et al. Elucidation of the MD-2/TLR4 interface required for signaling by lipid IVa. J Immunol. 2008;181:1245–1254. doi: 10.4049/jimmunol.181.2.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prohinar P, et al. Specific high affinity interactions of monomeric endotoxin.protein complexes with Toll-like receptor 4 ectodomain. J Biol Chem. 2007;282:1010–1017. doi: 10.1074/jbc.M609400200. [DOI] [PubMed] [Google Scholar]
- 30.Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 32.Nyman T, et al. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 33.Yoon SI, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi M, et al. Regulatory roles for MD-2 and TLR4 in ligand-induced receptor clustering. J Immunol. 2006;176:6211–6218. doi: 10.4049/jimmunol.176.10.6211. [DOI] [PubMed] [Google Scholar]
- 36.Resman N, et al. Essential roles of hydrophobic residues in both MD-2 and toll-like receptor 4 in activation by endotoxin. J Biol Chem. 2009;284:15052–15060. doi: 10.1074/jbc.M901429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.