Abstract
Ultraconserved elements (UCEs) are DNA sequences that are 100% identical (no base substitutions, insertions, or deletions) and located in syntenic positions in at least two genomes. Although hundreds of UCEs have been found in animal genomes, little is known about the incidence of ultraconservation in plant genomes. Using an alignment-free information-retrieval approach, we have comprehensively identified all long identical multispecies elements (LIMEs), which include both syntenic and nonsyntenic regions, of at least 100 identical base pairs shared by at least two genomes. Among six animal genomes, we found the previously known syntenic UCEs as well as previously undescribed nonsyntenic elements. In contrast, among six plant genomes, we only found nonsyntenic LIMEs. LIMEs can also be classified as either simple (repetitive) or complex (nonrepetitive), they may occur in multiple copies in a genome, and they are often spread across multiple chromosomes. Although complex LIMEs were found in both animal and plant genomes, they differed significantly in their composition and copy number. Further analyses of plant LIMEs revealed their functional diversity, encompassing elements found near rRNA and enzyme-coding genes, as well as those found in transposons and noncoding DNA. We conclude that despite the common presence of LIMEs in both animal and plant lineages, the evolutionary processes involved in the creation and maintenance of these elements differ in the two groups and are likely attributable to several mechanisms, including transfer of genetic material from organellar to nuclear genomes, de novo sequence manufacturing, and purifying selection.
Keywords: extreme conservation, repetitive elements
Analysis of animal genomes has uncovered regions of extreme sequence conservation that appear to have been preserved over periods approaching 300 million years (1, 2). Four hundred eighty-one ultraconserved elements (UCEs) of 200 bp or longer were identified by comparing the human, mouse, and rat genomes (3, 4). These elements, found in syntenic positions (where synteny is defined as a collinear arrangement of homologous sequences among a set of genomic regions), were characterized as being exonic, nonexonic, or possibly exonic (3). Later, UCEs shared between other genomes, including tetrapod and arthropod species, were identified (2, 5, 6). Many UCEs occur in noncoding regions and are thought to function as distal enhancers (7–9), transcriptional coactivators (10), or splicing regulators (11), or to associate with other regulatory factors (12–14). UCEs in exonic regions may be associated with RNA binding and splicing regulation (3, 15). The evolutionary mechanisms behind such extreme sequence conservation remain a mystery, although several hypotheses have been proposed (1, 16, 17). The regions containing one or more UCEs are thought to experience much stronger purifying selection than do conserved noncoding regions or protein-coding regions (1). The increased selective pressure is most likely attributable to a combination of a functional requirement for very specific DNA sequences and a high fitness cost for the absence of those sequences. The most basic approach for identifying UCEs in two or more genomes relies on constructing pairwise sequence alignments of large genomic regions (5, 6, 18). Although this approach is tractable for closely related genomes, it becomes both more computationally intensive and less accurate for more distantly related organisms.
Plant genomes differ from mammalian genomes in several ways that affect the identification of regions of extreme conservation (19). First, compared with mammals, plant genomes have undergone far more dynamic genomic evolution as a result of repeated polyploidy events (20). Furthermore, chromosomal changes, such as fractionation following polyploidy, crossovers, and mutations, make it harder to identify homologous regions between plant genomes (21, 22). Such extensive rearrangements render the current whole-genome alignment-based methods inapplicable for detecting identical sequences in plants. To the best of our knowledge, the only work on extreme sequence conservation in plants to date used a pattern-matching algorithm that identified a set of 25 elements of 100 bp or longer between the genomes of Arabidopsis thaliana and Oryza sativa (23). The small number of identical sequences, the fact that no sequence pairs were in syntenic positions, and the fact that one sequence was a part of a larger segment of mtDNA led to the hypothesis that at least some “ultraconservatism” in plants could be explained by horizontal transfer events (22).
Results
We have developed an information-retrieval based method to identify all long identical multispecies elements (LIMEs) shared by two or more genomes, given the element’s minimal length (Materials and Methods). The method is alignment-free, allowing us to detect both syntenic and nonsyntenic sequences. We used this method to identify and compare sequences of extreme conservations shared between a set animal genomes and a set of plant genomes (Fig. 1 and SI Appendix, Figs. S1–S5). Specifically, we first obtained a comprehensive set of LIMEs 100 bp or longer for six animal genomes: dog (Canis familiaris; Cf), chicken (Gallus gallus; Gg), human (Homo sapiens; Hs), mouse (Mus musculus; Mum), macaque (Macaca mulatta; Mam), and rat (Rattus norvegicus; Rn). We also obtained all LIMEs of 100 bp or longer among the six publicly available large (>100 Mbp) plant genomes: Arabidopsis (Arabidopsis thaliana; At), soybean (Glycine max; Gm), rice (Oryza sativa; Os), cottonwood (Populus trichocarpa; Pt), sorghum (Sorghum bicolor; Sb), and grape (Vitis vinifera; Vv).
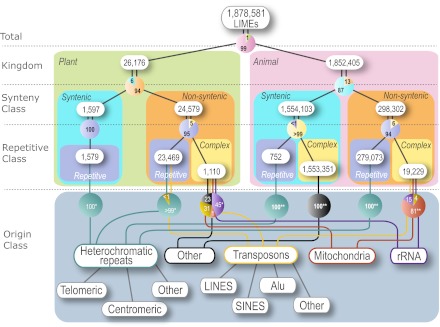
Fig. 1.
Structural taxonomy of plant and animal LIMEs. The plant genome set consists of Arabidopsis, soybean, rice, cottonwood, sorghum, and grape. The animal genome set is dog, chicken, human, mouse, macaque, and rat. LIMEs (≥100 bp) are identified for every pair of plant genomes and every pair of animal genomes, and categorized. A pie chart shows the percentage of contribution of each LIME category connected with the pie chart. Because of the lack of the annotation for all species involved, the last classification level, Origin Class, includes the percentages for Arabidopsis LIMEs in plants (*) and the percentages for the animal LIMEs in human, mouse, rat, and chicken (**); the absolute numbers are given in SI Appendix, Table S5. We have defined telomeric repeats as syntenic for clarity. LINEs, long interspersed elements; SINES, short interspersed elements.
The comparative analysis of flowering plant and animal LIMEs revealed key similarities and differences between the two groups (Fig. 1). Both groups include repetitive LIMEs, consisting of multiple copies of one or two repeated motifs, as well as nonrepetitive, or complex, LIMEs. Furthermore, each group has LIMEs that occur in multiple copies in a genome and are often spread across multiple chromosomes. Finally, animal and plant LIMEs are likely to owe their origins to several mechanisms, including purifying selection, transferring genetic material from organellar to nuclear genomes, and de novo sequence manufacturing; some of these mechanisms may be unique to plants.
LIMEs in Animal Genomes.
We first compared the complex LIMEs shared between the human, mouse, and rat genomes (2004 builds) found by our algorithm with the UCEs obtained by Bejerano et al. (3). We found that in addition to identifying all 481 previously reported UCEs, our method identified 12 previously undescribed elements of 200 bp or longer (more details are provided in Materials and Methods and SI Appendix, Table S1). Unexpectedly, 4 of those 12 elements were nonsyntenic (SI Appendix, Table S2), including two LIMEs originating from retrotransposition events (SI Appendix, sections S1 and S2). Overall, there were 1,572,580 unique complex elements of at least 100 bp in the animal set of six genomes: 19% (297,329) had multiple copies in a single genome, and 10% (157,723) had multiple copies in multiple genomes, including 95 having multiple copies in at least four genomes. These 95 were merged into just 12 “supersequences” based on overlaps in their genomic locations. A BLAST search of these elements against the nonredundant (NR) nucleotide database at the National Center for Biotechnology Information (NCBI) (24) revealed exact matches to snRNAs, such as human 7SL, RNU1-6, RNU1-9, and RNU6-1, as well as heterogeneous nuclear ribonucleoprotein A1 from horse (more details are provided in SI Appendix, section S1). SI Appendix, Fig. S6 shows the distribution of multicopy complex and repetitive LIMEs. Most of the complex LIMEs were shared between human and macaque (SI Appendix, section S3), whereas mouse had most of the repetitive LIMEs. Complex elements were often near each other and sometimes overlapped. For instance, in human, 92% (7,384,943 of 7,960,078) of the complex elements overlapped; as a result, they could be grouped into just 668 clusters (2 elements are assigned to the same cluster if they are within 60,000 bp). There were only 11 single-element clusters, whereas the largest cluster contained 295,876 elements.
There were 241 distinct motifs that made up the repetitive LIMEs in animals (SI Appendix, Table S3), and they ranged from 2 to 30 bp, with an average length of 8.2 bp (SD = 4.7 bp). There were 127 motifs that were shared by three species, 74 shared by four, 48 shared by five, and 28 shared by all six species. Although most repetitive elements overlapped, this was not universally the case. For instance, there were 8,331 nonoverlapping repetitive elements in animals that were dispersed across 90% (142 of 157) of the chromosomes, except for 15 chromosomes in chicken.
Of the complex LIMES shared by at least two animal genomes (Fig. 1), there were 1,120 (average length = 136.55 bp, SD = 41.60 bp) shared by all six genomes, with 76 LIMEs of length greater than 200 bp. Of those 76 LIMEs, 33 were nongenic in human, 43 were genic, and none shared more than 50% sequence identity with chicken when considering the surrounding genomic regions (±40,000 bp). In fact, 3 of the 76 LIMEs had only 2–3% sequence identity to chicken (an example is provided in SI Appendix, Fig. S7). This contrasts sharply with the results reported previously in animals, where UCEs were all from highly similar genomic regions. In fact, the term “ultraconserved,” arguably, does not apply in these cases.
LIMEs in Plant Genomes.
Using methods identical to those utilized for animal genomes, we determined the comprehensive set of LIMEs shared between six plant species (Fig. 1). Because extreme conservation between three or more plant species had never been addressed before, we focused on characterizing plant LIMEs in this work, determining their possible origins and comparing them with the animal LIMEs. Unlike animal genomes, repetitive LIMEs were prevalent in all six plant genomes (Fig. 2A). An average plant repetitive LIME was 143 bp long, which is shorter than an average complex LIME (175 bp; Fig. 3A). The relative ratios of repetitive LIMEs to complex LIMEs were similar across the plant genomes considered (Fig. 3B); the Arabidopsis genome was typical in its possession and distribution of repetitive and complex LIMEs. We detected 214 unique complex LIMEs shared by Arabidopsis and at least one of the remaining five genomes (Fig. 2 B and C), including 91 unique complex elements shared between Arabidopsis and rice, 3.64-fold more than had previously been identified (23). In Arabidopsis, 35 of the 91 complex LIMES are nonoverlapping and (when considering multiple copies) 81 overlap with other complex elements (SI Appendix, section S4), whereas in rice, 69 of the 91 complex LIMES are nonoverlapping and 72 overlap with other complex elements. The repetitive elements constituted the majority of Arabidopsis LIMEs [1,685 distinct LIMEs (∼88.7%)], but the repertoire of repeated motifs was surprisingly small; we found that a repetitive LIME contained copies of either one or two motifs from a total set of six motifs of 2–7 bp, with each occurring up to 323 times in tandem. The majority of Arabidopsis LIMEs were nongenic; of 26,367 unique locations of repetitive LIMEs, 4,015 corresponded to genic sequences and 22,352 to nongenic sequences; of the 305 locations of complex LIMEs, 169 were genic and 136 were nongenic. Using the Arabidopsis information resource annotation framework TAIR (25), we also categorized all genic LIMEs as exonic, partly exonic, or possibly intronic, based on their overlap with annotated gene models. We found 3,251 exonic, 713 partly exonic, and 220 possibly intronic locations of both repetitive and complex LIMEs.
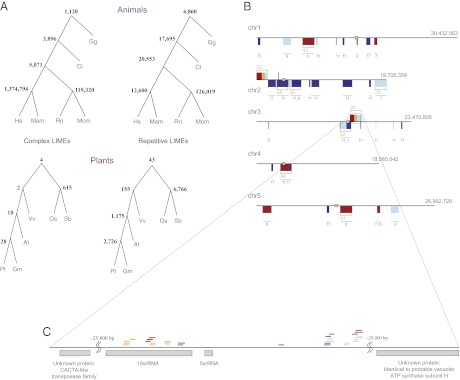
Fig. 2.
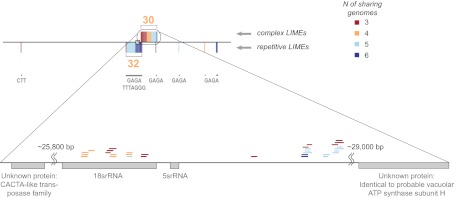
Plant LIMEs are remarkably diverse in their structure and function. (A) Phylogenetic trees of the six animal and six plant species for complex and repetitive LIMEs. Mam corresponds to Macaca mulatta, and Mum corresponds to Mus musculus. A node number (bold) is the number of elements common to each species in a subtree below. All LIMEs ≥100 bp are considered for each subtree. At, Arabidopsis thaliana; Gm, Glycine max (soybean); Hs, Homo sapiens (human); Mam, Macaca mulatta (macaque); Mum, Mus musculus (mouse); Os, Oryza sativa (rice); Pt, Populus trichocarpa (cottonwood); Rn, Rattus norvegicus (rat); Sb, Sorghum bicolor (sorghum); Vv, Vitis vinifera (grape). (B) LIMEs in the Arabidopsis (At) genome, depicted as colored ticks with complex LIMEs above and repetitive LIMEs below each chromosome (chr) sequence. Tick color corresponds to the number of genomes, including the At genome, sharing a LIME: red for three genomes, orange for four, light blue for five, and dark blue for six. When two LIMEs are 45 kbp or less apart, they are grouped in the same box. Once there are more than 20 LIMEs in such a box, the box size is unchanged but correct proportions of LIMEs shared by three, four, five, and six genomes are depicted by the relative thickness of the colored parts. Orange numbers specify the total number of LIMEs per box, and blue corresponds to the motif ID for one or multiple repetitive LIMEs. Identified centromere positions are shown as gray boxes. (C) Detailed representation of a chromosome 3 region that includes 2 LIMEs shared by all six genomes, and the nearest genes.
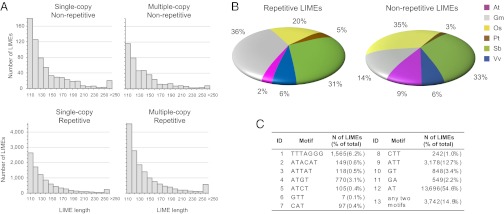
Fig. 3.
Each identified plant LIME could be classified into one of two basic structural classes: repetitive and complex LIMEs. (A) Distribution of LIME lengths in four groups of elements: single-copy complex, single-copy repetitive, multiple-copy complex, and multiple-copy repetitive. (B) Distribution of repetitive and complex LIMEs across six genomes (as percentage of total). At, Arabidopsis thaliana; Gm, Glycine max (soybean); Os, Oryza sativa (rice); Pt, Populus trichocarpa (cottonwood); Sb, Sorghum bicolor (sorghum); Vv, Vitis vinifera (grape). (C) Basic types of sequence motifs used by repetitive LIMEs. In total, there are 12 unique motifs 2–7 bp long.
Taxonomy of Plant LIMEs Based on Their Possible Origins.
Syntenic analysis using the Comparative Genomic platform CoGe (26) revealed that complex plant LIMEs are nonsyntenic (Fig. 1). This finding unexpectedly contrasts with the syntenic nature of the mammalian UCEs (3). The lack of synteny further supports our contention that some plant LIMEs are not inherited vertically. Indeed, we suggest there are three possible origins for the identical sequences found in our set of plant genomes: vertical inheritance, horizontal transfer, and de novo manufacturing. Although vertical inheritance of nuclear material is straightforward, detecting it can be confounded by extensive genome rearrangements. For instance, to determine whether the four overlapping LIMEs from Table 1 are conserved in species other than the six plants considered above, we used the shortest one (107 bp) in a BLAST search against the NR nucleotide database at the NCBI (24) and found exact copies of this LIME in the mature coding sequence of 18S (cytoplasmic), 26S (organellar), and 28S (cytoplasmic) rRNA genes of 76 eukaryotic organisms, including plants, animals, and fungi (more details are provided in SI Appendix, section S5).
Table 1.
Four LIMEs common to all six species and papaya
| LIME ID | Length | Species | Chromosome (contig) | No. copies |
| 1541 | 126 | Arabidopsis | 3 | 1 |
| Cottonwood | 14 | 1 | ||
| Grape | 6 | 1 | ||
| Papaya | (2112), (43833), (42612), (39182) | 4 | ||
| Rice | 2 | 1 | ||
| Sorghum | 1, 5 | 2 | ||
| Soybean | 13 | 89 | ||
| 1540 | 112 | Sorghum | 5 | 1 |
| 18704 | 114 | Soybean | 13 | 1 |
| 15791 | 107 | Sorghum | 1 | 1 |
| Soybean | 13 | 1 |
The four elements are considered unique because the two shortest elements are mapped to several additional locations in the sorghum genome. The second (LIME ID 1540), third (LIME ID 18704), and fourth (LIME ID 15791) elements are subsequences of the first element, and therefore are presented in all locations of the first LIME; for those three LIMEs, only locations distinct from the locations of the first LIME are shown.
Horizontally Inherited LIMEs.
The sequences of proposed horizontal inheritance detected by our algorithm could be of natural origin or artifactual. Some of the identified elements are likely the products of sequence assembly errors and/or bacterial sequence insertions (bacterial sequences were exclusively from Escherichia coli). On the other hand, we found several Arabidopsis repetitive elements associated with a transposon. A copy of a repetitive element containing the motif “GAGA” was found within an Arabidopsis gene annotated as “hAT-like transposase family” (TAIR gene ID AT5G28673); two other copies of this element were identified in genes annotated as “probable serine/threonine-protein kinase” (TAIR gene ID AT3G59410) and “unknown protein” (TAIR gene ID AT1G01725). Another copy of the same repetitive element, located on chromosome 2 of Arabidopsis, is classified as nongenic. SI Appendix, Fig. S8 shows the mapping of mitochondrial to nuclear genomes in Arabidopsis, rice, and sorghum. Arabidopsis has nine exonic LIMEs (SI Appendix, Table S4) that were derived from mitochondrial insertions. The cross-species genomic-to-genomic and mitochondrial-to-mitochondrial comparisons of these LIMEs revealed that the surrounding mitochondrial and nuclear sequences had rearranged and/or diverged, although still retaining these few elements throughout evolution (more details are provided in SI Appendix, section S6).
De Novo Sequence Manufacturing.
A process we refer to as “de novo sequence manufacturing” could be another possible source of identical cross-species sequences in plants. For example, telomeric repeats are manufactured by a known enzymatic mechanism (27), and these repeats certainly populate our collection of LIMEs. Strand slippage during DNA synthesis is another likely explanation for some of the repetitive elements identified. Likewise, gene conversion may underlie the LIMEs found among the rDNA genes. Similar to the previous description of Arabidopsis, although there were 25,066 unique repetitive LIMEs among the six genomes, these LIMEs were remarkably limited in the repeats they used. Thus, a repetitive LIME consisted of 1 or 2 short motifs; the set of all motifs used in LIMEs encompassed only 12 of the 1,699 possible 2- to 7-bp motifs (Fig. 3C). Moreover, only sorghum contained repetitive LIMEs of all 12 motifs, whereas other genomes used subsets of 5–11 motifs (Tables 2 and 3). On average, a repertoire of ∼7.8 unique motifs was used by repetitive LIMEs from one genome. Many repeats appeared to be microsatellites, consisting of motifs 2–6 bp long (28). The exceptions were the TTTAGGG (LIME label 1 in Fig. 2B) and GAGA, which are telomeric (29) and GAGA-binding (30) protein, respectively, and possibly two other motifs, ATACAT and ATTAT (Fig. 3C and SI Appendix, section S7).
Table 2.
Repeat motifs of repetitive LIMEs in plant genomes
| Motifs | At | Gm | Os | Pt | Sb | Vv |
| TTTAGGG | 1 | 1 | 1 | 1 | 1 | 1 |
| ATACAT | 0 | 0 | 1 | 0 | 1 | 0 |
| ATTAT | 0 | 0 | 1 | 0 | 1 | 0 |
| ATGT | 1 | 0 | 1 | 0 | 1 | 0 |
| ATCT | 0 | 0 | 1 | 0 | 1 | 0 |
| GTT | 0 | 1 | 1 | 1 | 1 | 1 |
| CAT | 1 | 1 | 0 | 0 | 1 | 0 |
| CTT | 1 | 1 | 1 | 0 | 1 | 0 |
| ATT | 1 | 1 | 1 | 0 | 1 | 1 |
| GT | 0 | 1 | 1 | 1 | 1 | 1 |
| GA | 1 | 1 | 1 | 1 | 1 | 1 |
| AT | 0 | 1 | 1 | 1 | 1 | 1 |
| Total | 6 | 8 | 11 | 5 | 12 | 6 |
All 12 distinct motifs contributing to LIMEs and their presence (1) or absence (0) in each of the six plant genomes are listed. At, Arabidopsis thaliana; Gm, Glycine max (soybean); Os, Oryza sativa (rice); Pt, Populus trichocarpa (cottonwood); Sb, Sorghum bicolor (sorghum); Vv, Vitis vinifera; (grape).
Table 3.
Distinct repeat motifs with a length of 2–7 bp shared between pairs of genomes that are found to contribute to the repetitive LIMEs
| At | Gm | Os | Pt | Sb | |
| Gm | 5 | ||||
| Os | 5 | 7 | |||
| Pt | 2 | 5 | 5 | ||
| Sb | 6 | 8 | 11 | 5 | |
| Vv | 3 | 6 | 6 | 5 | 6 |
At, Arabidopsis thaliana; Gm, Glycine max (soybean); Os, Oryza sativa (rice); Pt, Populus trichocarpa (cottonwood); Sb, Sorghum bicolor (sorghum); Vv, Vitis vinifera (grape).
Colocalization of LIMEs: Clusters and Superclusters in Plants.
Whether to consider elements individually or in groups depends on the question being asked. For instance, when studying sequence function, it is often beneficial to view elements individually, whereas when studying evolution, as we do now, it is beneficial to group nearby elements into a cluster that serves as a coselected functional unit. The animal UCEs, including the nonexonic elements, are often clustered in the genomes near transcription factors and genes associated with development (3); however, little is known about the colocalization of plant LIMEs. Although this property is expected for repetitive plant LIMEs, where one tandem repeat sequence could be a source of many repetitive LIMEs, we also found more overlapping than nonoverlapping complex LIMEs in four of the six plant genomes, with the exceptions being rice and sorghum (Fig. 4A and SI Appendix, section S8). The soybean genome, for example, contained 5,451 copies of 336 unique complex elements that could be grouped into just 47 clusters, where adjacent/overlapping elements were ≤60,000 bp apart. In Arabidopsis, the cluster of such neighboring LIMEs containing the 4 LIMEs shared by all six genomes was located in close proximity to the centromere of chromosome 3. On the other hand, the cluster in rice (chromosome 2) containing the same LIMEs was not located near the centromere or the telomere (SI Appendix, Fig. S1). Colocalization of LIMEs had its extremes: Soybean chromosome 13 (SI Appendix, Fig. S2B) contained the largest group of 3,062 neighboring LIMEs (the average distance between the starting nucleotides of 2 neighboring LIMEs for the first 3,061 LIMEs was only 291 bp). This number was surprisingly high, surpassing the number of neighboring LIMEs in the remaining five genomes by at least an order of magnitude; the rest of the soybean genome had 43 clusters with an average of 3.325 elements per cluster. Determining the origins of these abundant complex LIMEs in the region of the chromosome that is known for its unique association with the nucleolus organizer region (NOR) (31) could provide insights into differences between the soybean NOR and NORs of other species. For all six species, there were 631 complex clusters in total, with an average of ∼14 LIMEs per cluster (96.6%) and 306 complex LIMEs occurring alone (Fig. 4B). Also, there were 3,601 repetitive clusters (99.99%), with ∼1,007 LIMEs per cluster on average and 193 repetitive LIMEs occurring alone. A possible explanation for this clustering of LIMEs is horizontal gene/genome transfer events from organelle genomes.
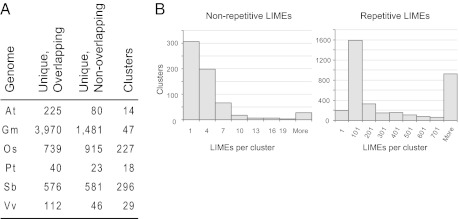
Fig. 4.
Plant LIMEs are often found overlapping or in close proximity to each other. (A) Numbers of complex LIMEs that (i) overlap with at least one complex LIME and (ii) do not overlap. Shown in the last column is the total number of complex LIME clusters, where each element in the cluster either overlaps with another element or is located within 60 kbp of another complex LIME. At, Arabidopsis thaliana; Gm, Glycine max (soybean); Os, Oryza sativa (rice); Pt, Populus trichocarpa (cottonwood); Sb, Sorghum bicolor (sorghum); Vv, Vitis vinifera (grape). (B) Distribution of cluster sizes among clusters containing repetitive and complex LIMEs.
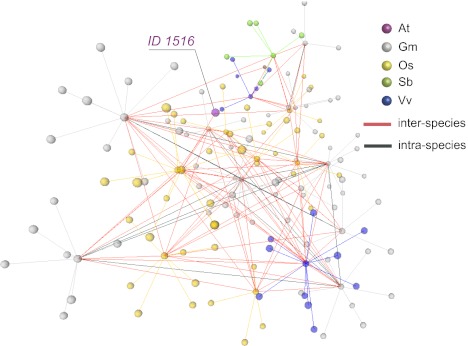
We next studied the relationship between the propensity of LIMEs to localize within the same cluster and to occur in multiple copies within the same genome and across multiple genomes. When constructing a network of clustered complex LIMEs, where two clusters were connected if they shared at least one common LIME, we found that the clusters were naturally grouped into 170 “superclusters,” where no 2 superclusters shared a single LIME (Fig. 5 and SI Appendix, section S9 and Fig. S9). When analyzing connectivity within superclusters, we found that LIMEs that belonged to the same cluster in one species were dispersed into multiple clusters in another species. For instance, in a supercluster that included a single complex LIME from Arabidopsis (LIME ID 1516), the average number of interspecies connections for one cluster was ∼3.4 (red edges in Fig. 5). Similarly, the intraspecies copies of a multicopy LIME often did not colocalize in the same cluster (dark green edges in Fig. 5 and SI Appendix, Fig. S10).
Fig. 5.
“Supercluster” of complex LIMEs that includes a single element from Arabidopsis (LIME ID 1516) and 24 clusters from four other genomes: soybean, rice, sorghum, and grape. The network of complex LIMEs from Arabidopsis (At; maroon node), soybean (Gm; gray nodes), rice (Os; gold nodes), sorghum (Sb; green nodes), and grape (Vv; blue nodes) is shown. All elements in one cluster are connected to a selected representative with the edges of the same color as the nodes. Clusters of LIMEs within one species are connected through the representative nodes with dark green edges if they share one or more multiple-copy complex LIMEs. Clusters sharing LIMEs across multiple species are connected through their representatives with red edges.
LIMEs in Plants vs. Animals.
Individual elements are defined as the longest common subsequence between two larger sequences. Our algorithm finds all such matching subsequences (≥100 bp) between genomes. The simplest way to quantify the elements is to count them individually. However, this leads to “double counting,” because many overlap (Materials and Methods). The structural taxonomy shown in Fig. 1 can be used to quantify them differently. It breaks down cross-species elements into two initial categories: repeated motifs and complex sequences. SI Appendix, Table S3 lists the 241 repeated motifs in the animal set and the 12 motifs in the plant set. To determine whether any of the repeated sequences were contained within mobile elements, we used the Repeat Masker server (32, 33), scanning the entire set of repetitive LIMEs. Among our LIMEs, we found homology only to several long interspersed elements (LINEs) and LTRs in mammals (1 LINE and 2 LTRs in human, 2 LINEs and 8 LTRs in rat as well as in mouse, and 1 LINE in dog); no homologous repeats for the chicken or plant LIMEs were found. Interestingly, nine repetitive LIMEs are shared between plants and animals. However, the LIME distribution is quite different between the two groups: Only a small minority of plant LIMEs have complex sequences [1,110 (4%)]. On the other hand, most of the elements in the animal set have complex sequences [1,572,580 (85%)]. If we count not the existence of an element but the total number of copies of it in each genome, these figures change to 0.24% and 60% for plants and animals, respectively. The number of copies of repetitive and complex elements also differs: 16,029 (64%) of repeated motif elements in plants and 151,091 (54%) in animals have multiple copies in at least one genome. For complex elements, the numbers are 435 (39%) and 455,052 (29%), respectively. In the plant set, there were 1,110 unique complex sequences of LIMEs shared by two genomes, 234 shared by three genomes, 144 shared by four genomes, 54 shared by five genomes, and 4 shared by all six genomes (Fig. 1 and SI Appendix, Figs. S1–S5). Exact copies of the shortest of the last four LIMEs were also found in 76 different organisms, including species from plants, animals, and fungi.
Discussion
Previous studies used synteny-based approaches to find UCEs in animals. However, these approaches were unable to find nonsyntenic ultraconserved regions in animals or any ultraconserved regions in plants. In this study, we developed and used a unique alignment-free information-retrieval approach to find a comprehensive set of LIMEs (both syntenic and nonsyntenic conserved regions over 100 bp in length) from two sets of genomes: six flowering plants and six vertebrates. Our comparison of LIMEs from the two groups reveals three major insights. First, although animal LIMEs are largely syntenic, plant LIMEs are exclusively nonsyntenic. Second, LIMEs can occur either in multiple or single copies in each genome and come in two types: simple repeated motifs and complex (nonrepetitive) sequences. Finally, the apparent extreme conservation may, in fact, result from several distinct processes.
Researchers have previously described a set of exclusively syntenic UCEs from animals. Although our method was able to add nonsyntenic elements to this collection, such nonsyntenic LIMEs are rare in animals. For instance, the only animal nonsyntenic LIMEs having multiple copies in at least four genomes were all derived from the snRNAs, which are known to retrotranspose. In contrast, plant LIMEs are all nonsyntenic, which explains why they had not been previously discovered by synteny-based searches.
Why would animal LIMES primarily be syntenic and plant LIMEs be all nonsyntenic? In many cases, the lack of synteny is attributable to the elements having been “created in place” rather than inherited from a common ancestor. Thus, the nonsyntenic nature of some LIMEs could be explained by their origin through the transfer of the genetic material from an organelle to the nuclear genome, or by the fact that these elements may be the parts of as yet unannotated mobile elements. Because plant mitochondrial genomes are large and evolve slowly (34), LIMEs can be created by the occasional insertion of copies of these genomes into the nuclear DNA. Although these insertions will eventually degrade through genetic drift, their relatively large size will give rise to LIMEs of reasonable size and longevity in the meantime. The other group of complex plant LIMEs derives from the rDNA genes of the 16S, 18S, 23S, 26S, and 45S subunits. We hypothesize that the high copy numbers of these genes and their propensity for concerted evolution will tend to homogenize these gene sequences. The intuition is that although there may be no bias to the gene conversion and duplicate processes, if one allele occurs in 99 of the 100 rDNA loci and the other, new, allele occurs at only one locus, most such new mutations will be removed by conversion or copy loss, lowering the overall rate of evolutionary change. Given that purifying selection is also acting on the rRNA genes, the combination of the two factors will tend to give rise to the observation of LIMEs. Note that the lack of synteny belies a true functional conservation in this case.
Although complex LIMEs were found in both animal and plant genomes, the two groups differ significantly in the types of LIMEs making up these sets. Specifically, the complex LIMEs in plants were found to be either the rRNA-associated or organellar insertions. Although LIMEs of these two types are also present in the animal LIMEs, they constitute only a small fraction of the animal UCEs: The remainder are the transposon, noncoding, and gene-coding LIMEs. These distinctions among LIMEs are further complicated by the fact that each element can occur in multiple copies, often across several chromosomes (Table 4 and SI Appendix, section S10). Although the multiple-copy complex LIMEs are found in similar proportions in animals and plants, the mechanisms behind them remain unclear in both groups of taxa. Some multiple-copy complex animal LIMEs can be explained by retrotransposition events involving snRNAs, whereas others can be the result of duplications. In plants, one might suspect that many of the multicopy LIMES were created by ancient genome duplications (i.e., paleopolyploidies). However, an analysis in rice and sorghum (SI Appendix, sections S11 and S12) suggests that at a minimum, polyploidy is not the only source of multiple-copy LIMEs. The occurrence of the multiple-copy repeat LIMEs, on the other hand, can likely be explained either by the action of the telomerase enzyme or by strand slippage during DNA replication. However, the mechanisms alone do not explain why only a small repertoire of 12 distinct repeat motifs in plants and 241 motifs in animals are found. In addition to the presence of the multiple-copy elements, the plant and animal genomes are similar in the structural organization of LIMEs and in the functions these elements are associated with. Thus, just as animal UCEs had already been shown to group in clusters (3), we find that plant LIMEs frequently formed compact clusters and that these clusters further formed closed networks with each other through shared LIMEs. Likewise, the functional annotation of plant and animal LIMEs is quite diverse, with LIMEs being found near rDNA genes, transposons, genes encoding enzymes, and noncoding DNA.
Table 4.
Numbers of single-copy and unique multiple-copy LIMEs
| Genome | Repetitive |
Nonrepetitive |
||
| Single-copy | Multiple-copy (average no. copies) | Single-copy | Multiple-copy (average no. copies) | |
| At | 993 | 692 (36.7) | 135 | 79 (2.2) |
| Gm | 11,826 | 11,053 (148.6) | 241 | 95 (54.8) |
| Os | 7,393 | 5,277 (65.5) | 628 | 212 (4.8) |
| Pt | 1,832 | 1,317 (18.2) | 61 | 1 (2.0) |
| Sb | 9,888 | 9,606 (159.7) | 678 | 114 (4.2) |
| Vv | 2,481 | 1,172 (19.1) | 126 | 9 (3.6) |
Additional copies of multiple-copy elements are not counted. Shown in brackets are average numbers of copies per unique multiple-copy element.
Exceptionally strong purifying selection has been proposed to underlie the extreme conservation of the complex animal LIMEs (1). However, this mechanism alone is insufficient to explain the more complete set of plant and animal LIMEs described here. For example, we have already invoked concerted evolution as a source of conservation among the ribosomal genes. In addition to contamination (for which we control), LIMEs might derive from other processes that deviate from strict vertical inheritance (an assumption of ultraconservation). We propose two other such alternate origins: horizontal transfer (from the organelle) and de novo manufacturing. However, even among the complex LIMEs, those elements that are mostly likely to be maintained by strong selection, plants and animals differ. The only complex plant LIMEs were in rRNA or the products of organellar insertion, whereas such elements constituted only a fraction of the complex animal LIMEs (SI Appendix, section S3). These large differences in the origins of the complex plant and animal LIMEs, as well as their 70-fold difference in numbers, suggest that the ultrapurifying selection seen in animal UCEs is essentially absent in plants, where UCEs, as classically defined, are rare to absent in all comparisons of both closely and distantly related taxa.
Materials and Methods
Information-Retrieval Method for Identification of LIMEs Between the Sets of Two or More Genomes.
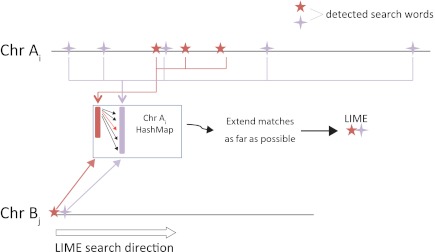
The original algorithm (35) was developed for “manual” searches of one-against-many genomes. That algorithm was designed to find exact matches to an input sequence and then rank the results according to input annotation terms. Identifying all-against-all exact matches in sets of plant and animal genomes required an extensive retooling of the algorithm, because the input sequences are entire chromosomes and the exact matches being sought are initially unknown. The approach described here extends the original algorithm to enable multiple genome-to-genome searches in a reasonably manageable time. In contrast, the running time of the original algorithm using a 48 central processing unit (CPU) cluster was expected to take 1 y for the set of six plant genomes and more than 3 y for the set of six animal genomes, and therefore was not feasible. The underlying information-retrieval methods in previous work and in this work do not depend on a sequence alignment; instead, they use a hash-mapping technique to identify exact matches between small sequence fragments and the entire chromosome sequence efficiently (Fig. 6). Each chromosome from each species is preprocessed into searchable hash tables (35). Each hash bin in a hash table includes every location of a unique 8-bp nucleotide sequence (key) within the chromosome. The hash bin(s) have been sorted based on the keys to facilitate retrieval of the correct hash bin for an input sequence of arbitrary length. The method can be applied to extract exact matches among an arbitrary number of genomes, by finding matches among all their subsets. The latter property will also guarantee finding all possible LIMEs, both syntenic and nonsyntenic, greater than or equal to a predefined length that are common to all n genomes. Alignment-based methods cannot make this guarantee because they require an initial alignment to pass a given threshold before a region is considered further.
Fig. 6.
Main step of the information-retrieval method for detecting identical sequences across multiple genomes. Identical sequences are detected between each pair of chromosomes (Chr), Ai and Bj, of two genomes, A and B. To find all LIMEs between a pair of chromosomes, the method first detects the matches of small (8 bp) search words using hash maps of substring locations, followed by extension of the matched regions to be at least 100 bp long.
The LIME detection algorithm is organized as follows. To initialize an input sequence (chromosome Ai from genome A in this example), we first partition the chromosome into nonoverlapping adjacent windows labeled W0, W1, …, Wx, where x = floor(|Ai|/M) and M = 45 is the window length. Choosing M = 45 allows for 45-fold fewer searches while still guaranteeing the desired result as discussed below. Chromosome Ai is traversed from W0 to Wx, and a small search word w, |w| = 8, is used to detect all locations with locally identical sequences (the specific choice of M and |w| values is made to ensure detection of all identical sequences of 100 bp or longer). Specifically, at each window Wk, (0 ≤ k < x), the first |w| bases are defined as
Word Wk(w) is then used as a key to search a hash map of substring locations in chromosome Bj of genome B (35). A result set, Rk, obtained from the search consists of a list of every location of Wk(w) within Bj. Every location of word Wk+1(w) is similarly retrieved, so that elements from its result set, Rk+1, can be compared with the elements from Rk. Proportionate locations within two adjacent windows, Wk and Wk+1, of chromosome Ai are always offset by M bases. If an exact match to window Wk is present in Bj, proportionate locations in Bj must also be offset by exactly M bases. All pairs of elements (a, b) from the two result sets, Rk and Rk+1, are identified, such that Rk+1[b] − Rk[a] = M. For the hits identified, each location in window Wk of Ai is compared with the corresponding location in Bj with a range from Rk[a] to Rk+1[b]. All windows between the two chromosomes A and B with the exact matches are recorded as identical. This is how matches are found without prior knowledge of their presence and without a previous sequence alignment. In addition, because matches can be greater than M bases, the identical windows are extended in both the upstream and downstream directions as far as possible until a mismatch is encountered. All matches greater than or equal to a predefined minimum length, min = 2M + |w| − 1, are recorded as LIMEs shared by A and B. The described algorithm guarantees finding all LIMEs of size 2M + |w| − 1 or longer, based on the fact that every subsequence s of Ai, |s| ≥ 2M + |w| −1, has at least two words that are the first words of adjacent windows in Ai and are subsequently used to search Bj.
Thus, the previous search algorithm (35) has been modified to perform additional processing of search results in such a way that still guarantees that all exact matches (≥100 bp) are found regardless of their location in either genome or prior knowledge of their presence. Furthermore, with a window size of M = 45, 45-fold fewer searches are required than by a simple brute force method, whereby every 8-bp sequence on one chromosome is used as input to search another chromosome. It took, on average, 5.52 h per chromosome-to-chromosome search between human and rat (roughly 5,688,000 individual searches) using a multicore Intel 64 architecture Xeon processor. By running 48 parallel processes with the same processors, the 504 (21 mouse × 24 human) pairs of chromosome-to-chromosome searches between human and rat took about 2.5 d. All the genome-to-genome searches in the animal set took about 3–4 wk, whereas the plant set was analyzed in just over 1 wk.
The obtained algorithm can be applied to extract LIMEs of an arbitrary number of genomes, by finding LIMEs of all genome subsets and using the fact that a LIME for genomes G1, G2, …, GN is also a LIME for all subsets of size (N − 1). The latter property guarantees finding all LIMEs that are common to all N genomes. Application of this method to retrieve the LIMEs for all possible subsets of the six plant genomes resulted in 2,917 pairwise chromosome comparisons. For the plant set, the most time-consuming step involved analysis of the soybean genome against five other genomes; it included processing 1,300 chromosome pairs and took ∼24 h on a 48-CPU parallel cluster. The complete list of plant LIMEs can be downloaded at http://korkinlab.org/datasets/limes/limes_data.html.
Synteny Analysis of LIMEs.
Genomic regions containing LIMEs were manually compared using CoGe’s Genome Evolution analysis (GEvo) tool (36) for high-resolution genomic comparisons. Synteny is inferred by identifying a collinear series of homologous gene pairs between two regions. With GEvo, genomic regions spanning 10 kb to 1 megabase were compared up and downstream of each LIME (spanning ∼20 genes); a minimum of 5 collinear independent (nontandemly duplicated) homologous genes were required in order for synteny to be positively identified.
Overlapping LIMEs.
The two initial categories of LIMEs (repeated motifs and complex sequence) in Fig. 1 are further subcategorized as either overlapping with other nearby elements or nonoverlapping. As expected, elements containing repeated motifs almost always overlap. However, there are 8,331 (0.08%) repeated-motif elements in the animal set and 285 (0.01%) elements in the plant set that do not overlap with other elements (all occurrences of the elements are considered when determining overlap). For complex sequences, 80.43% (7,068 of 8,788) of plant elements and 91.56% (14,106,012 of 15,405,907) of animal elements overlap with neighboring elements. This leaves 1,720 locations within the set of plant genomes that have nonoverlapping complex elements and 1,299,895 such locations in the animal set. Among the plants, cottonwood has the fewest such locations at 12 and rice has the most at 671, and among the animals, chicken has the fewest such locations at 1,879 and human has the most at 575,135.
Validation of the Framework on UCEs Between the Human, Mouse, and Rat.
Using the recent human, mouse, and rat genome builds (NCBI 36.1, 37.1, and 4.1, respectively), we identified 503 unique complex elements ≥200 bp in length. If one includes subsequences of these elements that map to distinct locations in at least one genome, the number rises to 619 elements. For comparison, Bejerano et al. (3) identified 481 UCEs ≥200 bp in length in 2004, using builds 34 (NCBI; human), 30 (NCBI; mouse) and 3.1 (Baylor Human Genome Sequencing Center; rat). Our reanalysis of these three builds identified 493 UCEs ≥200 bp in length. It also identified 405 unique nonsyntenic repetitive LIMEs (≥200 bp) that all consist of AT repeats located on human chromosomes 3 and 19; on mouse chromosomes 4, 8, and X; and on every rat chromosome except chromosome 19. Excluding these repetitive elements, there are 9 unique complex elements (and 3 subsequences) present in the more recent 2004 builds (SI Appendix, Table S1).
Statistical Model of Repeat Motif Common Between Six Plant Genomes.
The set of all distinct motifs forming the tandem repeat sequences was determined for each of the six genomes using Tandem Repeat Finder (37). Two motifs are defined as distinct if they are neither reverse complementary nor form highly similar repeat sequences (these sequences differ only in the N- and C-terminal tails of the first and/or last motifs in each tandem repeat sequence). For example, motifs TATA and ATAT are not distinct. There are 1,699 possible distinct motifs with a length of 2–7 bp; we found 228 motifs with a length of 2–7 bp in Arabidopsis, 576 in cottonwood, 550 in grape, 493 in rice, 680 in soybean, and 552 in sorghum. Pairs of plant genomes were found to share hundreds of the motifs. For instance, there were 365 motifs (2–7 bp) in common between rice and sorghum. However, only between 2 and 11 motifs shared by a pair of genomes were found to contribute to the repetitive LIMEs (Table 3). Moreover, we found that only 12 different motifs were used in total by all repetitive LIMEs shared between two or more plant genomes (Table 2). To estimate the probability of getting such a small set of common motifs by chance, a simple statistical model was used. For N genomes, we randomly selected 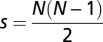 independent samples of distinct motifs from the population of 1,699 possible motifs. Each sample corresponds to an overlap of the short motifs between a pair of genomes occurring by chance. The total number of distinct motifs shared by all the samples is the size of a set
independent samples of distinct motifs from the population of 1,699 possible motifs. Each sample corresponds to an overlap of the short motifs between a pair of genomes occurring by chance. The total number of distinct motifs shared by all the samples is the size of a set 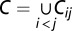 , where Cij is the sample of motifs common between genomes i and j. We then estimated the probability that the size of C is equal to i, P(|C| = i). Although, to the best of our knowledge, this problem is still open for a set of samples of arbitrary sizes, a simpler problem, where all s samples are of the same size, k, has been solved recently (38), with the probability determined:
, where Cij is the sample of motifs common between genomes i and j. We then estimated the probability that the size of C is equal to i, P(|C| = i). Although, to the best of our knowledge, this problem is still open for a set of samples of arbitrary sizes, a simpler problem, where all s samples are of the same size, k, has been solved recently (38), with the probability determined:
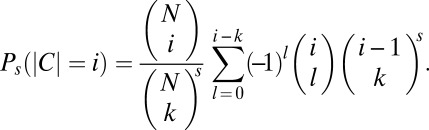 |
We use the result from that simpler problem to estimate the upper bound of P(|C| = i), where i = 12. Because only 1 of 15 pairs of genomes shares 2 motifs, and the remaining 14 pairs share between 3 and 11 motifs each, the upper bound of P(|C| = i) was estimated using the union set of s = 15 samples, each of size k = 3 [note that the probability, Ps(|C| = i), is decreasing with the increasing value of k from 3 to 11]. We found that the probability that the union set of 15 motif samples, each containing 3 distinct motifs, consists of 12 distinct motifs is smaller than 3 × 10−69.
Supplementary Material
Acknowledgments
We thank S. Kolennikov and S. Chakraborty for discussions and D. Xu for feedback on the manuscript. J.R. is supported by the National Institutes of Health. E.L., J.C.P., M.F., C.-R.S., and D.K. are supported by the US National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C-t.W. is a guest editor invited by the Editorial Board.
See Author Summary on page 7150 (volume 109, number 19).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121356109/-/DCSupplemental.
References
- 1.Katzman S, et al. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- 2.Stephen S, Pheasant M, Makunin IV, Mattick JS. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol Biol Evol. 2008;25:402–408. doi: 10.1093/molbev/msm268. [DOI] [PubMed] [Google Scholar]
- 3.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 4.Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- 5.Glazov EA, Pheasant M, McGraw EA, Bejerano G, Mattick JS. Ultraconserved elements in insect genomes: A highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005;15:800–808. doi: 10.1101/gr.3545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 8.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 9.Paparidis Z, et al. Ultraconserved non-coding sequence element controls a subset of spatiotemporal GLI3 expression. Dev Growth Differ. 2007;49:543–553. doi: 10.1111/j.1440-169X.2007.00954.x. [DOI] [PubMed] [Google Scholar]
- 10.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 13.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 15.Sandelin A, et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kryukov GV, Schmidt S, Sunyaev S. Small fitness effect of mutations in highly conserved non-coding regions. Hum Mol Genet. 2005;14:2221–2229. doi: 10.1093/hmg/ddi226. [DOI] [PubMed] [Google Scholar]
- 17.Keightley PD, Lercher MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brudno M, et al. Automated whole-genome multiple alignment of rat, mouse, and human. Genome Res. 2004;14:685–692. doi: 10.1101/gr.2067704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson AH, et al. Comparative genomics of plant chromosomes. Plant Cell. 2000;12:1523–1540. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- 21.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2):135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Freeling M, Subramaniam S. Conserved noncoding sequences (CNSs) in higher plants. Curr Opin Plant Biol. 2009;12(2):126–132. doi: 10.1016/j.pbi.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Zheng WX, Zhang CT. Ultraconserved elements between the genomes of the plants Arabidopsis thaliana and rice. J Biomol Struct Dyn. 2008;26(1):1–8. doi: 10.1080/07391102.2008.10507218. [DOI] [PubMed] [Google Scholar]
- 24.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33(Database issue):D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008;36(Database issue):D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyons E, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szostak JW, Blackburn EH. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982;29:245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- 28.Hancock JM. Microsatellites and other simple sequences: Genomic context and mutational mechanisms. In: Goldstein DB, Schlötterer C, editors. Microsatellites. Evolution and Applications. New York: Oxford Univy Press; 1999. pp. 1–9. [Google Scholar]
- 29.Adams SP, et al. Loss and recovery of Arabidopsis-type telomere repeat sequences 5′-(TTTAGGG)(n)-3′ in the evolution of a major radiation of flowering plants. Proc Biol Sci. 2001;268:1541–1546. doi: 10.1098/rspb.2001.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangwan I, O’Brian MR. Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. Plant Physiol. 2002;129:1788–1794. doi: 10.1104/pp.002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffor MC, Vodkin LO, Singh RJ, Hymowitz T. Fluorescent in situ hybridization to soybean metaphase chromosomes. Plant Mol Biol. 1991;17:101–109. doi: 10.1007/BF00036810. [DOI] [PubMed] [Google Scholar]
- 32.Smit AFA, Hubley R, Green P. Seattle, WA: Institute for Systems Biology; 1996–2004. RepeatMasker Open-3.0. Available at www.repeatmasker.org, accessed March 7, 2012. [Google Scholar]
- 33.Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 34.Galtier N. The intriguing evolutionary dynamics of plant mitochondrial DNA. BMC Biol. 2011;9:61. doi: 10.1186/1741-7007-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reneker J, Shyu CR. Refined repetitive sequence searches utilizing a fast hash function and cross species information retrievals. BMC Bioinformatics. 2005;6:111. doi: 10.1186/1471-2105-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 37.Gelfand Y, Rodriguez A, Benson G. TRDB—The Tandem Repeats Database. Nucleic Acids Res. 2007;35(Database issue):D80–D87. doi: 10.1093/nar/gkl1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barot M, de la Peña J. Estimating the size of a union of random subsets of fixed cardinality. Elemente der Mathematik. 2001;56(4):163–169. [Google Scholar]