Abstract
Plants must effectively defend against biotic and abiotic stresses to survive in nature. However, this defense is costly and is often accompanied by significant growth inhibition. How plants coordinate the fluctuating growth-defense dynamics is not well understood and remains a fundamental question. Jasmonate (JA) and gibberellic acid (GA) are important plant hormones that mediate defense and growth, respectively. Binding of bioactive JA or GA ligands to cognate receptors leads to proteasome-dependent degradation of specific transcriptional repressors (the JAZ or DELLA family of proteins), which, at the resting state, represses cognate transcription factors involved in defense (e.g., MYCs) or growth [e.g. phytochrome interacting factors (PIFs)]. In this study, we found that the coi1 JA receptor mutants of rice (a domesticated monocot crop) and Arabidopsis (a model dicot plant) both exhibit hallmark phenotypes of GA-hypersensitive mutants. JA delays GA-mediated DELLA protein degradation, and the della mutant is less sensitive to JA for growth inhibition. Overexpression of a selected group of JAZ repressors in Arabidopsis plants partially phenocopies GA-associated phenotypes of the coi1 mutant, and JAZ9 inhibits RGA (a DELLA protein) interaction with transcription factor PIF3. Importantly, the pif quadruple (pifq) mutant no longer responds to JA-induced growth inhibition, and overexpression of PIF3 could partially overcome JA-induced growth inhibition. Thus, a molecular cascade involving the COI1–JAZ–DELLA–PIF signaling module, by which angiosperm plants prioritize JA-mediated defense over growth, has been elucidated.
Keywords: disease resistance, plant growth, plant immunity, light response, plant defense
Sessile plants have evolved a dynamic regulatory network to adapt to the daily and seasonally fluctuating environment. Jasmonate (JA) is a lipid-derived plant hormone that regulates developmental processes, including pollen development, tendril coiling, fruit ripening and senescence, as well as response to biotic and abiotic stress (1–3). The F-box protein coronatine insensitive 1 (COI1), a component of the SCF E3 ubiquitin ligase, has been identified as a principal component of a receptor of JA in Arabidopsis and other plants (4–7), and the JA ZIM-domain (JAZ) family proteins are key regulators of JA signaling that repress transcription of JA-responsive genes through interaction with transcription factors, such as MYC2 (8–10). This transcriptional repression requires novel interactor of JAZ (NINJA) and TOPLESS corepressor proteins (11). Bioactive JA, the jasmonoyl-isoleucine conjugate, promotes physical interaction between COI1 and JAZ proteins that results in degradation of JAZs by the 26S proteasome, leading to initiation of JA responses (9, 10). Therefore, the SCFCOI1–JAZ protein complex acts as a core site of JA perception. As a regulatory loop, JA also activates JAZ gene transcription, leading to the down-regulation of JA action and the dynamic nature of JA response (12).
Gibberellic acids (GAs) are plant growth-promoting hormones that play important roles in diverse aspects of plant growth and development, such as stem elongation, leaf expansion, flowering, seed development, and germination (13–17). The DELLA family proteins are key regulators of GA signaling that repress transcription of GA-responsive genes through interaction with growth-promoting transcription factors, such as phytochrome interacting factors (PIFs) (18, 19). Bioactive GAs bind to the GA insensitive dwarf1 (GID1) receptor, which, in turn, interacts with DELLA proteins (five members in Arabidopsis and a single SLR1 protein in rice) (16, 20). The GA-GID1–bound DELLA/SLR1 proteins are recognized by an F-box protein (SLY1 in Arabidopsis and GID2 in rice), resulting in proteasome-dependent degradation of DELLA/SLR1 repressors. Degradation of DELLA/SLR1 proteins derepresses the transcriptional activities of downstream transcription factors, including PIFs (16).
Activation of JA defense signaling is known to severely restrict plant growth, representing a prominent example of growth–defense tradeoff in plants. There have been several reports of crosstalk between JA and GA signaling pathways in Arabidopsis, mostly documenting the antagonistic effect of GA on JA signaling (1). A quadruple della mutant (which lacks four of the five Arabidopsis DELLA proteins) was shown to be partially insensitive to gene induction by JA, whereas the constitutively active dominant DELLA mutant gai was found to be sensitized for JA-responsive gene induction, implicating DELLAs in JA signaling and/or perception (21). Cheng et al. (22) found that GA promotes JA biosynthesis, thereby inducing the expression of MYB21, MYB24, and MYB57 to promote stamen filament elongation. Most recently, it has been shown that DELLA repressors promote JA signaling through physically interacting with JAZ1 (23), suggesting a mechanism for GA-mediated down-regulation of JA defense responses. However, it remains unknown how JA could inhibit plant growth. In this study, through analysis of rice and Arabidopsis, we have elucidated a molecular cascade by which JA antagonizes GA signaling that explains how monocot and dicot plants prioritize JA defense over growth.
Results
Knockdown of Rice COI1 Genes Decreases JA Response.
As a model monocot, rice (Oryza sativa L.) has been commonly used to study hormone signaling as well as defense responses in cereal crops. A number of studies have been conducted to dissect JA signaling and function in defense response and developmental process in rice (24–27). However, the role of COI1 in mediating JA signal perception is still unclear in rice. The rice genome contains two closely related COI1 genes, OsCOI1a (Os01g0853400) and OsCOI1b (Os05g0449500), which share 83% and 82% sequence identity at the DNA and protein levels, respectively. To determine the function of COI1 in rice, a double-strand RNAi construct containing the conserved sequence of OsCOI1a and OsCOI1b (Fig. S1A) was introduced into the model variety Nipponbare. More than 20 independent RNAi lines were produced (Fig. S1B). The transcript levels of both OsCOI1a and OsCOI1b were significantly reduced in these RNAi lines as detected by RNA blot and quantitative RT-PCR (qRT-PCR) analyses (Fig. 1 A and B), indicating that OsCOI1 expression was effectively knocked down by RNAi.
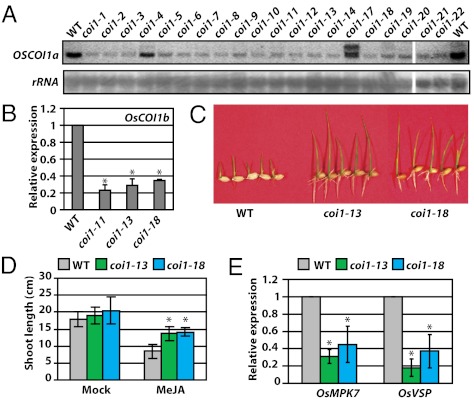
Fig. 1.
Generation of OsCOI1 RNAi transgenic rice with reduced JA sensitivity. (A) Suppression of OsCOI1a expression in transgenic RNAi lines as shown by RNA blot analysis. A fragment of 825 to 1,244 nt of OsCOI1 was used as the probe, and 25S rRNA was used as the loading control. (B) Suppression of OsCOI1b expression in transgenic RNAi lines as shown by qRT-PCR. (C and D) The representative RNAi lines coi1-13 and coi1-18 show reduced sensitivity to MeJA treatment. Seedlings were grown in one-half MS medium with 0.6% agar supplemented with or without 20 μM MeJA. Pictures were taken at day 7 and shoot length was measured at day 12. Data shown in D are the means from 12 plants. Error bars represent SD. Asterisks indicate significant difference between WT and coi1 mutants based on Student's t test (P < 0.01). (E) Reduced expression of JA-response genes OsMPK7 and OsVSP in coi1-13 and -18 plants revealed by qRT-PCR. Data shown in C and E are the means from two independent experiments. Error bars represent SD. Asterisks indicate the significant difference between WT and coi1 mutants based on Student's t test (P < 0.01).
JA sensitivity was investigated in two stable RNAi lines, coi1-13 and coi1-18, that carry a single copy of the transgene. In this study, we used methyl JA (MeJA), which is converted to JA and then to bioactive jasmonoyl-isoleucine in plants (3), to treat rice and Arabidopsis. As expected, the transgenic lines were much less sensitive to MeJA than the WT plants in the growth-inhibition assay (Fig. 1 C and D). Furthermore, JA-responsive genes such as OsVSP and OsMPK7 exhibited reduced expression in the OsCOI1-RNAi plants (Fig. 1E). These results demonstrate that OsCOI1 is required for JA signaling in rice.
OsCOI1-RNAi Plants Display Phenotypes Similar to Those of GA Overproduction.
Intriguingly, when grown in the greenhouse or the paddy field, the coi1-13 and coi1-18 plants consistently showed increased plant height in comparison with the WT plants, a phenotype that mainly resulted from elongated internodes (Fig. 2 A–C). This elongated phenotype of the OsCOI1-RNAi plants is similar to that of the rice eui1 mutants, which contain a loss-of-function mutation in the P450 monooxygenase CYP714D protein that catalyzes the 16α,17-epoxidation reaction of GA deactivation (28, 29). Another similarity between the rice coi1 lines and the eui1 mutants was that they produce longer grains than the WT plants (Fig. S2). Plant growth is controlled by cell division and cell elongation. Cell length in the uppermost internode of the coi1-18 plants was found to be significantly increased in comparison with that of the WT plants (Fig. 2 D and E), indicating that increased plant height is mainly caused by cell elongation instead of cell division, a GA-related feature (30). Whole transcriptomic analysis of coi1-13 plants revealed that basal expression of several GA-related genes, including GA2ox, GA20ox, and OsWRKY71, was altered in the OsCOI1-RNAi rice (Fig. S1C).
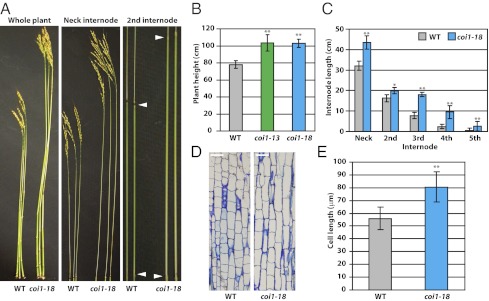
Fig. 2.
Morphological phenotypes of coi1-18 plants. (A) Images of WT (Nipponbare) and coi1-18 plants show plant heights and internode lengths. (B) Quantification of heights of the WT, coi1-13, and coi1-18 plants. More than 30 plants of each line were analyzed. The difference between the control and transgenic plants is significant (**P < 0.001, Student's t test). (C) Lengths of individual internodes in the WT and coi1-18 plants. Each internode of coi1-18 was longer than the counterpart of Nipponbare (*P < 0.01 and **P < 0.001, Student's t test). (D) Microscopic sections of the elongating zone of the uppermost internodes from Nipponbare and coi1-18 grown in the isolated paddy field. (Scale bar, 40 μm.) (E) Cell lengths at the base of the elongating zone of the uppermost internodes in the WT and coi1-18 plants. The cell of coi1-18 is much longer than that of Nipponbare (**P < 0.001, Student's t test).
GA signaling regulates diverse biological processes, including α-amylase release, during seed germination in rice (31). We examined the effect of MeJA on the GA induction of α-amylase activity in embryoless seeds, and found that coincubation with MeJA strongly suppressed the GA induction of α-amylase (Fig. S3A). Consequently, the seed germination rate was significantly decreased with MeJA treatment (Fig. S3B). In contrast, the seeds of coi1-13 and coi1-18 had significantly higher levels of α-amylase activity and germinated at a much quicker rate than those of the WT (Fig. S3 C and D). These results indicated that the modulation of GA signaling by JA occurs not only in plant growth but also during seed germination. Taken together, these results suggest that interruption of JA signaling in the coi1 mutants augments the GA signal pathway in rice.
OsCOI1-RNAi Plants Are Hypersensitive to GA and Hyposensitive to GA Biosynthesis Inhibitor.
To further confirm the alteration of GA signaling in the OsCOI1-RNAi rice plants, the growth of rice seedlings was examined in semisolid one-half Murashige–Skoog (MS) medium supplemented with different concentrations of GA3. The coi1-18 plants exhibited more sensitivity to exogenous application of GA3 in comparison with the WT plants (Fig. S4A). Consistent with their increased GA sensitivity, the coi1-18 plants exhibited reduced sensitivity to the GA biosynthesis inhibitor uniconazole compared with the WT plants (Fig. S4B). Therefore, the OsCOI1-RNAi plants were hypersensitive to exogenous GA and hyposensitive to GA biosynthesis inhibitor.
To examine the effect of the OsCOI1 silencing on the endogenous GA levels, the bioactive GAs, GA1 and GA4, were measured in the RNAi and WT plants. In contrast to the eui1 mutants, which accumulate extremely high levels (30- to 100-fold) of GA1 and GA4 (28), the coi1-18 plants accumulated only slightly higher levels of GA4 (3.8 fold) than the WT plants in the elongating uppermost internode (Table S1). The levels of GA1 were similar in coi1-18 and WT plants. The modest change in the GA4 level may be correlated to the differential expression of several GA metabolism genes in OsCOI1-RNAi plants (Fig. S1C). However, overall the defect in JA signaling does not appear to dramatically affect bioactive GA biosynthesis/accumulation in rice, even though OsCOI1-RNAi plants exhibited eui1-like growth phenotypes. Consistent with this observation, no significant difference in the transcript levels of EUI1 was found between coi1-18 and the WT (Fig. S5). Taken together, these results strongly suggest that the increased plant height and cellular elongation of the OsCOI1-RNAi plants is mainly a result of the hypersensitivity to GA.
Elongation of OsCOI1-RNAi Plants Is Inhibited by Attenuating GA Signaling.
EUI1 overexpression resulted in a series of GA-deficient phenotypes, with drastic reduction of the bioactive GAs (28) and accumulation or stabilization of the DELLA protein SLR1 (32). We crossed the EUI1-overexpression plants (Eui1-OX) to the coi1-18 plant (Fig. 3D). The homozygous Eui1-OX/coi1-18 plants showed greatly reduced plant height (Fig. 3 A–C) and reduced cell size in the uppermost internode (Fig. 3 E and F), similar to Eui1-OX plants. In addition, the longer grain phenotype of the coi1-18 plants was also reverted to that of the WT (Fig. 3 G and H). We also crossed coi1-18 with the GA receptor gid1-1 mutant. Again, the gid1-1/coi1-18 double mutant exhibited a dwarf phenotype like gid1-1 (Fig. S6). This result demonstrated that the GA receptor gene GID1 is required for the function of OsCOI1 in the GA pathway. These results suggest that the OsCOI1-RNAi morphology is dependent on the GA signaling pathway.
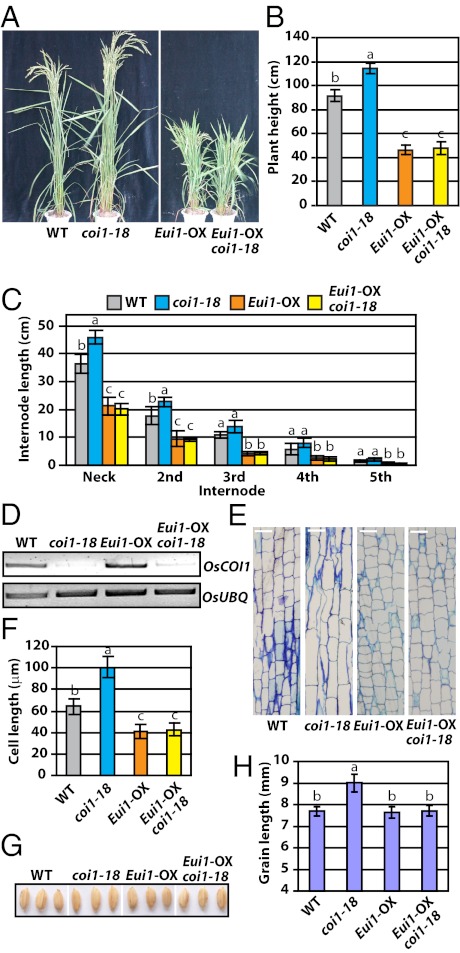
Fig. 3.
The GA-deficiency mutation reverses the phenotype of coi1-18 plants. (A) Morphological phenotype of Eui1-OX/coi1-18. (B) The average plant height of WT, coi1-18, Eui1-OX, and Eui1-OX/coi1-18 plants. (C) The length of each internode of coi1-18 decreased in the Eui1-OX/coi1-18 plants. (D) Expression of OsCOI1 in Eui1-OX/coi1-18. (E and F) Cell lengths at the bases of the uppermost internodes in coi1-18, Eui1-OX/coi1-18, and Eui1-OX plants. (Scale bar, 40 μm.) (G and H) Grain lengths of coi1-18, Eui1-OX/coi1-18, and Eui1-OX plants. Letters on the columns in B, C, F, and H indicate significant differences determined by Tukey–Kramer multiple comparison test (P < 0.05).
JA Antagonizes GA Signaling Pathway by Delaying GA-Induced SLR1 Degradation.
We next wanted to determine the level of SLR1, a rice DELLA protein that functions as a key repressor of the GA signaling pathway (33). However, it was difficult to detect SLR1 in the coi1-18 or the WT plant because of the low basal level of SLR1. Instead, we found that SLR1 accumulated in the Eui1-OX/coi1-18 plant at a level comparable to the Eui1-OX plant (Fig. 4A). Therefore, the degradation dynamics of SLR1 protein was examined in Eui1-OX/coi1-18 in the presence of exogenous GA3. When transferred into the medium with 100 μM GA3 for 30 min, SLR1 was significantly degraded in the Eui1-OX/coi1-18 plants, whereas it was degraded only slightly even after 2 to 3 h with GA treatment in the Eui1-OX plants (Fig. 4A). This result suggested that JA signaling antagonizes GA-mediated reduction of the DELLA protein.
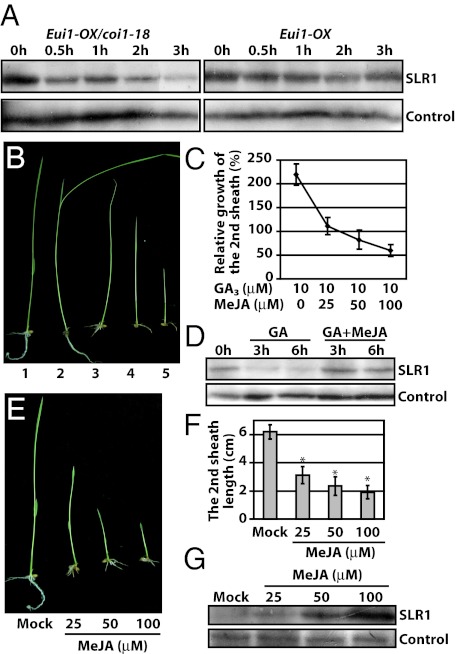
Fig. 4.
Levels of the rice DELLA protein SLR1 and the antagonistic effect of MeJA on GA-mediated plant growth. (A) The GA-mediated degradation of the DELLA protein SLR1 was promoted in the coi1-18 plants. The 10-d-old seedlings grown on one-half MS plates with 0.6% agar were transferred to liquid one-half MS medium with 100 μM GA3, and the SLR1 protein was detected with an SLR1 antibody at the indicated time points. (B and C) MeJA inhibited GA-induced shoot elongation in WT plants: 1, control plant; and 2 to 5, representative plants treated with GA3 and MeJA in the same order as in Fig. 4C. The relative growth is indicated by the length of second leaf sheath after being treated with 10 μM GA3 and various concentrations of MeJA. (D) MeJA delayed SLR1 degradation induced by GA3. The concentration of GA3 and MeJA used were 10 μM and 100 μM, respectively. (E and F) MeJA inhibited WT rice seedling growth and second sheath elongation in a dose-dependent pattern. Asterisks indicate significant difference between mock and treatments (P < 0.01, Student's t test). (G) MeJA treatment promoted the accumulation of SLR1 in a dose-dependent manner in WT plants. The rice plants were grown on one-half MS plates with 0.6% agar supplemented with MeJA (final concentration indicated on Top).
Having shown that turning down the JA pathway could increase the GA signaling output, we next examined the possibility that turning on JA signaling might antagonize the GA signaling pathway. Indeed, whereas growing WT rice seedlings in the presence of 10 μM GA3 leads to elongation of the second leaf sheath by approximately 120%, addition of MeJA greatly reduced the GA-triggered elongation in a dose-dependent manner (Fig. 4 B and C). Furthermore, in the presence of MeJA, GA-induced SLR1 degradation was significantly inhibited as long as 6 h after treatment (Fig. 4D). Consistent with this observation, seedling growth was inhibited by MeJA in a dose-dependent manner, with decreased shoot length (Fig. 4 E and F). In addition, the SLR1 protein accumulated in the plants grown in the medium supplemented with MeJA (Fig. 4G), whereas no change was observed in the SLR1 transcript level (Fig. S7). Finally, the growth inhibition effect of MeJA was significantly suppressed in the SLR1 loss-of-function mutant slr1 in comparison with the WT (Fig. 5), further supporting that JA-mediated growth inhibition is in part dependent on the DELLA repressor. These results collectively demonstrated that JA represses rice growth through antagonizing GA signaling at least partly via affecting the level of the DELLA protein SLR1.
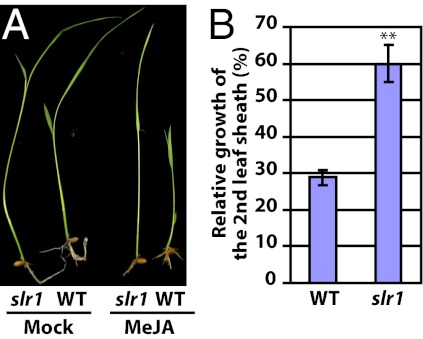
Fig. 5.
The rice slr1 mutant is insensitive to MeJA. (A) WT and slr1 seedling were grown on one-half MS plates with 0.6% agar supplemented with or without 25 μM MeJA. (B) Relative growth of slr1 and WT plants is indicated by percentages of the second leaf sheath lengths with MeJA treatment compared with those without MeJA treatment (**P < 0.001, Student's t test).
Arabidopsis coi1 Mutant Also Exhibits GA-Related Phenotypes.
The significant GA hypersensitivity phenotypes of the OsCOI1 RNAi lines was somewhat unexpected because such phenotypes were not previously reported for the Arabidopsis coi1 mutants (6, 34). We therefore looked for GA-related phenotypes in the Arabidopsis coi1 mutant plants. We found that Arabidopsis coi1 plants have several robust phenotypes that resemble GA hypersensitivity, including longer hypocotyls and petioles under low-intensity light condition and early flowering (Fig. 6). Moreover, transgenic overexpression of JAZ repressors, which mimics the effect of coi1 mutations, was found to phenocopy the coi1 mutant. Among eight AtJAZ genes (AtJAZ1, 3, 4, 5, 6, 9, 10, and 11) we were able to overexpress, AtJAZ1, 3, 4, 9, 10, and 11 produced the early flowering phenotype, but, interestingly, AtJAZ5 and 6 overexpression plants did not (Fig. S8). We also checked AtJAZ9 overexpression plants for GA-mediated germination response and found that they were more resistant to the GA biosynthesis inhibitor paclobutrazol (Fig. S9), which is a GA-hypersensitivity phenotype.
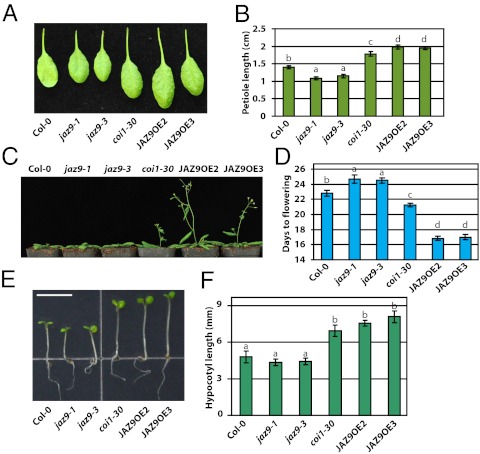
Fig. 6.
GA-associated phenotypes of the Arabidopsis coi1 mutant and JAZ9 overexpression plants. Image (A) and quantification (B) of petioles of coi1-30 and JAZ9 overexpression plants. The plants were grown in a long-day growth chamber (16 h 120 μmol m−2⋅s−1 light/8 h dark, 22 °C/18 °C). The third true leaves of 21-d-old plants were imaged, and their petiole lengths were measured. Image of 28-d-old plants (C) and quantification (D) of early flowering phenotypes of JAZ9 overexpression lines and coi1-30 plants. The plants were grown under the same conditions as in A. Image (E) and quantification (F) of hypocotyls of coi1-30 and JAZ9 overexpression plants when grown under 10 μmol m−2⋅s−1 continuous white light at 22 °C for 6 d. (Scale bar, 5 mm.) Data shown in B, D, and F are the means from 12 plants. Error bars represent SD. Letters on columns indicate significant differences (P < 0.05, Tukey–Kramer multiple comparison test).
Next, we investigated whether, like in rice, JA could antagonize GA signaling by affecting the level of DELLA proteins in Arabidopsis. A well characterized DELLA protein, RGA, was monitored in these experiments. Consistent with what was observed in rice, when Arabidopsis seedlings were continuously treated with JA, the RGA protein level increased, whereas the RGA transcript level did not change (Fig. 7). As internal controls, JA induced degradation of JAZ9 and expression of a known JA-responsive gene, AOS (Fig. 7). Taken together, these results collectively show that disruption of JA perception and signaling affects GA phenotypes in Arabidopsis and that JA negatively regulates GA responses through modulating the level of DELLA repressors in rice and Arabidopsis.
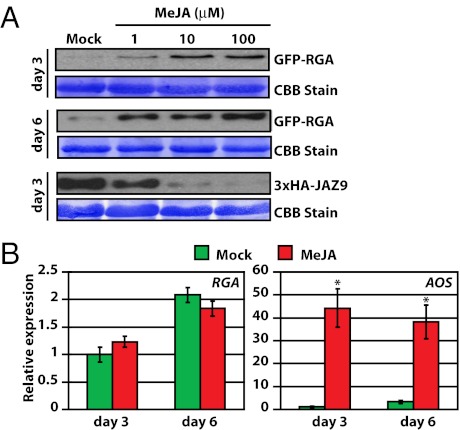
Fig. 7.
MeJA increases the level of the Arabidopsis DELLA protein RGA. Seedlings were grown under 10 μmol m−2⋅s−1 continuous white light at 22 °C. (A) Increased accumulation of the RGA protein in Arabidopsis seedlings treated with different concentrations of MeJA for 6 d. RGA and JAZ9 (positive control) were detected by anti-GFP and anti-HA antibody, respectively. (B) MeJA treatment does not affect the RGA transcript level in Arabidopsis. Arabidopsis seedlings were treated with 0.1% ethanol (mock) or 100 μM MeJA for 6 d. Total RNA was purified and used for qRT-PCR analysis. Data shown are the means of three biological replicates. Error bars represent SD. JA-inducible expression of AOS was used as a positive control for JA treatment. Asterisks indicate significant difference between mock and MeJA treatment (P < 0.01, Student's t test).
JAZ Repressors Directly Interfere with DELLA–PIF Interaction.
DELLA proteins have been shown to interact and repress growth-promoting transcription factors, such as PIFs in Arabidopsis (18, 19). Interestingly, the DELLA proteins were recently found to also interact with AtJAZ1 in Arabidopsis (23). By using multiple methods, we independently observed multiple JAZ–DELLA interactions in plant or yeast, and found that, in the case of the JAZ9–GAI interaction, the N terminus of JAZ9 and the GRAS domain of GAI are important for interaction in yeast* (Fig. 8 A and B and Fig. S10). Although Hou et al. focused their study on how GA antagonizes JA signaling through the AtJAZ1–DELLA interaction, we noticed a striking correlation between the ability of AtJAZ overexpression to confer early flowering (Fig. S8) and physical interaction with DELLA proteins: AtJAZ1, 3, 4, 9, 10, and 11, but not AtJAZ5 and 6, interacted with DELLA proteins and produced the early flowering phenotype (Fig. S10). We therefore investigated the intriguing possibility that AtJAZ repressors may impede the DELLA–PIF interaction. We first confirmed the interaction between the GRAS domain of RGA and PIF3 in a yeast two-hybrid assay (Fig. 8C). We found that expression of AtJAZ9 inhibited the RGA–PIF3 interaction in yeast three-hybrid assays (Fig. 8C), without affecting the protein levels of RGA and PIF3 (Fig. 8D). The effect of JAZ9 on the RGA-PIF3 interaction could also be observed in plant cells by using Nicotiana tabacum-based transient expression assays. Again, RGA interacted with PIF3 in this system; however, coexpression with AtJAZ9 could effectively prevent RGA–PIF3 interaction (Fig. 8E).
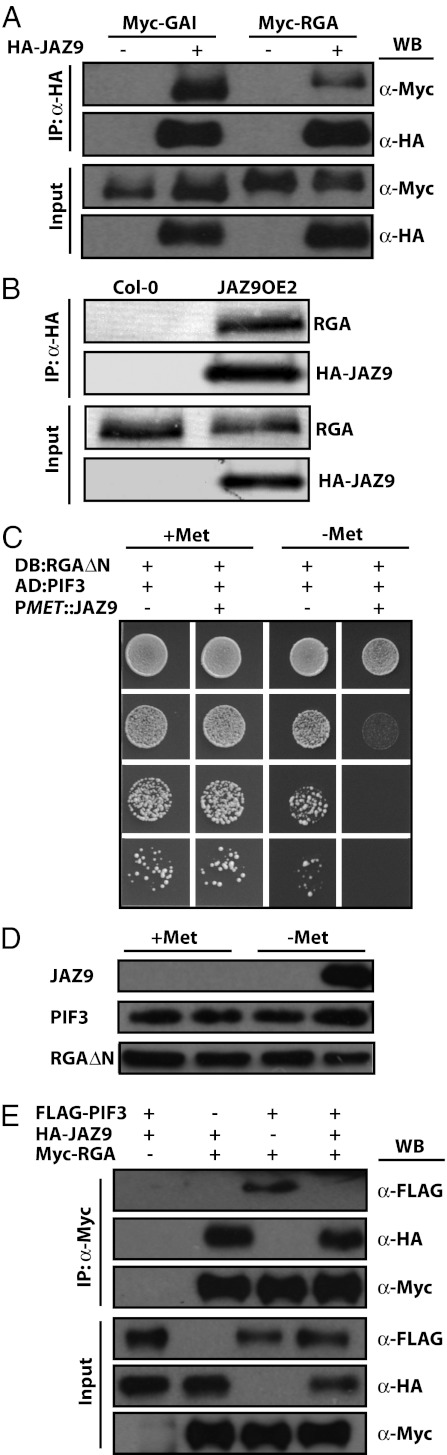
Fig. 8.
JAZ9 interferes with the interaction between RGA and PIF3. (A) 3xHA-JAZ9 interacts with 9xMyc-GAI or 9xMyc-RGA protein when expressed transiently in N. tabacum leaves. Protein extracts were immunoprecipitated with an anti-HA antibody and analyzed by Western blot with an anti-Myc antibody. (B) JAZ9 interacts with RGA protein in 3xHA-JAZ9 transgenic Arabidopsis plants. Protein extracts from 12-d-old seedlings were immunoprecipitated with an anti-HA antibody and analyzed by Western blot with an anti-RGA antibody. (C) JAZ9 inhibits the interaction between RGA and PIF3 in yeast. The activity of the reporter gene HIS3, which indicates the interaction between RGA and PIF3, is greatly reduced [indicated by reduced growth on medium lacking histidine (-His)] in the presence of JAZ9 [induced in medium without methionine (-Met)]. (D) Western blot shows that all proteins analyzed in the Y3H assay (C) were expressed as expected. (E) 3xHA-JAZ9 interferes with the interaction between 9xMyc-RGA and 3xFLAG-PIF3 when transiently expressed in N. tabacum leaves. Protein extracts were immunoprecipitated with an anti-Myc antibody and analyzed by Western blot with anti-FLAG, anti-HA, or anti-Myc antibody.
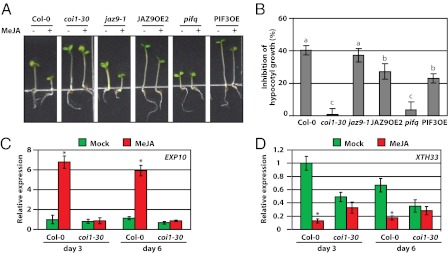
Our results suggest that JAZ-mediated interference with the DELLA–PIF interaction is a key mechanism that modulates plant growth. To obtain genetic evidence for or against this possibility, we analyzed the responses of pif mutants and PIF3 overexpression plants to JA treatment. We found that the pif quadruple mutant (pifq) grew more slowly compared with WT plants, and were no longer able to respond to JA-mediated inhibition of hypocotyl growth (Fig. 9 A and B). This result suggests that PIFs are likely the main, if not the only, growth-promoting transcription factors that are targeted by JA-induced growth inhibition. More interestingly, overexpression of PIF3 alone was sufficient to partially overcome JA-induced inhibition of hypocotyl growth (Fig. 9 A and B). Our results contrast with those from a recent report that showed that PIF4 transgenic overexpression plants exhibited enhanced JA-induced growth inhibition (23). Also, although the della quadruple mutant (dellaq) showed only a slightly lower sensitivity to JA-mediated inhibition of hypocotyl growth in the study by Hou et al. (23), under our experimental conditions, the della quintuple mutant (gai-t6/rga-t2/rgl1-1/rgl2-1/rgl3-1) was almost completely insensitive to JA-induced hypocotyl inhibition (Fig. S11). Finally, we found that expression of two examined DELLA/PIF-regulated genes—expansin 10 (EXP10, At1G26770) and xyloglucan:xyloglucosyl transferase 33 (XTH33, At1G10550) (35–37)—was altered in a predicted manner upon JA treatment: JA up-regulates the expression of EXP10, which is down-regulated by PIFs, whereas JA down-regulated XTH33, which is up-regulated by PIFs (Fig. 9 C and D). Taken together, our results strongly suggest that JAZ-mediated interference with the DELLA–PIF interaction is a critical part of a mechanism by which JA antagonizes GA signaling in modulating growth.
Fig. 9.
JA sensitivity of PIF-overexpressing plants and pif mutants. Seedlings were grown on MS medium with or without 10 μM of MeJA under 10 μmol m−2⋅s−1 continuous white light at 22 °C for 6 d. Image (A) and quantification (B) of the effect of MeJA on Arabidopsis hypocotyl elongation. The hypocotyl lengths were measured and the inhibition of hypocotyl growth was calculated as (1 − treated / untreated) × 100%. Data shown are the means from 16 seedlings. Error bars represent SD. Letters on columns indicate significant differences (P < 0.05, Tukey–Kramer multiple comparison test). (C and D) MeJA has antagonizing effects on the expression of XTH33 (PIF-up-regulated) and EXP10 (down-regulated) genes in Arabidopsis. Total RNAs were purified and used for qRT-PCR analysis. Data shown are the means of three biological replicates. Error bars represent SD. Asterisks indicate significant difference between mock and MeJA treatment (P < 0.05, Student's t test).
Discussion
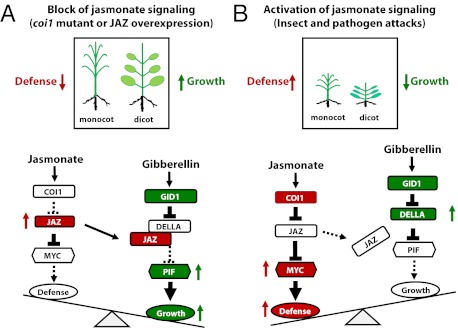
The growth–defense conflict is a widely known phenomenon in plants, although the underlying molecular mechanism is not well characterized. JA is an important plant hormone that plays a prominent role in plant defense against diverse pathogens and herbivores (3, 38). Despite rapid progress on dissecting the JA signaling pathway in recent years, a mechanistic explanation for how plants effectively balance growth and defense in response to the activation of JA signaling has remained elusive. In this study, we show that modulation of the level of DELLA repressors and interference with DELLA interaction with growth-promoting PIF transcription factors are two key mechanisms underlying JA-mediated growth inhibition in monocot rice and dicot Arabidopsis, illustrating a potentially widely conserved strategy by which angiosperm plants coordinate a major form of growth/defense tradeoff. Future research shall address whether the two mechanisms function independently of each other and how JA signaling modulates the level of DELLA proteins.
The DELLA proteins were first identified as key repressors of the GA pathway (39), and were subsequently shown to impact other hormone pathways such as auxin, abscisic acid, and ethylene (36, 40–42); plant photomorphogenesis (18, 19, 43); and plant survival under abiotic stress (44–46). Our results further support the notion that DELLA proteins act as key regulators/switches in integrating hormone and environmental signals and in fine-tuning plant growth and stress responses (47), which are critical for plant survival under harsh conditions (44, 46). We have provided clear evidence that JA treatment increases SLR1 levels in rice and RGA in Arabidopsis, which are predicted to result in the repression of plant growth. Conversely, we found that JAZ9 could effectively interrupt RGA–PIF3 interaction, suggesting that, in the absence of JA signaling, some DELLA repressors could be titrated out by JAZ proteins, which would allow more PIF transcription factors to activate growth programs. This mechanism could explain the GA-hypersensitivity phenotypes observed in the coi1 mutants of rice and Arabidopsis and in transgenic Arabidopsis plants overexpressing those JAZ proteins (e.g., AtJAZ1, 3, 4, 9, 10, and 11) that interact with DELLA proteins. It can also explain why not only the della quintuple mutant was largely insensitive to JA-induced growth inhibition, as expected, but also why overexpression of PIF3 could partially counter JA inhibition of growth (Fig. 9A). In short, we have provided experimental evidence for a signaling cascade, involving the COI1–JAZ–DELLA–PIF signaling module, that underlies the growth inhibition during JA defense activation.
In this study, we noticed interesting differences between rice, a domesticated monocot crop, and Arabidopsis, a wild dicot, in that prominent GA phenotypes of coi1 mutants are displayed under different conditions for rice and Arabidopsis. Whereas OsCOI1-RNAi plants exhibit exaggerated stem elongation and other GA-related phenotypes under strong light conditions in the greenhouse and in the field, Atcoi1 plants show most obvious GA phenotypes under dim light conditions (10 μmol m−2⋅s−1 continuous white light; Materials and Methods), but not under other growth conditions previously reported (6, 34). Therefore, although the core JA and GA pathways are likely conserved in monocot and dicot plants, divergence in JA and GA signaling and/or the crosstalk between JA and GA signaling in dicot and monocot species might exist. Recently, Robson et al. (48) found that the coi1 mutant flowered earlier and developed longer hypocotyls under low red/far-red light conditions than the WT. It is possible that these phenotypes could also be related to GA phenotypes studied here because light and GA signals integrate to regulate Arabidopsis growth (18, 19, 44), with DELLA proteins functioning in plant photomorphogenesis (43). We therefore propose that DELLA-mediated integration of JA, GA, and light signaling may give rise to a fundamental framework and needed flexibility in JA-induced growth–defense tradeoff in adaptation to and/or reflecting extraordinarily diverse growth habitats and domestication histories of angiosperm plants.
Materials and Methods
Plant Materials and Growth Condition.
Rice plants (cv. Nipponbare) were grown in a greenhouse or in the isolated paddy field. All Arabidopsis plants described here were derived from Col-0 except for the della mutant, which is in Landsberg erecta (Ler) background. The jaz9-1, dellaq (gai-t6/rga-t2/rgl1-1/rgl2-1/rgl3-1), and pifq (pif1-1/pif3-3/pif4-2/pif5-3) mutants, as well as the PRGA::GFP-RGA and PIF3OE transgenic lines, have been previously described (9, 18, 35, 49, 50). The jaz9-3 (SM_3_34031; Fig. S12) and coi1-30 (SALK_035548; Fig. S13) mutants were characterized in the present study. All Arabidopsis seeds were ordered from the Arabidopsis Biological Resource Center.
Arabidopsis seeds were stratified for 3 d at 4 °C before planting. Surface-sterilized seeds were sown on MS medium containing 0.8% agar and 5 mM Mes (pH 5.8), and placed in a growth chamber with 10 μmol m−2⋅s−1 continuous cool-white fluorescent light at 22 °C or in a long-day growth chamber with a 16-h day (120 μmol m−2⋅s−1 cool-white fluorescent light, 22 °C) and 8-h night (18 °C) cycle. The soil-grown plants were placed in the long-day growth chamber.
Transgenic Expression.
For generation of OsCOI1-RNAi transgenic rice, two fragments of the OsCOI1 genes were amplified by using the primer pairs OsCOI1-F1/R1 and OsCOI1-F2/R2 (Dataset S1), respectively. The fragments were inversely inserted into the pCAMBIA1300S that contained a double 35S promoter and a terminator. The resulting OSCOI1 RNAi construct was introduced into the model variety rice Nipponbare (Oryza sativa L. ssp. japonica) by using Agrobacterium-mediated transformation. Independent RNAi rice lines were analyzed and confirmed by Southern and Northern blot analyses, as well as qRT-PCR assays. All transgenic plants were grown in a greenhouse or in the isolated paddy field for measurement of plant height and other morphological traits.
For production of transgenic Arabidopsis lines expressing P35S::3×HA-AtJAZ9-8×His, the coding sequence of AtJAZ9, excluding the stop codon, was amplified by PCR by using PfuUltra II DNA polymerase (Agilent Technologies) and Arabidopsis cDNA, which was obtained from a 28-d-old Col-0 plant by using the AtJAZ9-F and -R primer set (Dataset S1). The PCR product was first cloned into vector pGEM-T easy (Promega), and then moved into a binary vector pJYP003 (SI Materials and Methods) to create a 3×HA-AtJAZ9-8×His fusion construct. Col-0 plants were transformed by Agrobacterium-mediated transformation. Primary transformants were selected based on BASTA resistance, and the JAZ9 expression level was determined by Western blotting. Transgenic lines with a 3:1 (resistant:sensitive) segregation ratio for BASTA resistance were selected, and several homozygous lines were identified in the T3 generation.
The Gateway entry clones containing GAI, RGA, or PIF3 were identified from the REGIA Arabidopsis transcription factor library (51). Inserts were transferred into pJYP006 or pJYP012 by LR recombination (Life Technologies) to create P35S::9×Myc-GAI, P35S::9×Myc-RGA, and P35S:3×FLAG-PIF3, which were used for transient expression in tobacco leaves as previously described (52).
Hormone Treatment and Growth Assay.
For rice, the seeds were sterilized and incubated on one-half MS medium with 0.6% agar and supplemented with different concentrations of GA3 and MeJA. Seedling (i.e., shoot) growth and the lengths of the second sheath were measured 12 d after treatment. For Western blotting, 10-d-old seedlings grown in one-half MS medium with 0.6% agar were transferred to liquid one-half MS medium supplemented with 100 μM GA3 with or without 100 μM MeJA. Samples were harvested at different time points and frozen at −80 °C for RNA and protein extraction.
Arabidopsis seeds used for growth assays were harvested on the same day from plants grown side by side. Seedlings were grown on MS plates for 4 d before being transferred onto soil. Seedlings of homozygous coi1-30 plants were selected on MS plates containing 10 μM MeJA (Sigma-Aldrich). Plants were kept in a long-day growth chamber unless indicated otherwise. Flowering time was determined when floral buds became visible at the center of rosette. Petiole lengths of the third true leaves were measured on day 21. At least 16 plants of each line were assessed.
Arabidopsis seedlings were also grown on plates with or without 10 μM MeJA under continuous light for 6 d. Seedlings were then placed on a new plate and scanned at a resolution of 600 dpi. The hypocotyl length was measured by using ImageJ software (National Institutes of Health).
Endogenous GA Assay in Rice.
The elongating uppermost internodes of the transgenic and WT plants were harvested and lyophilized at −20 °C. GAs were extracted, and GA1 and GA4 were assayed by LC-MS with internal standards as described previously (53).
RNA Blot and Transcript Analysis.
Total RNA was isolated from rice leaf tissues by using TRIzol reagent according to the manufacturer’s protocol (Invitrogen). RNA gel blotting was performed by using standard protocol with PerfectHyb buffer (Sigma), and relative gene expression was quantified by using a PhosphorImager (Amersham Biosciences). For qRT-PCR, total RNA was first treated with DNase I and the first-strand cDNA was then synthesized by using the oligo dT primer and SuperScript II reverse transcriptase (Invitrogen). Rice ubiquitin 1 gene (UBQ1; Os06g0681400) was used as an internal control to normalize samples. Quantitative PCR was performed on the Mx3000P real-time PCR system (Agilent Technologies) with a QuantiTect SYBR Green PCR kit (Qiagen). Semiquantitative RT-PCR was conducted by using the SuperScript III First-Strand Synthesis System (Invitrogen). The primers used to detect the transcripts of the target genes are listed in Dataset S1.
For analysis of Arabidopsis transcripts, total RNA was extracted by using an Ambion ToTALLY RNA Total RNA Isolation Kit (Life Technologies) according to the manufacturer’s manual. After DNase I (Roche) treatment, RNA was further purified by using an RNeasy Mini Kit (Qiagen). First-strand cDNA was synthesized by using M-MLV reverse transcriptase (Invitrogen) and oligo(dT) as primers. actin2 (AT3G18780), cap-binding protein 20 (CBP20; AT5G44200), protein phosphatase 2A subunit A3 (PP2AA3; AT1G13320), and ubiquitin-conjugating enzyme 21 (UBC21; AT5G25760) (54) were used as internal controls to normalize target gene expression by GeNORM method (55). Quantitative PCR was performed on an ABI7500 Fast Real-time PCR System with Fast SYBR Master Mix (Applied Biosystems) according to the manufacturer’s protocol. The primers used to detect specific transcripts are listed on Dataset S1.
Protein Extraction, Quantification, and Immunoblots.
Total proteins were extracted using plant protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, and 1 mM DTT) with 1% protease inhibitor for plant cell and tissue extract (Sigma-Aldrich) and 100 μM MG132 (Cayman Chemicals). Protein content was quantified by using the Protein DC assay kit (Bio-Rad). Equal amounts of protein were subjected to SDS/PAGE followed by Western blotting analysis. Immunodetection of GFP-RGA, HA-JAZ9, RGA, Myc-RGA, Myc-GAI, FLAG-PIF3, and SLR1were performed by using anti-GFP antisera (Abcam), anti-HA antibody (Roche Applied Science), anti-RGA antisera (49), anti-Myc antisera (Abcam), anti-FLAG antibody (Sigma-Aldrich), and anti-SLR1 (33), respectively. Corresponding HRP conjugated secondary antibodies and SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) were used for detection.
Coimmunoprecipitation Assay.
Total proteins were extracted from Arabidopsis seedlings or tobacco leaves by using lysis buffer (50 mM Tris-HCl, pH 7.5, 75 mM NaCl, 0.2% Triton X-100, 5 mM EDTA, 5 mM EGTA, 1 mM DTT, 1% Sigma Protease inhibitor mixture,100 μM MG132, 10 mM NaF, and 2 mM Na3VO4). The immunocomplexes were captured by anti-HA or anti-Myc agarose beads (Sigma-Aldrich), washed, and released by 2× SDS sampling buffer, and were then subjected to SDS/PAGE followed by further Western blotting analysis.
Rice Microarray Assay.
Whole transcriptomic analysis was performed with the Affymetrix GeneChip Rice Genome Array, representing 51,279 transcripts with three biological replicates. Raw data were analyzed with Affymetrix GeneChip Operating Software (GeneSpring, version 11.0) using Affymetrix default analysis settings and global scaling as normalization method. The GA, JA, and defense-related gene was analyzed by MAPMAN. The microarray data has been deposited into the National Center for Biotechnology Information Gene Expression Omnibus database (accession nos. GSE29577 and GSM732294–GSM732299).
Assays for α-Amylase Activity.
The release of α-amylase was assayed as described by Zhang et al. (29). The sterilized embryoless half seeds were incubated on agar plates containing 0.2% starch with 1 μM GA3 or 1 μM GA3 and 50 μM MeJA for 3 d at 28 °C in darkness. These plates were exposed to iodine vapor for a few minutes. The reaction between starch and iodine turned the agar plates a blue-purple color. The agar around half seeds with α-amylase activity remained colorless because of hydrolysis of starch by α-amylase. The α-amylase protein was extracted from deembryonated half seeds imbibed in 1.0 μM of GA3 solution in the dark at 28 °C for 2 d (56). The α-amylase activity was assayed by quantifying reducing sugar released from substrate starch (57).
Microscopic Observation.
Rice internodes and sheaths were sampled for resin sectioning. The samples were first fixed in FAA [3.7% (vol/vol) formaldehyde, 5% (vol/vol) acetic acid, 50% (vol/vol) ethanol], followed by dehydration through a graded ethanol series. The samples were embedded in resin and polymerized at 58 °C. Sections (8–10 μm) were examined under a microscope (DMLB; Leica) and documented by photography.
Yeast Three-Hybrid Assay.
The coding sequence corresponding to the GRAS domain of RGA was amplified by PCR by using the RGAdN-F and -R primer set (Dataset S1) and cloned into pGBridge (SI Materials and Methods) to create a Gal4DB–Myc–RGAΔN construct. The AtJAZ9 was cloned into pET42a-3×FLAG (SI Materials and Methods) to create a 3×FLAG-JAZ9 cassette, which was further transferred into pBridge (Clontech) to create PMET::3×FLAG-JAZ9. The PMET::3×FLAG-JAZ9 insert was transferred into the Gal4DB–Myc–RGAΔN fusion plasmid to create the yeast three-hybrid (Y3H) bait vector. The AtPIF3 was cloned into pDEST-GADT7 (58) by LR recombination to create the prey vector. The bait and prey vectors were transformed into the yeast strain AH109 (Clontech). Y3H assays were performed following the manufacturer’s protocol (Clontech).
Statistic Analysis.
Unless indicated otherwise, one-way ANOVA was performed for all data sets. Tukey–Kramer multiple comparison test was used to compare the means of the treatments at an α level of 0.05.
Supplementary Material
Acknowledgments
We thank Dr. M. Matsuoka (Nagoya University) for the gid1-1 mutant and the antibody against SLR1, Dr. Q. Qian (China National Rice Research Institute) for the slender rice 1 mutant, Dr. J.-M. Li for useful discussion on the research, Dr. P. Quail (University of California) for pifq mutant and PIF3 antibody, H. Zhang, R. K. Kalia, and J. Withers for critical reading of the manuscript, and J.-Q. Li and X.-Y. Gao for microscopy assistance. This work was supported by Natural Science Foundation of China Grants 90817102 and 30730064 (to Z.H.), National Key Basic Research and Development Program Grant 2011CB100700 (to Z.H.), US Department of Agriculture National Research Initiative Grant 2003-35319-17873 (to Y.Y.), National Science Foundation Plant Genome Research Program Grant DBI-0922747 (to Y.Y.), National Institutes of Health Grant R01AI068718 (to S.Y.H. for experiments with jasmonate signaling), and US Department of Energy (Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science) Grant DE-FG02-91ER20021 (to S.Y.H. for experiments with growth and GA signaling). S.Y.H. is a Howard Hughes Medical Institute and Gordon and Betty Moore Foundation Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 7152 (volume 109, number 19).
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE29577 and GSM732294–GSM732299).
*Yao J, Withers J, He SY, Plant Biology 2011, August 6–10, 2011, Minneapolis, MN, abstr P16037.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201616109/-/DCSupplemental.
References
- 1.Kazan K, Manners JM. The interplay between light and jasmonate signalling during defence and development. J Exp Bot. 2011;62:4087–4100. doi: 10.1093/jxb/err142. [DOI] [PubMed] [Google Scholar]
- 2.Kazan K, Manners JM. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012;17:22–31. doi: 10.1016/j.tplants.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Browse J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 4.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 5.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 7.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 10.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 11.Pauwels L, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farmer EE. Plant biology: Jasmonate perception machines. Nature. 2007;448:659–660. doi: 10.1038/448659a. [DOI] [PubMed] [Google Scholar]
- 13.Hedden P, Phillips AL. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 14.Schwechheimer C. Understanding gibberellic acid signaling—are we there yet? Curr Opin Plant Biol. 2008;11:9–15. doi: 10.1016/j.pbi.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 16.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21:1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 20.Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol. 2007;58:183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- 21.Navarro L, et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 22.Cheng H, et al. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5:e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Kim EH, et al. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009;149:1751–1760. doi: 10.1104/pp.108.134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou G, et al. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009;60:638–648. doi: 10.1111/j.1365-313X.2009.03988.x. [DOI] [PubMed] [Google Scholar]
- 26.Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011;157:305–316. doi: 10.1104/pp.111.177576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei C, Qi M, Sheng G, Yang Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe Interact. 2006;19:1127–1137. doi: 10.1094/MPMI-19-1127. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006;18:442–456. doi: 10.1105/tpc.105.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res. 2008;18:412–421. doi: 10.1038/cr.2008.28. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 2011;67:342–353. doi: 10.1111/j.1365-313X.2011.04596.x. [DOI] [PubMed] [Google Scholar]
- 31.Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo A, et al. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 2006;47:181–191. doi: 10.1093/pcp/pci233. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda A, et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Turner JG. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE. 2008;3:e3699. doi: 10.1371/journal.pone.0003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zentella R, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 38.Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 39.Peng J, et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu X, Harberd NP. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;421:740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- 41.Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell. 2003;15:2816–2825. doi: 10.1105/tpc.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Achard P, et al. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA. 2007;104:6484–6489. doi: 10.1073/pnas.0610717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Achard P, et al. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007;143:1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 45.Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA. 2008;105:16814–16819. doi: 10.1073/pnas.0807821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol. 2008;18:656–660. doi: 10.1016/j.cub.2008.04.034. [DOI] [PubMed] [Google Scholar]
- 47.Davière J-M, de Lucas M, Prat S. Transcriptional factor interaction: A central step in DELLA function. Curr Opin Genet Dev. 2008;18:295–303. doi: 10.1016/j.gde.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Robson F, et al. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell. 2010;22:1143–1160. doi: 10.1105/tpc.109.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silverstone AL, et al. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park E, et al. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004;45:968–975. doi: 10.1093/pcp/pch125. [DOI] [PubMed] [Google Scholar]
- 51.Paz-Ares J. Regia Consortium REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp Funct Genomics. 2002;3:102–108. doi: 10.1002/cfg.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 53.Xiao LT, Wang SG. In: in Plant Physiology Experimental Technique. Xiao LT, Wang SG, editors. Beijing: China Agricultural Press; 2005. pp. 169–172. [Google Scholar]
- 54.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Washio K, Ishikawa K. Structure and expression during the germination of rice seeds of the gene for a carboxypeptidase. Plant Mol Biol. 1992;19:631–640. doi: 10.1007/BF00026789. [DOI] [PubMed] [Google Scholar]
- 57.Shaw J-F, Lin F-P, Chen S-C, Chen H-C. Purification and properties of an extracellular a-amylase from Thermus sp. Bot Bull Acad Sin. 1995;36:195–200. [Google Scholar]
- 58.Rossignol P, Collier S, Bush M, Shaw P, Doonan JH. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J Cell Sci. 2007;120:3678–3687. doi: 10.1242/jcs.004119. [DOI] [PubMed] [Google Scholar]