Abstract
Despite long-standing interest in the origin and maintenance of species diversity, little is known about historical drivers of species assemblage structure at large spatiotemporal scales. Here, we use global species distribution data, a dated genus-level phylogeny, and paleo-reconstructions of biomes and climate to examine Cenozoic imprints on the phylogenetic structure of regional species assemblages of palms (Arecaceae), a species-rich plant family characteristic of tropical ecosystems. We find a strong imprint on phylogenetic clustering due to geographic isolation and in situ diversification, especially in the Neotropics and on islands with spectacular palm radiations (e.g., Madagascar, Hawaii, and Cuba). Phylogenetic overdispersion on mainlands and islands corresponds to biotic interchange areas. Differences in the degree of phylogenetic clustering among biogeographic realms are related to differential losses of tropical rainforests during the Cenozoic, but not to the cumulative area of tropical rainforest over geological time. A largely random phylogenetic assemblage structure in Africa coincides with severe losses of rainforest area, especially after the Miocene. More recent events also appear to be influential: phylogenetic clustering increases with increasing intensity of Quaternary glacial-interglacial climatic oscillations in South America and, to a lesser extent, Africa, indicating that specific clades perform better in climatically unstable regions. Our results suggest that continental isolation (in combination with limited long-distance dispersal) and changing climate and habitat loss throughout the Cenozoic have had strong impacts on the phylogenetic structure of regional species assemblages in the tropics.
Keywords: biodiversity, biogeography, climate change, evolution, extinction
Despite long-standing interest, the mechanisms behind the origin and maintenance of high tropical biodiversity remain elusive (1, 2). Much recent macroecological research has focused on explaining large-scale species richness gradients by contemporary climate (3–5), but the importance of evolutionary diversification and past environments has also been highlighted (2, 6–8). Over the past decades it has become increasingly clear that local community structure depends on both local processes and large-scale factors that influence regional species diversity (9, 10). Regional species assemblages are jointly shaped by within-region diversification and dispersal between regions, the former being constrained by time for speciation (7, 11) and by climatic or other factors influencing net diversification rates (12, 13), and the latter by the formation and disappearance of dispersal barriers (14, 15), time for dispersal (16), and phylogenetic niche conservatism (17). Understanding the historical assembly and present-day structure of regional species assemblages thus requires integration of ecological, paleogeographic, and phylogenetic information (10).
Increasing availability of phylogenies has ignited interest in the phylogenetic structure of species assemblages (18–20). Numerous studies have used this approach to examine the assembly of local communities (21–24). However, measures of assemblage phylogenetic structure may also reveal large-scale biogeographic processes (25, 26), although this approach has rarely been taken in biogeography (27). Several consequences of large-scale biogeographic processes can be predicted for the phylogenetic structure of species assemblages (Table 1). First, strong dispersal barriers should lead to in situ diversification within major biogeographic realms or continents. As a consequence, regional species assemblages are expected to consist predominantly of species that are relatively closely related (hypothesis H1). Second, it has been suggested that lineages inhabiting large areas over extended geological time experience higher speciation rates and lower extinction rates than lineages inhabiting small areas (“time-integrated area effect”) (12). The resulting higher net diversification should lead to phylogenetically clustered assemblages at regional scales (35) (hypothesis H2). Third, the contraction of major habitat types (“biomes”) during the Cenozoic should have resulted in severe extinction (31, 32). If net diversification decreases with the loss of biome area, most lineage divergences should be ancient and we could expect a tendency toward random phylogenetic structuring of species assemblages (hypothesis H3). Finally, the frequency and magnitude of Quaternary climatic oscillations has also been suggested as a major driver of assemblage structure, specifically for species richness and endemism (8, 36, 37). Although never assessed on a global scale, this could also have major consequences for the phylogenetic structure of species assemblages, for example, by favoring clades that perform well in such variable environments (hypothesis H4) (33).
Table 1.
Four core hypotheses in historical biogeography and their predictions for the phylogenetic structure of regional species assemblages
| Hypotheses | Prediction | References |
| H1: The long-term geographic isolation of continents and biogeographic realms has caused dispersal limitation and in situ diversification. | Phylogenetic clustering within continents and biogeographic realms increases with an increasing spatial extent of the sampling pool (e.g., from continental to hemispheric and global extent). | 20, 23, 28, 29 |
| H2: Large habitat areas over deep geological time increase speciation rates and decrease extinction rates [“time-integrated species-area effect” sensu (12)]. | Regions with large time-integrated areas of suitable habitat show higher phylogenetic clustering than regions where suitable habitat is limited over deep geological time. | 12, 29, 30 |
| H3: Strong loss of biome area over Cenozoic time scales leads to decreasing diversification rates (less speciation, more extinction). | Regions with high initial, but strongly decreasing, biome area—and thus diversification rates—will show less phylogenetic clustering than areas with constant or increasing diversification rates. | 31, 32 |
| H4: Areas with high Quaternary climate change harbor assemblages of survivors and/or species that were able to recolonize such areas after local extinction. | Areas with high paleoclimatic amplitudes (low stability) are characterized by phylogenetic clustering because assemblages predominantly consist of species with phylogenetically conserved traits that enable them to survive in or quickly recolonize such environments. | 33, 34 |
Here, we combine community phylogenetic and macroecological methods to test these four core hypotheses in historical biogeography (Table 1). We use a unique dataset of global species distributions and a dated phylogeny of an important tropical plant lineage: the palm family (Arecaceae). Palms are diverse (>2,400 species in 183 genera), are a characteristic element of tropical ecosystems worldwide (38), and have served as a model system in geographical ecology (39) and rainforest evolution (40). Global distribution patterns in palms are well documented (37, 41), and phylogenetic relationships within the family are well understood (42). Most present-day palm diversity has evolved during the Cenozoic (40). For these reasons, palms are ideally suited to study the evolutionary imprints of Cenozoic history on tropical biotic assemblages at a global scale. Using the Net Relatedness Index (NRI) (18), we quantify the phylogenetic structure of palm assemblages and relate it to paleo-reconstructions of climate and biomes across most of the Cenozoic. By quantifying to what extent species that co-occur are more (NRI > 0) or less (NRI < 0) closely related than expected by random sampling from a species pool, NRI can reveal ecological and evolutionary mechanisms of assemblage structure. Scaling sampling pools to different spatial extents provides insights into the critical scales at which the assembly processes operate (26).
Results
Phylogenetic Structure with a Global Sampling Pool.
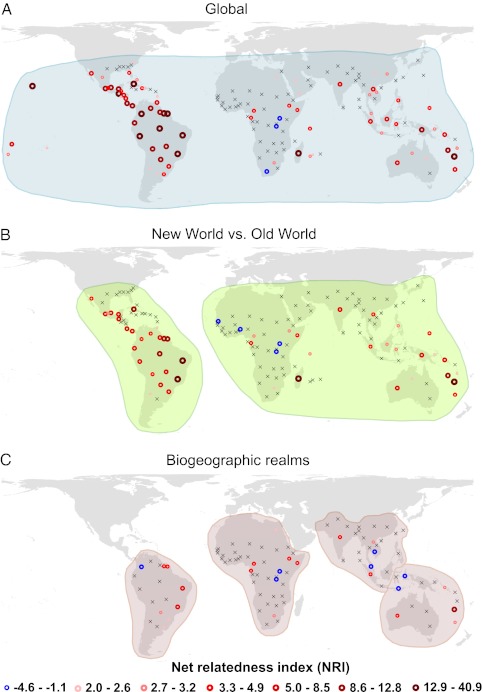
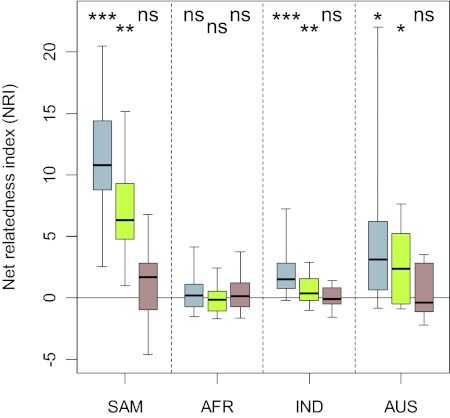
Across regional assemblages, NRI values calculated with a global sampling pool (Fig. 1A) ranged from −1.5 to 40.9 (median = 2.0). The global mean was significantly > 0 (SI Appendix, Table S1), indicating an overall predominance of phylogenetic clustering. A number of islands stood out with remarkably high NRI values (SI Appendix, Table S2; Fig. 1A), including Madagascar (NRI = 40.9), New Caledonia (NRI = 22.0), Hawaii (NRI = 20.7), and Cuba (NRI = 18.1). However, the majority of islands had small or intermediate NRI values (median = 2.6), and there was no general statistical difference in mean NRI between islands and continental geographic units at a global scale [t = −1.4, df = 150, P = 0.160, data ln(x + 2) transformed]. NRI values calculated with a global sampling pool (blue boxes in Fig. 2) were on average significantly larger than zero in South America, Indomalaya, and Australasia, indicating phylogenetic clustering. In contrast, mean NRI did not statistically differ from zero in Africa, indicating an overall random phylogenetic structure. Moreover, Africa harbored the only three regional assemblages (Uganda, Burundi, and the Cape Province) with phylogenetic overdispersion (Fig. 1A) using a global sampling pool.
Fig. 1.
The phylogenetic structure of palm assemblages given species pools restricted to (A) global (shaded blue), (B) hemispheric (New World vs. Old World) (shaded green), and (C) biogeographic realm (shaded brown) extent. The NRI is plotted for the mass centroid of each sample unit (“botanical country”). Blue circles indicate significantly (P < 0.05) negative NRI values (phylogenetic overdispersion). Red circles represent significantly positive NRI values (phylogenetic clustering) with darker red and larger circles indicating increases in NRI (quantile classification based on values in A). An “x” indicates nonsignificant NRI values. Maps are in Behrmann projection.
Fig. 2.
Effect of sampling pool extent on phylogenetic structure of palm assemblages in South America (SAM), Africa (AFR), Indomalaya (IND), and Australasia (AUS). Box plots summarize values of the NRI within each biogeographic realm for a given sampling pool. Colors of sampling pool extent as in Fig. 1. Statistical significance of whether mean NRI differs from zero is given above each box plot (ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; see SI Appendix, Table S1 for details of the tests). Boxes represent the interquartile range (IQR), horizontal lines within the boxes represent medians, and whiskers extend to 1.5 times the IQR.
Geographic Isolation and Sampling Pool Scaling (H1).
Restricting the sampling pool to a given hemisphere or biogeographic realm caused significant changes in NRI (SI Appendix, Fig. S1 and Table S3). NRI values in South America, Indomalaya, and Australasia decreased consistently with a decreasing sampling pool extent (Fig. 2), indicating a strong effect of geographic isolation (with limited dispersal among regions) and in situ diversification within three of four realms (supporting hypothesis H1). The strongest decrease in NRI was observed in South America (Fig. 2). Africa, in contrast, showed a predominantly random phylogenetic structure independent of the sampling pool extent (Fig. 2). Across realms, changes in the spatial distribution of NRI values were minor between global and hemispheric sampling pools (Fig. 1 A and B). However, geographically localized phylogenetic structuring became evident when reducing the sampling pool extent to realms (Fig. 1C). Notably, phylogenetic overdispersion emerged in biogeographic contact zones (Colombia, Wallacea).
Tropical Rainforest Distribution Through Time (H2 and H3).
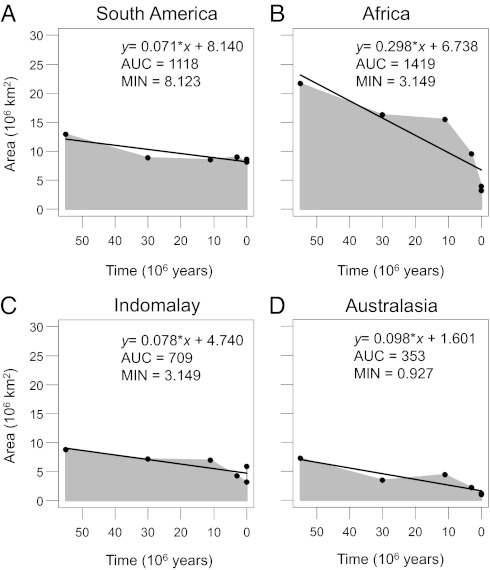
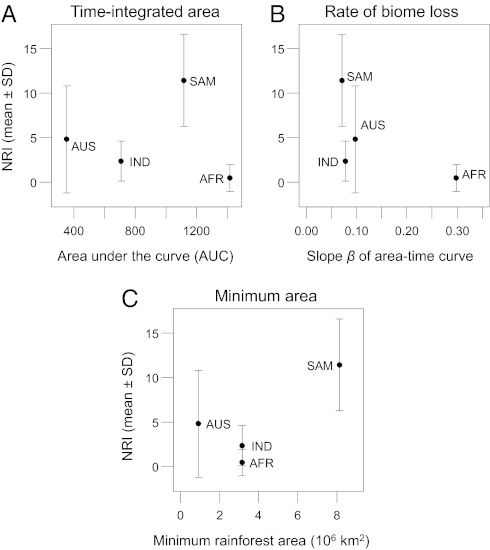
We quantified the historical extent of tropical rainforests throughout the Cenozoic (Fig. 3). All four biogeographic realms have harbored large rainforest areas since the Eocene, with Africa having by far the highest time-integrated area [area under the curve (AUC)] (Fig. 3). However, in disagreement with hypothesis H2, mean NRI values of regional palm assemblages did not consistently increase with AUC (Fig. 4A). Rainforest area decreased throughout the Cenozoic in all realms; this loss was much more dramatic in Africa (>18 × 106 km2 loss) than in Australasia (6 × 106 km2), South America (4 × 106 km2), and Indomalaya (3 × 106 km2). In three of four regions, rainforest losses were most pronounced after the Miocene (11 Mya) (Fig. 3 B–D), especially in Africa (Fig. 3B). In agreement with hypothesis H3, the realm with the highest rate of biome loss (Africa) showed the lowest mean NRI (Fig. 4B). Also consistent with hypothesis H3, the realm with the largest minimum Cenozoic rainforest area (South America) showed the highest mean NRI (Fig. 4C).
Fig. 3.
Area-time plots representing the area of tropical rainforest over geological time: (A) South America, (B) Africa, (C) Indomalaya, and (D) Australasia. The area under the curve (AUC) is illustrated in gray and measures the time-integrated area of this biome from the Eocene to the present (106 km2/55 Mya). Black lines indicate a simple linear regression. The steepness (i.e., negative slope β) of these regression lines (given in the regression formulas) approximates the rate of biome loss over geological time. The minimum area (MIN, in 106 km2) during the Cenozoic is also provided.
Fig. 4.
The relationship between net relatedness (NRI) of palm assemblages and habitat availability over geological time (AFR, Africa; AUS, Australasia; IND, Indomalay; SAM, South America). Habitat availability was specified as (A) area under the curve (AUC) of area-time plots, (B) rate of biome loss (measured as slope β of a simple linear regression of area vs. time), and (C) minimum area of tropical rainforest during the Cenozoic. Compare with Fig. 3.
Quaternary Climate Change (H4).
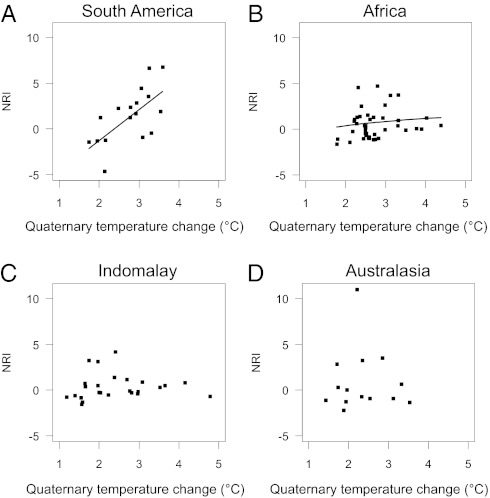
Climatic oscillations (temperature anomalies) during the Quaternary were strongest at the northern range boundary of the palm family (SI Appendix, Fig. S2). Within realms, Quaternary climate change peaked in southern and eastern South America, eastern Africa, northern Indomalaya, and Australia (SI Appendix, Fig. S2). In contrast to continental sites, islands generally showed low temperature anomalies. Globally, the relationship between temperature anomaly and phylogenetic structure of palm assemblages was not significant (SI Appendix, Fig. S3). However, at the realm scale, South American and African palm assemblages showed increasing NRI with increasing temperature anomaly, with no significant relationship in Indomalaya and Australasia (Fig. 5; SI Appendix, Table S4). The Quaternary climate change effects in South America and Africa remained in multiple-predictor models when accounting for covariation with present-day environment (SI Appendix, Table S5). Furthermore, the commonly observed increase of species richness with contemporary precipitation was not reflected in NRI (SI Appendix, Fig. S4).
Fig. 5.
The effect of Quaternary climatic oscillations on the net relatedness index (NRI, calculated with a realm sampling pool): (A) South America, (B) Africa, (C) Indomalaya, and (D) Australasia. Quaternary climatic oscillations were quantified as the change in mean annual temperature (anomaly) between the Last Glacial Maximum (∼0.021 Mya) and the present (in °C; SI Appendix, Fig. S2). See SI Appendix, Tables S4 and S5, for statistical results.
Discussion
Diversification in Isolation (H1).
The overall phylogenetic clustering of regional palm assemblages in South America, Indomalaya, and Australasia (Fig. 2) is consistent with hypothesis H1 and reflects that many higher-level palm taxa are endemic to continents, biogeographic realms, or islands (38, 39) (SI Appendix, Table S6). This provides evidence for a strong role of in situ diversification within biogeographic realms—and limited dispersal between them—in the formation of regional biota. Limited long-distance dispersal in palms is evident from few species having seeds suitable for oceanic drift, few palm genera being represented on both sides of the Atlantic, and a high degree of endemism (SI Appendix, SI Text S1). The decrease of NRI with sampling pool extent is further consistent with a strong dispersal limitation at higher taxonomic levels (39, 43) (SI Appendix, Table S6). Phylogenetic clustering might further be influenced by dispersal limitation at finer spatial scales. Specifically, frugivorous understory birds with low dispersal capacities might have promoted palm speciation in the undergrowth of tropical rainforests (44) (SI Appendix, SI Text S1). On islands, outstandingly high NRI values coincide with spectacular in situ radiations (SI Appendix, Table S2), for example, Dypsis on Madagascar (45), Pritchardia on Hawaii (46), Coccothrinax on Cuba (38), and three subtribes, Archontophoenicinae, Basseliniinae, and Clinospermatinae on New Caledonia (47). However, there are also many islands with low-to-medium NRI values, probably because these islands are rather small and too recently colonized for major radiations to happen.
Availability of Time-Integrated Area (H2).
We used the distribution of tropical rainforests to characterize habitat availability for palms over geological time because palms are used as paleo-indicators for this biome (31, 32, 48, 49) and their present-day distribution, species richness, and net diversification rate are related to humid megathermal climates (37, 39, 40, 50). Time-integrated biome area has been suggested as an important driver of differences in species diversity between tropical and temperate regions (12). This effect should be reflected in assemblage phylogenetic structure (hypothesis H2). For South America, the pronounced phylogenetic clustering is consistent with a relatively large and stable rainforest area over geological time (Fig. 4A). However, our results for Africa suggest that phylogenetic assemblage structure at a realm scale can be more sensitive to temporal dynamics in biome area than to absolute time-integrated biome area (Fig. 4), indicating that the latter is insufficient to explain major differences between realms.
Biome Loss over Geological Time (H3).
The major continental-scale differences in phylogenetic assemblage structure seem to be partly driven by Cenozoic change of biome area (Fig. 4 B and C). Africa stands out with the strongest biome loss (Fig. 4B), i.e., a very large extent of tropical rainforest in the Mid-Eocene, which subsequently declined and substantially so from the Mid-Miocene onward (Fig. 3B). The African fossil record of palms indicates severe Tertiary extinction (51). It is therefore likely that decreasing speciation and increasing extinction rates due to dramatic rainforest decline have caused an overall random phylogenetic structure of African palm assemblages (Fig. 2). Interestingly, there is a paucity of small-fruited palms in Africa (SI Appendix, Fig. S5), as can functionally be expected from its Cenozoic drying (SI Appendix, SI Text S1). In South America, the strong phylogenetic clustering might further be related to a relatively large minimum area of tropical rainforest (Fig. 4C), suggesting a smaller bottleneck effect in this realm. In addition, the upheaval of the Andes and other paleogeographical reorganizations in the Miocene and Pliocene (52, 53) might also play an important role in the Neotropics.
Quaternary Survival and Recolonization (H4).
Our results show that Quaternary climatic oscillations can affect the phylogenetic structure of tropical species assemblages (hypothesis H4). For palm assemblages in South America, the strong observed relationship between NRI and Quaternary temperature anomaly (Fig. 5A) could be driven by tribe Cocoseae (SI Appendix, Fig. S6), which dominates assemblages in eastern South America. These areas are drier and more seasonal than the Amazon basin and characterized by high Quaternary temperature oscillations (SI Appendix, Fig. S2). We suggest that strong Quaternary climate oscillations have favored the survival and diversification of palm lineages adapted to dry and seasonal climates (e.g., Cocoseae in South America; cf. SI Appendix, Fig. S6) and have prevented the survival of, or colonization by, species from lineages adapted to warm, wet rainforest environments (cf. 33). A similar filtering might explain the relationship between NRI and Quaternary climate change in Africa. The absence of such an effect in Indomalaya and Australasia might be due to these regions’ predominance of islands where Quaternary climate change was less severe (SI Appendix, Fig. S2) than on continents, possibly due to oceanic buffering (54) and the expansion of rainforests in these areas during glacials (55).
Conclusions.
Our study demonstrates that broad-scale patterns in phylogenetic assemblage structure are consistent with differences in long-term historical drivers: continental isolation in combination with limited long-distance dispersal, Cenozoic habitat loss, and Quaternary climate instability. The relative importance of Quaternary and deep-time climate change depends on spatial scale (global, within vs. between realms), geographic settings (barriers and degree of isolation, continents vs. islands), and the unique history of biogeographic realms (e.g., dramatic biome changes in Africa, Quaternary climate change effects in South America). In addition to the four major biogeographic hypotheses tested here, phylogenetic assemblage structure might provide additional insights into other large-scale biogeographic processes: for example, phylogenetic overdispersion might indicate areas of biotic interchange (e.g., Colombia, Wallacea), and high levels of phylogenetic clustering might be facilitated through biotic interactions (e.g., via dispersers, herbivores, pollinators, and pathogens). Measures of phylogenetic assemblage structure such as NRI can thus provide insights into biogeographic and evolutionary processes at the assemblage level. We see great potential for better understanding the origins of tropical biodiversity by integrating geological, geographic, and paleoclimatic reconstructions with phylogenetic and species distribution data at large spatiotemporal scales.
Materials and Methods
Assemblage Data.
Presence and absence of all palm species (n = 2,440) in all level 3 geographic units (“botanical countries”) of the World Geographical Scheme for Recording Plant Distributions (56) was obtained from the World Checklist of Palms (41). The units are often countries, but very small countries are omitted whereas very large countries are subdivided according to states or provinces. We used an updated version of the World Checklist of Palms (downloaded on March 9, 2009 from http://apps.kew.org/wcsp) excluding introduced occurrences.
Phylogeny.
We used a dated phylogeny of the 183 palm genera (40) (SI Appendix, Fig. S7), which is based on a recent supertree, the most extensive phylogenetic study of the family published to date (42). The phylogeny was dated using a Bayesian relaxed molecular clock approach with uncorrelated rates and calibrated using four palm fossil taxa (40). Below the genus level, species were appended as polytomies with a divergence age arbitrarily set at two-thirds the stem node age of the genus. A sensitivity analysis indicated that our results are not dependent on this arbitrarily set mean divergence age within genera (SI Appendix, Fig. S8).
Phylogenetic Assemblage Structure.
We calculated the NRI (18) for each assemblage with more than one palm species (n = 151). NRI measures how mean phylogenetic distance (MPD) between all species pairs in the assemblage deviates from random. The random expectation is computed from “null” assemblages that are randomly sampled from a predefined species pool. NRI is calculated as:
where MPDobs is the observed MPD of a given assemblage measured in million years, meanMPDrnd the mean of the MPD values of the null assemblages, and sdMPDrnd the SD of the MPD values of the null assemblages. Values near zero indicate phylogenetically random assemblages, and deviations indicate overdispersion (<0) or clustering (>0) (18, 24). NRI was calculated with “picante” (57) in R (58) using the function ses.mpd(). We simulated null assemblages using the “taxa.labels” null model (n = 999 randomizations), which randomizes taxon labels on the phylogeny for the species included in the sampling pool. We tested three different spatial extents for the sampling pools: global (all species in the phylogeny), hemispheric (Old World vs. New World species), and continental/biogeographic realm (South America, continental Africa, Indomalaya, and Australasia). Note that we used NRI and not the nearest taxon index (NTI) (18) because NTI represents mainly recent clustering or overdispersion (near the tips of the phylogeny) and therefore is less relevant for assessing deep-time hypotheses.
Biome Reconstructions and Time-Integrated Area.
We used paleogeographic vegetation and biome reconstructions to estimate the area and distribution of tropical rainforests during the Cenozoic (SI Appendix, SI Text S2 and Table S7). We obtained biome estimates for the Eocene (∼ 55 Mya), Oligocene (∼30 Mya), Miocene (∼ 11 Mya), Middle Pliocene (∼ 3 Mya), Last Glacial Maximum (LGM) (0.021 Mya), and the present (SI Appendix, Table S8). We digitized the distribution of rainforest for all time steps and plotted rainforest area against geological time [area-time plots sensu (12)]. We then estimated the AUC (12), the rate of area loss (measured as the slope β of a simple linear regression of area vs. time), and the minimum area during the Cenozoic as summary statistics for each biogeographic realm (SI Appendix, Table S8).
Quaternary Climate Change.
We compiled two paleoclimatic reconstructions for the LGM (0.021 Mya), namely from the Community Climate System Model version 3 (CCSM3) and the Model for Interdisciplinary Research on Climate version 3.2 (MIROC3.2) (available at http://pmip2.lsce.ipsl.fr/) (59). We used the mean of the anomaly between LGM and present-day annual temperature across the two paleoclimatic simulations (CCSM3, MIROC3.2) to represent Quaternary climatic oscillations (37). These temperature anomalies cover almost the full Quaternary (past 2.6 Mya) temperature range with a geographic pattern that is representative for at least a large portion of the period (36). We used nonspatial as well as spatial regression models (60) to relate NRI to Quaternary climate change (SI Appendix, Tables S4 and S5).
Supplementary Material
Acknowledgments
We thank Ulrich Salzmann and Sandy Harrison for information on biome reconstructions; Paul Fine for discussion of the time-integrated species-area effect; Peder K. Bøcher for assistance with geographic information system analysis; and John Dransfield, Rafaël Govaerts, and the Royal Botanic Gardens for providing the World Checklist of Palms. We further acknowledge the international modeling groups for providing the LGM climate data and the Laboratoire des Sciences du Climat et de l’Environnement for collecting and archiving them. The data archive of the Paleoclimate Modeling Intercomparison Project (PMIP2) is supported by Commissariat à l’Energie Atomique, Centre National de la Recherche Scientifique, the European Union project MOTIF (EVK2-CT-2002-00153), and the Programme National d’Etude de la Dynamique du Climat. This work was supported by Villum Kahn Rasmussen Foundation Grant VKR09b-141 (to J.-C.S.), Danish Council for Independent Research–Natural Sciences Grant 10-083348 (to H.B.) and starting independent researcher Grant 11-106163 (to W.D.K.), European Community Seventh Framework Programme Grant 212631 (to H.B.), and the Aarhus University Research Foundation (postdoctoral stipend to W.L.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120467109/-/DCSupplemental.
References
- 1.Hill JL, Hill RA. Why are tropical rain forests so species rich? Classifying, reviewing and evaluating theories. Prog Phys Geogr. 2001;25:326–354. [Google Scholar]
- 2.Mittelbach GG, et al. Evolution and the latitudinal diversity gradient: Speciation, extinction and biogeography. Ecol Lett. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 3.Hawkins BA, et al. Energy, water, and broad-scale geographic patterns of species richness. Ecology. 2003;84:3105–3117. [Google Scholar]
- 4.Allen AP, Brown JH, Gillooly JF. Global biodiversity, biochemical kinetics, and the energetic-equivalence rule. Science. 2002;297:1545–1548. doi: 10.1126/science.1072380. [DOI] [PubMed] [Google Scholar]
- 5.Currie DJ, et al. Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol Lett. 2004;7:1121–1134. [Google Scholar]
- 6.Dynesius M, Jansson R. Evolutionary consequences of changes in species’ geographical distributions driven by Milankovitch climate oscillations. Proc Natl Acad Sci USA. 2000;97:9115–9120. doi: 10.1073/pnas.97.16.9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiens JJ. The causes of species richness patterns across space, time, and clades and the role of “ecological limits”. Q Rev Biol. 2011;86:75–96. doi: 10.1086/659883. [DOI] [PubMed] [Google Scholar]
- 8.Sandel B, et al. The influence of Late Quaternary climate-change velocity on species endemism. Science. 2011;334:660–664. doi: 10.1126/science.1210173. [DOI] [PubMed] [Google Scholar]
- 9.Zobel M. The relative role of species pools in determining plant species richness: An alternative explanation of species coexistence? Trends Ecol Evol. 1997;12:266–269. doi: 10.1016/s0169-5347(97)01096-3. [DOI] [PubMed] [Google Scholar]
- 10.Ricklefs RE. Phylogenetic perspectives on patterns of regional and local species richness. In: Bermingham E, Dick CW, Moritz C, editors. Tropical Rainforests: Past, Present and Future. Chicago, London: University of Chicago Press; 2005. pp. 16–40. [Google Scholar]
- 11.Stephens PR, Wiens JJ. Explaining species richness from continents to communities: The time-for-speciation effect in emydid turtles. Am Nat. 2003;161:112–128. doi: 10.1086/345091. [DOI] [PubMed] [Google Scholar]
- 12.Fine PVA, Ree RH. Evidence for a time-integrated species-area effect on the latitudinal gradient in tree diversity. Am Nat. 2006;168:796–804. doi: 10.1086/508635. [DOI] [PubMed] [Google Scholar]
- 13.Svenning JC, Borchsenius F, Bjorholm S, Balslev H. High tropical net diversification drives the New World latitudinal gradient in palm (Arecaceae) species richness. J Biogeogr. 2008;35:394–406. [Google Scholar]
- 14.Renner S. Plant dispersal across the tropical Atlantic by wind and sea currents. Int J Plant Sci. 2004;165:S23–S33. [Google Scholar]
- 15.Webb SD. The Great American Biotic Interchange: Patterns and processes. Ann Mo Bot Gard. 2006;93:245–257. [Google Scholar]
- 16.Paul JR, Morton C, Taylor CM, Tonsor SJ. Evolutionary time for dispersal limits the extent but not the occupancy of species’ potential ranges in the tropical plant genus Psychotria (Rubiaceae) Am Nat. 2009;173:188–199. doi: 10.1086/595762. [DOI] [PubMed] [Google Scholar]
- 17.Wiens JJ, Donoghue MJ. Historical biogeography, ecology and species richness. Trends Ecol Evol. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 19.Emerson BC, Gillespie RG. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol Evol. 2008;23:619–630. doi: 10.1016/j.tree.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. The merging of community ecology and phylogenetic biology. Ecol Lett. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- 21.Cooper N, Rodríguez J, Purvis A. A common tendency for phylogenetic overdispersion in mammalian assemblages. Proc Biol Sci. 2008;275:2031–2037. doi: 10.1098/rspb.2008.0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamilar JM, Guidi LM. The phylogenetic structure of primate communities: Variation within and across continents. J Biogeogr. 2010;37:801–813. [Google Scholar]
- 23.Swenson NG, Enquist BJ, Pither J, Thompson J, Zimmerman JK. The problem and promise of scale dependency in community phylogenetics. Ecology. 2006;87:2418–2424. doi: 10.1890/0012-9658(2006)87[2418:tpapos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 24.Webb CO. Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 25.Pennington RT, Richardson JE, Lavin M. Insights into the historical construction of species-rich biomes from dated plant phylogenies, neutral ecological theory and phylogenetic community structure. New Phytol. 2006;172:605–616. doi: 10.1111/j.1469-8137.2006.01902.x. [DOI] [PubMed] [Google Scholar]
- 26.Vamosi SM, Heard SB, Vamosi JC, Webb CO. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol Ecol. 2009;18:572–592. doi: 10.1111/j.1365-294X.2008.04001.x. [DOI] [PubMed] [Google Scholar]
- 27.Cardillo M. Phylogenetic structure of mammal assemblages at large geographical scales: Linking phylogenetic community ecology with macroecology. Philos Trans R Soc Lond B Biol Sci. 2011;366:2545–2553. doi: 10.1098/rstb.2011.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crisp MD, Trewick SA, Cook LG. Hypothesis testing in biogeography. Trends Ecol Evol. 2011;26:66–72. doi: 10.1016/j.tree.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Lomolino MV, Riddle BR, Whittaker RJ, Brown JH. Biogeography. Sunderland, MA: Sinauer Associates; 2010. [Google Scholar]
- 30.Rosenzweig ML. Species Diversity in Space and Time. Cambridge, UK: Cambridge University Press; 1995. [Google Scholar]
- 31.Morley RJ. Origin and Evolution of Tropical Rain Forests. Chichester, UK: John Wiley & Sons; 2000. [Google Scholar]
- 32.Morley RJ. Cretaceous and Tertiary climate change and the past distribution of megathermal rainforests. In: Bush MB, Flenley JR, editors. Tropical Rainforest Responses to Climate Change. Berlin: Springer; 2007. pp. 1–31. [Google Scholar]
- 33.Hortal J, et al. Ice age climate, evolutionary constraints and diversity patterns of European dung beetles. Ecol Lett. 2011;14:741–748. doi: 10.1111/j.1461-0248.2011.01634.x. [DOI] [PubMed] [Google Scholar]
- 34.Jansson R, Dynesius M. The fate of clades in a world of recurrent climatic change: Milankovitch oscillations and evolution. Annu Rev Ecol Syst. 2002;33:741–777. [Google Scholar]
- 35.Davies TJ, Buckley LB. Phylogenetic diversity as a window into the evolutionary and biogeographic histories of present-day richness gradients for mammals. Philos Trans R Soc Lond B Biol Sci. 2011;366:2414–2425. doi: 10.1098/rstb.2011.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansson R. Global patterns in endemism explained by past climatic change. Proc Biol Sci. 2003;270:583–590. doi: 10.1098/rspb.2002.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kissling WD, et al. Quaternary and pre-Quaternary historical legacies in the global distribution of a major tropical plant lineage. Glob Ecol Biogeogr. 2012 10.1111/j.1466-8238.2011.00728.x. [Google Scholar]
- 38.Dransfield J, et al. Genera Palmarum: The Evolution and Classification of Palms. Kew, UK: Royal Botanical Gardens; 2008. [Google Scholar]
- 39.Eiserhardt WL, Svenning J-C, Kissling WD, Balslev H. Geographical ecology of the palms (Arecaceae): Determinants of diversity and distributions across spatial scales. Ann Bot. 2011;108:1391–1416. doi: 10.1093/aob/mcr146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Couvreur TLP, Forest F, Baker WJ. Origin and global diversification patterns of tropical rain forests: Inferences from a complete genus-level phylogeny of palms. BMC Biol. 2011;9:44. doi: 10.1186/1741-7007-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Govaerts R, Dransfield J. World Checklist of Palms. UK: Royal Botanic Gardens, Kew; 2005. [Google Scholar]
- 42.Baker WJ, et al. Complete generic-level phylogenetic analyses of palms (Arecaceae) with comparisons of supertree and supermatrix approaches. Syst Biol. 2009;58:240–256. doi: 10.1093/sysbio/syp021. [DOI] [PubMed] [Google Scholar]
- 43.Bjorholm S, Svenning J-C, Baker WJ, Skov F, Balslev H. Historical legacies in the geographical diversity patterns of New World palm (Arecaceae) subfamilies. Bot J Linn Soc. 2006;151:113–125. [Google Scholar]
- 44.Givnish TJ. Ecology of plant speciation. Taxon. 2010;59:1326–1366. [Google Scholar]
- 45.Dransfield J, Beentje H. The Palms of Madagascar. Kew, UK: Royal Botanic Gardens and International Palm Society; 1995. [Google Scholar]
- 46.Hodel DR. A review of the genus Pritchardia. Palms. 2007;51:S1–S53. [Google Scholar]
- 47.Pintaud J-C, Baker WJ. A revision of the palm genera (Arecaceae) of New Caledonia. Kew Bull. 2008;63:61–73. [Google Scholar]
- 48.Willis KJ, McElwain JC. The Evolution of Plants. Oxford: Oxford University Press; 2002. [Google Scholar]
- 49.Greenwood DR, Wing SL. Eocene continental climates and latitudinal temperature gradients. Geology. 1995;23:1044–1048. [Google Scholar]
- 50.Svenning J-C, Borchsenius F, Bjorholm S, Balslev H. High tropical net diversification drives the New World latitudinal gradient in palm (Arecaceae) species richness. J Biogeogr. 2008;35:394–406. [Google Scholar]
- 51.Pan AD, Jacobs BF, Dransfield J, Baker WJ. The fossil history of palms (Arecaceae) in Africa and new records from the Late Oligocene (28-27 Mya) of north-western Ethiopia. Bot J Linn Soc. 2006;151:69–81. [Google Scholar]
- 52.Rull V. Neotropical biodiversity: Timing and potential drivers. Trends Ecol Evol. 2011;26:508–513. doi: 10.1016/j.tree.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 53.Hoorn C, et al. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science. 2010;330:927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- 54.Cronk QCB. Islands: Stability, diversity, conservation. Biodivers Conserv. 1997;6:477–493. [Google Scholar]
- 55.Cannon CH, Morley RJ, Bush ABG. The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc Natl Acad Sci USA. 2009;106:11188–11193. doi: 10.1073/pnas.0809865106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brummitt RK. World Geographical Scheme for Recording Plant Distributions. 2nd Ed. Pittsburgh: Hunt Institute for Botanical Documentation Carnegie Mellon University; 2001. [Google Scholar]
- 57.Kembel SW, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 58.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 59.Braconnot P, et al. Results of PMIP2 coupled simulations of the Mid-Holocene and Last Glacial Maximum—Part 1: Experiments and large-scale features. Climate of the Past. 2007;3:261–277. [Google Scholar]
- 60.Kissling WD, Carl G. Spatial autocorrelation and the selection of simultaneous autoregressive models. Glob Ecol Biogeogr. 2008;17:59–71. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.