Abstract
Plastid transcription is mediated by two distinct types of RNA polymerases (RNAPs), bacterial-type RNAP (PEP) and phage-type RNAP (NEP). Recent genomic and proteomic studies revealed that higher plants have lost most prokaryotic transcription regulators and have acquired eukaryotic-type proteins during plant evolution. However, in vivo dynamics of chloroplast RNA polymerases and eukaryotic-type plastid nucleoid proteins have not been directly characterized experimentally. Here, we examine the association of the α-subunit of PEP and eukaryotic-type protein, plastid transcriptionally active chromosome 3 (pTAC3) with transcribed regions in vivo by using chloroplast chromatin immunoprecipitation (cpChIP) assays. PEP α-subunit preferentially associates with PEP promoters of photosynthesis and rRNA genes, but not with NEP promoter regions, suggesting selective and accurate recognition of PEP promoters by PEP. The cpChIP assays further demonstrate that the peak of PEP association occurs at the promoter-proximal region and declines gradually along the transcribed region. pTAC3 is a putative DNA-binding protein that is localized to chloroplast nucleoids and is essential for PEP-dependent transcription. Density gradient and immunoprecipitation analyses of PEP revealed that pTAC3 is associated with the PEP complex. Interestingly, pTAC3 associates with the PEP complex not only during transcription initiation, but also during elongation and termination. These results suggest that pTAC3 is an essential component of the chloroplast PEP complex. In addition, we demonstrate that light-dependent chloroplast transcription is mediated by light-induced association of the PEP–pTAC3 complex with promoters. This study illustrates unique dynamics of PEP and its associated protein pTAC3 during light-dependent transcription in chloroplasts.
Plastids are DNA-containing organelles unique to plant cells and are thought to have originated from an ancestral cyanobacterial endosymbiont. Whereas cyanobacteria contain over 3,000 genes, the plastid genome in higher plants consists of small, circular, double-stranded DNA (120–150 kbp) encoding ∼120 genes for photosynthesis and gene expression machineries (1, 2), indicating massive transfer of chloroplast genes to nuclear genome during evolution (3). Vascular plants have evolved a complex transcriptional network that is mediated by two types of RNA polymerases (RNAPs): cyanobacterium-derived plastid-encoded plastid RNA polymerase (PEP) and nuclear-encoded phage-type RNA polymerase (NEP). PEP is composed of four catalytic subunits and a promoter recognition subunit, σ-factor (4). Genes for PEP core subunits, α, β, β′, and β′′ were retained in plastid genomes as rpoA, rpoB, rpoC1, and rpoC2 during plant evolution, but genes for σ-factors involved in transcription initiation, have been transferred to the nuclear genome (5), which allows the nucleus to control PEP transcription initiation in response to developmental and environmental cues (recently reviewed in ref. 6). PEP is responsible for transcription of photosynthesis genes in chloroplast in response to light. On the other hand, housekeeping genes encoding PEP core subunits and ribosomal proteins are transcribed by the phage-type NEP (7, 8). Thus, two types of RNAP have distinct roles in chloroplast transcription in higher plants.
It has been proposed that light-dependent initiation of transcription by PEP is controlled by light-induced expression of nuclear-encoded plastid σ-factors (6). Contrarily, several evidences suggest that phosphorylated PEP may tightly bind to the promoter region to arrest transcription in the dark (9–11). Dark-induced phosphorylation of PEP and/or σ-factors by redox-regulated plastid transcription kinase (PTK) may be a key step in light-dependent plastid gene transcription. Thus, molecular mechanism of light-dependent transcription in chloroplasts still remains controversial.
Plastid DNAs are densely packed into protein–DNA complexes called “plastid nucleoids,” as well as bacterial nucleoids. However, higher plants and moss have lost prokaryotic major nucleoid proteins including Hu during evolution (12, 13), whereas several eukaryotic-type proteins such as PEND (14), MFP1 (15), SiR (16), and CND41 (17) have been identified as major components of nucleoids in higher plants. Furthermore, recent proteome analysis identified eukaryotic-type chloroplast nucleoid proteins including putative DNA/RNA-binding proteins as components of a chloroplast-derived DNA–protein complex termed pTAC (plastid transcriptionally active chromosome) (18) and a blue native (BN)-PAGE separated basic PEP complex (19). These findings suggest that chloroplasts have lost most of prokaryotic nucleoid proteins involved in DNA packaging, replication, transcription, and translation and acquired eukaryotic-type chloroplast nucleoid proteins during evolution (20). Because vascular plants lack prokaryotic transcription regulators such as DNA-binding proteins and transcription elongation factors except for σ-factors, chloroplast transcription might be mediated by a unique hybrid system of prokaryotic-type RNA polymerase and eukaryotic-type accessory factors, However, the role of the nonprokryotic nucleoid proteins in plastid transcription remains largely unknown.
Earlier studies on pTAC proteins and PEP-associated proteins (PAPs) have focused mainly on in planta analyses using mutant and transgenic plants [e.g., pTAC2, pTAC6, and pTAC12 (18); PAP3/pTAC10, PAP6/FLN1, and PAP7/pTAC14 (19); ET1 (21); Trx-z (22)]. However, mutations in plastid transcription-related genes occasionally give rise to drastic pleiotropic phenotypes such as albino or pale green plants due to reduced accumulation of plastid rRNA and tRNAs, which are mainly transcribed by PEP, and impaired translation of chloroplast-encoded essential proteins. Thus, it is sometimes difficult to characterize the molecular function of pTAC proteins in chloroplast transcription using null mutants. Chromatin immunoprecipitation (ChIP) assays have been widely used to study protein–DNA interactions and map the localization of proteins to specific DNA sequences in the cell nucleus and bacterial nucleoids. However, very little work has been done using ChIP to analyze the association of PEP and its accessory proteins with transcribed regions in chloroplast genomes. Here, we examine the association of the α-subunit of PEP and pTAC3 with transcribed regions in vivo by using chloroplast chromatin immunoprecipitation (cpChIP) assays. A pTAC3 has been identified in various PEP-containing chloroplast fractions (18, 19, 23, 24). The pTAC3 contains a SAP DNA-binding domain (Pfam 02037), which is found in matrix attachment region binding protein (25) (Fig. 1A). In this study, cpChIP assays demonstrate that pTAC3 associates with the PEP complex not only during transcription initiation, but also during elongation and termination. In addition, we demonstrate light-dependent association of the PEP–pTAC3 complex with PEP promoters. This study provides unique insight into the role of the eukaryotic-type PEP accessory protein pTAC3 in PEP-dependent transcription in chloroplasts.
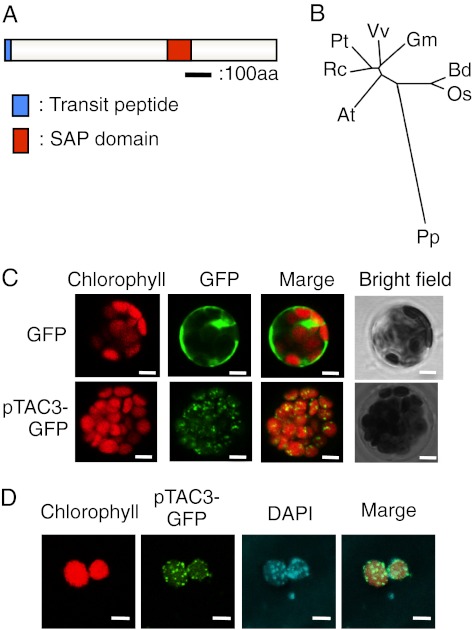
Fig. 1.
pTAC3 is a chloroplast nucleoid protein. (A) Schematic representation of pTAC3 proteins. Black bar corresponds to 100 aa in length. Blue and red boxes represent the chloroplast transit peptide and SAP domain, respectively. (B) Phylogenic tree of pTAC3 homolog proteins. Phylogenetic analysis was conducted using ClustalW based on amino acid sequences of pTAC3 proteins including At, Arabidopsis thaliana; Rc, Ricinus communis; Gm, Glycine max; Vv, Vitis vinifera; Pt, Populus trichocarpa; Os, Oryza sativa; Bd, Brachypodium distachyon; Pp, Physcomitrella patens. (C) Localization analysis of pTAC3–GFP in a protoplast transient expression system. Protoplasts prepared from wild-type A. thaliana leaves were transformed with expression plasmids harboring the GFP gene (control) or the full-length AtpTAC3 fused to GFP at its N terminus under the control of the CaMV 35S promoter. GFP fluorescence and chlorophyll autofluorescence of transformed protoplasts were observed by confocal microscopy. (Scale bar, 5 μm.) (D) DAPI staining of protoplasts expressing pTAC3–GFP. Protoplasts were disrupted by osmotic pressure, stained with DAPI, and observed by confocal microscopy. (Scale bar, 5 μm.)
Results
pTAC3 Is a Land Plant-Specific Protein That Is Localized to Plastid Nucleoids.
To investigate whether pTAC3 is an evolutionally acquired component of plastid transcription, we examined its evolutionary history. BLAST and position-specific iterative (PSI)-BLAST searches revealed that pTAC3 is conserved among land plants including moss and higher plants, but not in cyanobacteria and algae (Fig. 1B and Fig. S1), suggesting a functional conservation in embryophytes. TargetP and PSORT, subcellular localization prediction programs, suggest that AtpTAC3 (At3g04260) contains a putative transit peptide of 29 amino acid residues at the N terminus. To examine the subcellular localization of pTAC3, we expressed GFP-tagged pTAC3 in Arabidopsis. Full-length AtpTAC3 cDNA was fused to the GFP coding region and transiently expressed under the control of the constitutive CaMV35S promoter in protoplasts prepared from Arabidopsis mesophyll cells (Fig. 1C). It has been reported that GFP-fused plastid DNA-binding proteins such as PEND localized to plastid nucleoids (26). As shown in Fig. 1D, GFP-tagged AtpTAC3 appeared in small dot-like structures that were observed throughout chloroplasts and colocalized with DAPI-stained nucleoid DNA, suggesting that pTAC3 is a chloroplast nucleoid-associated protein.
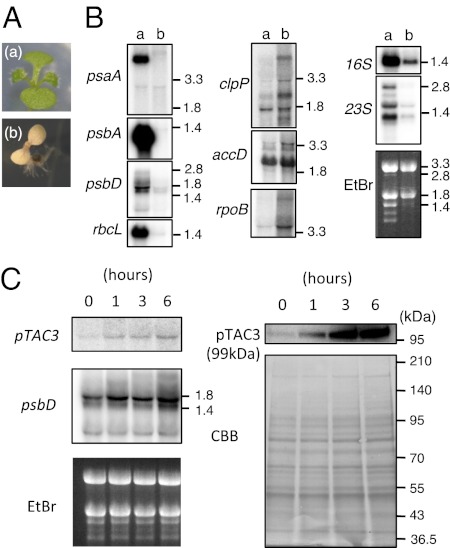
Homozygous T-DNA insertion mutants of AtpTAC3 [SALK110045 (ptac3-1)] displayed an albino phenotype (Fig. 2A and Fig. S2 A and B). To analyze the role of pTAC3 in plastid transcription, we examined global plastid gene expression patterns in ptac3-1 by using a plastid DNA macroarray. Expression of PEP-dependent photosynthesis genes, which are actively transcribed in chloroplasts, is significantly reduced in 8-d-old ptac3-1 mutant plants (Fig. S3). By contrast, knockout of pTAC3 resulted in increased accumulation of several low abundance gene transcripts, including NEP-dependent genes such as accD and PEP core subunit genes (rpoA, and the rpoB-C1-C2 operon). RNA gel blot analysis confirmed the reduced accumulation of PEP-dependent photosynthesis and rrn transcripts, and the up-regulation of NEP-dependent transcripts in ptac3-1 (Fig. 2B). This is a typical plastid gene expression pattern of mutant plants with impaired PEP transcription. Furthermore, we analyzed the developmental expression of AtpTAC3 during greening of etiolated seedlings. Wild-type Arabidopsis plants were grown in continuous darkness for 4 d and subsequently exposed to light for 1, 3, or 6 h. Both AtpTAC3 mRNA and AtpTAC3 protein are induced by light within 1 h. (Fig. 2C). The transcript levels of psbD, which are a marker for light-dependent plastid transcription, are also increased by light. These data suggest that pTAC3 may play a crucial role in PEP-dependent transcription and be essential for light-induced chloroplast development.
Fig. 2.
Analysis of pTAC3-deficient mutants. (A) Phenotype of ptac3-1. WT (a) and ptac3-1 (b) were grown on 1/2 MS medium containing 1% (wt/vol) sucrose for 8 d. (B) RNA gel blotting analysis of plastid gene expression in WT and ptac3-1. Five micrograms of RNA extracted from 8-d-old WT (a) or ptac3-1 (b) was separated on 1% (wt/vol) agarose gels, and blots were hybridized with 32P-labeled probes. Sizes determined by the position of 16S, 18S, mature 23S, and 25S ribodomal RNA, which correspond to 1.4, 1.8, 2.8, and 3.3 kbp in length, respectively. (C) RNA gel blotting (Left) and Western blotting analysis (Right) of pTAC3 expression during greening of etiolated seedlings. Wild-type Arabidopsis plants were grown in continuous darkness for 4 d, and then the etiolated plants were exposed to light for 1, 3, or 6 h. Total RNA and protein were extracted and analyzed by RNA gel blotting and Western blotting, respectively. EtBr and CBB staining image are shown below. psbD light-dependent expression is also shown.
cpChIP Analysis of pTAC3 and PEP Association with Wheat Chloroplast DNA.
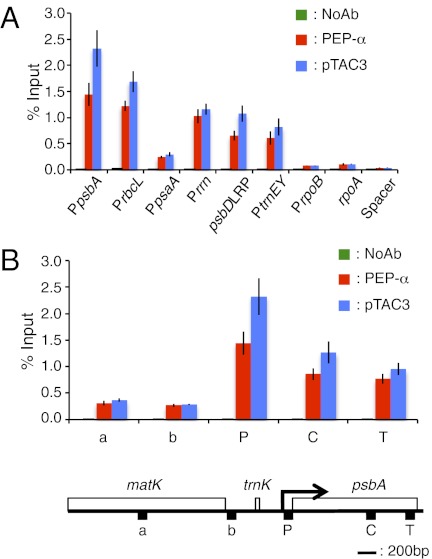
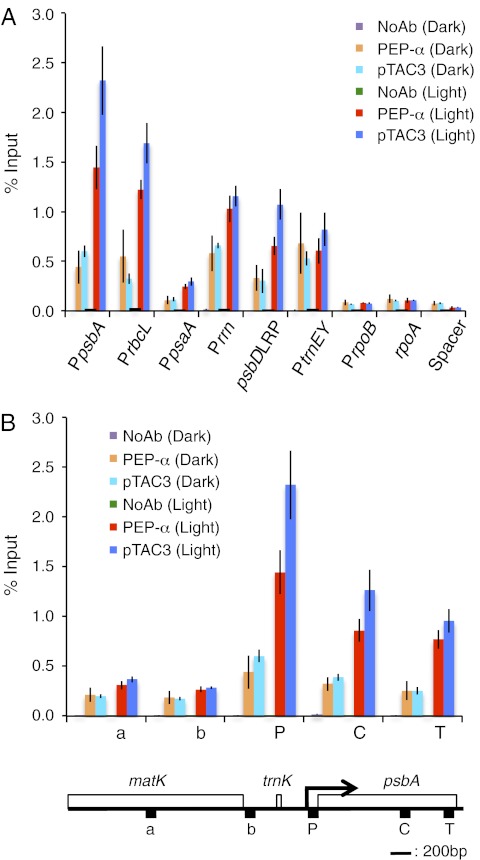
To understand the role of pTAC3 in PEP-dependent transcription in chloroplasts, we designed a modified cpChIP assay using wheat chloroplasts. First we evaluated the association of PEP α-subunit with specific regions of plastid DNA, including PEP promoters (psbA, rbcL, psaA, rrn16, psbDLRP, and trnEY), a NEP promoter (rpoB), the coding region of a NEP-dependent gene (rpoA), and the noncoding region between rps12 and rrn16 (Spacer) (Fig. S4 and Table S1). The immunoprecipitated DNA was analyzed by quantitative PCR and quantified using standard curves based on a dilution series of input samples. The data were analyzed as a percentage of the input sample (details in Materials and Methods). The cpChIP assay showed binding of PEP α-subunit to the promoter regions of PEP-dependent photosynthesis genes and the rrn operon, but not to those of NEP-dependent genes or the spacer region in vivo (Fig. 3A). By contrast, the cpChIP assay using an antibody against the non–DNA-binding chloroplast protein, glutamime synthetase 2 (GS2) resulted in a lower cpChIP-QPCR signal at the psbA promoter comparable to that at the rpoB promoter (Fig. S5). These results indicate that the cpChIP analysis can be used to detect the specific association of DNA-binding protein with chloroplast DNA. Furthermore, cpChIP signal of PEP α-subunit characteristics vary markedly among PEP promoters, suggesting that the relative association of PEP with promoters may be dependent on promoter architecture. The highest signal was detected at the psbA promoter, which is known as the most active promoter in the chloroplast genome. We further examined the distribution of PEP α-subunit along the psbA gene including the promoter, coding, and termination regions (Fig. 3B). The peak association with PEP α-subunit was localized in the promoter region, and the level decreased slightly toward the termination region of psbA, whereas the signal in the trnK-matK region, which is located upstream of psbA, was significantly lower. Less intensive ChIP signals in the coding region compared with the promoter region were also observed in rbcL and the rrn operon (Fig. S6), suggesting that PEP density decreases gradually along the transcription unit. This cpChIP assay provides the first direct evidence that PEP exclusively recognizes PEP promoters and initiates transcription in vivo, suggesting that PEP–σ complexes have high binding affinity to PEP promoters.
Fig. 3.
Association of PEP α-subunit and pTAC3 with chloroplast DNA. (A) Association of PEP α-subunit and pTAC3 with PEP promoter regions (PpsbA, PrbcL, PpsaA, Prrn16, psbDLRP, PtrnEY), a NEP promoter region (PrpoB), the coding region of the rpoA gene (rpoA), and a noncoding spacer region located between rps12 and rrn16 (Spacer) was analyzed by ChIP assay. Chloroplasts were prepared from wheat seedlings grown for 5 d in the light and subjected to ChIP assays using antibodies against PEP α-subunit and pTAC3. NoAb, no antibody control. Enriched DNA was quantified by qPCR. The amount of immunoprecipitated DNA in each sample is presented as a percentage of the total input chromatin. Mean values and SDs of three independent experiments are shown. (B) Spatial association of α-subunit and pTAC3 along the psbA transcription unit. Data are shown as in A. Schematic gene map of the matK-psbA region is shown below. Horizontal black bar represents 200 bp. Arrow indicates the transcription start site of the psbA gene and the direction of transcription. DNA regions corresponding to the psbA promoter (P), coding region (C), terminator (T), and two units (a and b) in loci upstream of psbA are shown.
We also examined the association of pTAC3 with chloroplast DNA via cpChIP assays. As in the case of PEP α-subunit, pTAC3 preferentially binds to promoter regions of PEP-dependent genes, but not of NEP-dependent genes, suggesting a role for pTAC3 in the PEP complex (Fig. 3A and Fig. S3). We further examined the local patterns of spatial association of pTAC3 with the psbA transcription unit (Fig. 3B). We found that pTAC3 binds not only to the promoter region but also the transcription elongation region during transcription, suggesting that pTAC3 as well as PEP is associated with chloroplast DNA along the psbA transcription unit. Thus, it is unlikely that pTAC3 binds to specific sequences in the psbA promoter; rather it may be an important component of the PEP complex in chloroplasts.
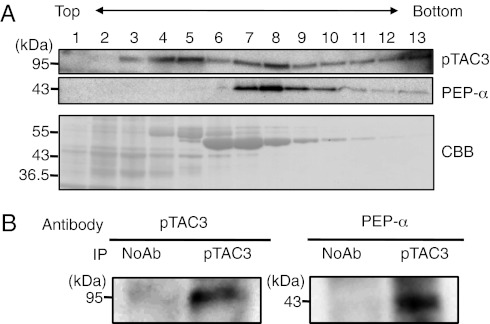
To examine further the interaction between pTAC3 and PEP, we isolated the PEP complex by glycerol density gradient centrifugation and probed for the presence of pTAC3 by Western blotting. The molecular weight of the PEP complex from wheat chloroplasts was estimated as around 700 kDa, as reported previously in mustard (27). pTAC3 was detected in two fractions, corresponding to a peak containing PEP α-subunit (fraction 8) and a lower molecular weight fraction (roughly estimated to 200–400 kDa; fractions 4 and 5) (Fig. 4A). These results suggest that pTAC3 in wheat chloroplasts is associated mainly with the PEP complex, although a portion of the pTAC3 may form another complex without PEP. Furthermore, immunoprecipitation assays using pTAC3 antibody with wheat chloroplast extracts demonstrated the presence of α-subunit in the pTAC3 immunoprecipitated complex (Fig. 4B). These results demonstrate that pTAC3 has a direct function in PEP transcription.
Fig. 4.
Analysis of the pTAC3 protein complex. (A) Separation of the PEP complex by glycerol density gradient centrifugation. Total chloroplast proteins prepared from wheat grown under continuous light for 6 d were loaded onto a 10–30% (vol/vol) glycerol density gradient and separated by centrifugation. Thirteen fractions were collected from top to bottom and analyzed by immunoblotting with anti-PEP α-subunit or pTAC3 antibodies. Gel staining with CBB is also shown. (B) Immunoprecipitation analysis of the pTAC3 complex. Total wheat chloroplast extracts were subjected to immunoprecipitation with pTAC3 antibody or without (NoAb) and analyzed by immunoblotting with anti-PEP α-subunit and pTAC3 antibodies.
Light-Dependent Association of the PEP–pTAC3 Complex with Chloroplast DNA.
PEP transcription activity is greatly stimulated by light in mature wheat chloroplasts (28). We therefore examined the light-dependent association of PEP with promoter regions of several plastid genes in vivo. Chloroplasts were prepared from wheat seedlings grown for 5 d in the light and then dark adapted for 24 h or seedlings reilluminated for 6 h after the 24-h dark adaptation. Immunoblot analysis revealed the constitutive accumulation of both PEP α-subunit and pTAC3 upon light illumination in wheat seedlings (Fig. S7), suggesting that the expression level of the PEP complex is not affected by light. On the other hand, the cpChIP assays showed that the relative amount of PEP α-subunit associated with the promoter region of photosynthesis genes including psbA, psbD, psaA, and rbcL, and ribosomal RNA rrn16, was two- to fivefold higher in the illuminated chloroplasts than in the dark-adapted chloroplasts (Fig. 5A). Amounts of chloroplast DNA in input samples between both conditions were not significantly different (Fig. S8). Thus, the light-dependent cpChIP signals would be indicative of light-dependent association of PEP α-subunit and pTAC3 to chloroplast DNA. Moreover, we could not detect tight binding of PEP α-subunit to the promoter region of photosynthesis genes in the dark-adapted chloroplasts, suggesting a limited role for transcription arrest mediated by dark-induced phosphorylation of PEP subunits and σ-factors. In addition to promoter regions, we also detected light-dependent association of PEP α-subunit with coding and termination regions of psbA (Fig. 5B). These results demonstrate that recruitment of PEP to its target promoters is dependent on light. On the other hand, light-dependent association of PEP with NEP-dependent genes including rpoA and rpoB and the spacer region was not detected. Furthermore, ChIP analysis of pTAC3 showed that light accelerated the association of pTAC3 with not only the PEP promoter region but also the coding region in the psbA transcription unit (Fig. 5 A and B) similar to the distribution pattern of PEP α-subunit, suggesting that pTAC3 associates with PEP-dependent transcribed regions as a component of a large PEP complex in a light-dependent manner.
Fig. 5.
Analysis of light-dependent association of PEP α-subunit and pTAC3 with chloroplast DNA. (A) Association of PEP α-subunit and pTAC3 with chloroplast DNA in response to light. Chloroplasts were prepared from wheat seedlings grown for 5 d in the light and then dark adapted for 24 h (dark) or seedlings reilluminated for 6 h after the 24-h dark adaptation (light). ChIP was performed to determine the association level of PEP α-subunit and pTAC3 with PEP promoter regions (PpsbA, PrbcL, PpsaA, Prrn16, psbDLRP, and PtrnEY), a NEP promoter (PrpoB), the coding region of the rpoA gene (rpoA), and a noncoding region located between rps12 and rrn16 (Spacer), using anti-PEP α-subunit or pTAC3 antibodies or no antibody (NoAb). The immunoprecipitated DNA was analyzed by quantitative PCR and quantified via standard curves based on a dilution series of input samples. The amount of immunoprecipitated DNA in each sample is presented as a percentage of the total input chromatin. Mean values and SDs of three independent experiments are shown. (B) Association level of PEP α-subunit and pTAC3 with regions of the psbA transcription unit in response to light. Data are presented as in A. Schematic gene map of the matK-psbA region is shown as in Fig. 3B.
Discussion
This study reveals light-dependent associations of PEP and pTAC3 with chloroplast DNA in vivo using cpChIP assays. ChIP assays have been widely used to detect specific binding sites for transcription factors (29), distribution patterns of several modified histones (30), and trafficking of RNAP and its associated proteins on genomic DNA (31). In chloroplasts, a few studies have shown the association of endogenous (Whirly1) and recombinant (LacI) transcription factors with chloroplast promoters in vivo by using ChIP assays (32, 33). However, in vivo dynamics of chloroplast RNA polymerases and/or their associated proteins have not been directly characterized experimentally. Unlike cyanobacteria, the chloroplasts of higher plants have two types of RNA polymerase, PEP and NEP. Transcriptome analyses of PEP- or NEP-deficient mutants and in vitro transcription analyses using isolated PEP and NEP have shown that PEP and NEP preferentially initiate transcription from bacterial-type and phage-type promoters, respectively. Here, we provide direct evidence that PEP exclusively associates in vivo with PEP promoters of photosynthesis genes and the rRNA operon, but not with NEP promoters by using cpChIP assay (Fig. 3), suggesting selective and accurate recognition of PEP promoters by PEP.
It has been shown that several PEP promoters including psbA, psbD LRP, rbcL, psaA, and rrn P1 are regulated by unique cis-elements, termed as the TATA-box, PGT-box, and AAG-box, CDF1-binding site, region U, and RUA, respectively, which are located upstream of or within the core promoter (28, 34–37) and recognized by promoter-specific transcription factors. However, sequence specific DNA-binding proteins have not been identified and characterized in chloroplasts of higher plants. Chloroplast protein pTAC3 contains a putative DNA-binding SAP domain. Here we show that pTAC3 is localized in chloroplast nucleoids and is essential for PEP-dependent transcription, suggesting that pTAC3 may be a chloroplast transcription factor that regulates PEP-dependent transcription. Thus, we further searched for pTAC3-binding regions on plastid DNA by using cpChIP assays. We found that pTAC3 binds to the transcribed regions of all PEP-dependent genes examined (psbA, psaA, psbD, rbcL, and rrn promoters), but not to a specific cis-element in a particular promoter, suggesting that pTAC3 is a PEP-associated general, rather than sequence-specific transcription factor. In contrast to PEP-dependent transcribed loci, pTAC3 associates weakly with NEP-dependent transcribed loci, suggesting that pTAC3 does not associate with the NEP transcription complex. However, chloroplasts exhibit very low NEP-dependent transcription activity. NEP proteins are thought to be present at very low levels in leaf chloroplasts, because they were not detected in the proteomic analysis of whole chloroplast proteins and pTAC fractions. Thus, there remains a possibility that the cpChIP assay could not detect the association of pTAC3 with NEP-dependent transcribed loci, due to the low levels of accumulation of NEP in chloroplasts.
In bacteria, it is proposed that RNAP accessory proteins modulate the promoter accessibility to RNAP by altering the 3D structure of the nucleoid. The nucleoid proteins factor for inversion stimulation (FIS) and histone-like nucleoid structuring protein (H-NS) are associated with RNAP and bind chromosomal DNA at many regions (38). Furthermore, FIS and H-NS were copurified with RNAP subunits and ribosomes in Escherichia coli (39). Similarly, cpChIP assays demonstrated that pTAC3 associates with PEP-dependent transcribed regions of photosynthesis and rRNA genes together with PEP α-subunit. Density gradient and immunoprecipitation analyses of PEP demonstrate that pTAC3 is an essential component of PEP in chloroplasts. Furthermore, pTAC3 is crucial for PEP activity and its expression increases dramatically during chloroplast development, as PEP activity increases. Taken together, these results suggest that pTAC3 is an essential component of the PEP complex and critically involved in chloroplast differentiation from immature plastids. Considering the eukaryotic origin of the SAP domain as a DNA-binding motif, our findings suggest that pTAC3 was acquired from host cells during plant evolution to enhance and/or maintain transcription of photosynthesis genes in higher plants.
Our ChIP analysis of PEP α-subunit shows that PEP preferentially associates with the promoter region rather than the coding region (Fig. 3B and Fig. S3), suggesting that the PEP complex remains at the promoter-proximal region. E. coli ChIP analysis using specific antibodies for RNAP core subunits revealed that there is also preferential association of the RNAP core with the promoter-proximal region of transcribed genes and decreased RNAP-binding density downstream from the promoter (31). The accumulation of RNAP at the promoter-proximal region in E. coli is controlled mainly by interactions between the elongating complex of RNAP and elongation regulators, such as NusA, NusG, and Rho. The Arabidopsis genome encodes only one putative bacterial-type elongation factor, pTAC13 (At3g09210) containing a KOW domain, which is characteristic for members of the NusG protein family (40). Furthermore, maize ET1, which resembles the eukaryotic elongation factor TFIIS, was identified as a component of the chloroplast transcriptionally active fraction, and plants deficient in ET1 showed an aberrant chloroplast development phenotype and decreased PEP-dependent transcription (21), suggesting that bacterial-type pTAC13 and eukaryotic-type ET1 may be involved in PEP elongation. In this study, we demonstrate that pTAC3 is associated with PEP through transcription initiation and elongation steps, suggesting a possible role for pTAC3 in the regulation of PEP transcription elongation in chloroplasts.
Light-dependent activation of PEP transcription may be controlled by PTK and plastid redox signaling. It has been proposed that phosphorylated PEP and/or σ-like factors tightly bind to promoters to arrest transcription under dark conditions (11). If PEP is trapped at promoter regions in the dark, ChIP signals at PEP promoters would not decrease in dark-adapted leaves. The present data, however, showed that ChIP signals at both promoters and coding regions of PEP-dependent photosynthesis and rRNA genes were reduced in dark-adapted wheat seedlings, suggesting that PEP dissociates from chloroplast genomic DNA in the dark. Thus, it is likely that light regulates the association of the PEP–pTAC3 complex with the promoter region possibly through the light-dependent expression of σ-factors, rather than via regulation of σ-factor phosphorylation by PTK.
In conclusion, here we present a characterization of in vivo dynamics of chloroplast RNA polymerase PEP and eukaryotic-type nucleoid protein pTAC3 with chloroplast DNA by using cpChIP analysis. We show that PEP α-subunit preferentially associates with the promoter-proximal regions of PEP-dependent photosynthesis and rRNA genes in a light-dependent manner, but not to NEP promoters, suggesting that light-dependent chloroplast transcription is mediated by accurate recognition of PEP promoters by PEP in response to light. Furthermore, we show that eukaryotic-type nucleoid protein pTAC3 is essential for PEP activity and chloroplast development and associates with the PEP complex not only during transcription initiation, but also during elongation and termination. As pTAC3 has been acquired early during land plant evolution, understanding the molecular function of pTAC3 should not only provide insights into the mechanisms enabling developmental regulation of plastid transcription but also offer a unique way of understanding plant evolution.
Materials and Methods
Plant Material.
The Arabidopsis ptac3-1 (SALK_110045) mutant (Columbia ecotype) was identified from the T-DNA insertion mutant line collection generated at the Salk Institute. For expression analysis, wild-type (Columbia) and ptac3-1 were grown on half-strength MS medium containing 1% (wt/vol) sucrose under 16-h light and 8-h dark conditions (normal light; 50–60 μmol ⋅ m−2s−1). Wheat (Triticum aestivum) seeds were grown on vermiculite at 25 °C under continuous white light (50 μmol ⋅ m−2s−1) for 5–6 d. Details regarding the identification of the mutant allele of pTAC3 are included in SI Materials and Methods.
Chloroplast ChIP Assay.
Detailed procedures for cpChIP assays are included in SI Materials and Methods.
Plastid Gene Expression Analysis.
Details for RNA extraction, RNA gel blotting analysis, and construction and analysis of plastid DNA tiling macroarrays are included in SI Materials and Methods.
Localization Analysis by Transient Expression in Protoplasts.
Protoplast transient expression assays were performed as described previously (41). Details for construction of pTAC3–GFP expression plasmids are included in SI Materials and Methods.
Protein Analysis.
Details regarding antibody materials, protein extraction from isolated chloroplasts, protein complex separation via glycerol density gradient centrifugation, immunoprecipitation, and immunoblotting analysis are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. M. H. Sato for helpful discussions, Mr. Y. Motomura for technical assistance, and the Salk Institute for providing Arabidopsis insertion mutants. This work was supported by Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid 21007485 (to Y.Y.), 22004564 (to T.S.), and 2020060 (to Y.N.), and by the Private University Strategic Research Foundation Support Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119403109/-/DCSupplemental.
References
- 1.Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko T, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement) DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 3.Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu J, Bogorad L. Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci USA. 1990;87:1531–1535. doi: 10.1073/pnas.87.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allison LA. The role of sigma factors in plastid transcription. Biochimie. 2000;82:537–548. doi: 10.1016/s0300-9084(00)00611-8. [DOI] [PubMed] [Google Scholar]
- 6.Lerbs-Mache S. Function of plastid sigma factors in higher plants: Regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol. 2011;76:235–249. doi: 10.1007/s11103-010-9714-4. [DOI] [PubMed] [Google Scholar]
- 7.Hedtke B, Börner T, Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;277:809–811. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- 8.Hajdukiewicz PT, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiller K, Eisermann A, Link G. The chloroplast transcription apparatus from mustard (Sinapis alba L.). Evidence for three different transcription factors which resemble bacterial sigma factors. Eur J Biochem. 1991;198:93–99. doi: 10.1111/j.1432-1033.1991.tb15990.x. [DOI] [PubMed] [Google Scholar]
- 10.Tiller K, Link G. Sigma-like transcription factors from mustard (Sinapis alba L.) etioplast are similar in size to, but functionally distinct from, their chloroplast counterparts. Plant Mol Biol. 1993;21:503–513. doi: 10.1007/BF00028807. [DOI] [PubMed] [Google Scholar]
- 11.Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L.) EMBO J. 1993;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi T, et al. Detection and localization of a chloroplast-encoded HU-like protein that organizes chloroplast nucleoids. Plant Cell. 2002;14:1579–1589. doi: 10.1105/tpc.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karcher D, Köster D, Schadach A, Klevesath A, Bock R. The Chlamydomonas chloroplast HLP protein is required for nucleoid organization and genome maintenance. Mol Plant. 2009;2:1223–1232. doi: 10.1093/mp/ssp083. [DOI] [PubMed] [Google Scholar]
- 14.Sato N, Albrieux C, Joyard J, Douce R, Kuroiwa T. Detection and characterization of a plastid envelope DNA-binding protein which may anchor plastid nucleoids. EMBO J. 1993;12:555–561. doi: 10.1002/j.1460-2075.1993.tb05687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong SY, Rose A, Meier I. MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 2003;31:5175–5185. doi: 10.1093/nar/gkg693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chi-Ham CL, Keaton MA, Cannon GC, Heinhorst S. The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol Biol. 2002;49:621–631. doi: 10.1023/a:1015500431421. [DOI] [PubMed] [Google Scholar]
- 17.Murakami S, Kondo Y, Nakano T, Sato F. Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett. 2000;468:15–18. doi: 10.1016/s0014-5793(00)01186-8. [DOI] [PubMed] [Google Scholar]
- 18.Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 2011;157:1043–1055. doi: 10.1104/pp.111.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato N. Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 2001;6:151–155. doi: 10.1016/s1360-1385(01)01888-x. [DOI] [PubMed] [Google Scholar]
- 21.da Costa e Silva O, et al. The Etched1 gene of Zea mays (L.) encodes a zinc ribbon protein that belongs to the transcriptionally active chromosome (TAC) of plastids and is similar to the transcription factor TFIIS. Plant J. 2004;38:923–939. doi: 10.1111/j.1365-313X.2004.02094.x. [DOI] [PubMed] [Google Scholar]
- 22.Arsova B, et al. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell. 2010;22:1498–1515. doi: 10.1105/tpc.109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki JY, et al. Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J. 2004;40:164–172. doi: 10.1111/j.1365-313X.2004.02195.x. [DOI] [PubMed] [Google Scholar]
- 24.Schröter Y, Steiner S, Matthäi K, Pfannschmidt T. Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics. 2010;10:2191–2204. doi: 10.1002/pmic.200900678. [DOI] [PubMed] [Google Scholar]
- 25.Aravind L, Koonin EV. SAP: a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 26.Terasawa K, Sato N. Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 2005;46:649–660. doi: 10.1093/pcp/pci070. [DOI] [PubMed] [Google Scholar]
- 27.Pfannschmidt T, Link G. Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol. 1994;25:69–81. doi: 10.1007/BF00024199. [DOI] [PubMed] [Google Scholar]
- 28.Satoh J, et al. Developmental stage-specific multi-subunit plastid RNA polymerases (PEP) in wheat. Plant J. 1999;18:407–415. doi: 10.1046/j.1365-313x.1999.00465.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 31.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008;36:5152–5165. doi: 10.1093/nar/gkn492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell CA, Gray JC. Binding of lac repressor-GFP fusion protein to lac operator sites inserted in the tobacco chloroplast genome examined by chromatin immunoprecipitation. Nucleic Acids Res. 2010;38:e145. doi: 10.1093/nar/gkq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satoh J, Baba K, Nakahira Y, Shiina T, Toyoshima Y. Characterization of dynamics of the psbD light-induced transcription in mature wheat chloroplasts. Plant Mol Biol. 1997;33:267–278. doi: 10.1023/a:1005799001271. [DOI] [PubMed] [Google Scholar]
- 35.Lam E, Hanley-Bowdoin L, Chua NH. Characterization of a chloroplast sequence-specific DNA binding factor. J Biol Chem. 1988;263:8288–8293. [PubMed] [Google Scholar]
- 36.Cheng MC, Wu SP, Chen LF, Chen SC. Identification and purification of a spinach chloroplast DNA-binding protein that interacts specifically with the plastid psaA-psaB-rps14 promoter region. Planta. 1997;203:373–380. doi: 10.1007/s004250050203. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki JY, Sriraman P, Svab Z, Maliga P. Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA polymerase in tobacco and other higher plants. Plant Cell. 2003;15:195–205. doi: 10.1105/tpc.007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 40.Steiner T, Kaiser JT, Marinkoviç S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsunoyama Y, et al. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc Natl Acad Sci USA. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.