Summary
In this issue of Dev. Cell, Haskin et al. describe a molecular link between an apoptotic cell receptor, a branch of the unfolded protein response, and innate immunity. The finding that the plasma membrane phagocytic receptor Ced-1 promotes the expression of unfolded response genes suggests a novel regulatory mechanism for the control of this primitive immune system that may couple the presence of pathogen with the production of secreted antimicrobial effectors.
The innate immune system is the first line of defense against pathogens. Germline encoded pattern recognition receptors (PRRs), and their recognition of highly conserved microbe-associated molecular patterns (MAMPs), enable the innate system to discriminate between different types of pathogens and also to generate an appropriate immune response (Akira et al., 2006). Once activated, these pattern recognition receptors initiate signaling cascades that lead to the rapid induction of appropriate anti-microbial effectors. Because these patterns are present both on pathogenic and non-pathogenic microbes, other signals must exist to alert the organism to the infection. There is increasing evidence for a two-step model that involves the recognition of both non-self and of pathogen derived ‘danger signals’ (Matzinger, 2002). These damage-associated molecular patterns (DAMPs) are thought to include components of dying or injured cells which act as a signal to alert cells to a pathogenic encounter. The innate immune system of many organisms includes both specialized mechanisms as well as intrinsic mechanisms which give all cells the capacity to respond to infection. These intrinsic responses have only recently been recognized as an essential component of immunity and include apoptosis, autophagy, as well as the production of some classes of anti-microbial effectors. How the innate immune system is orchestrated to detect and respond to pathogenic microbes while inhabited by large numbers of commensals is unclear.
C. elegans is a free-living soil-dwelling nematode that consumes and is colonized by live bacteria, some of which are pathogenic to this organism. Recent genetic and functional genomic studies have revealed some of the mechanisms at play that control microbial infection of this organism (Kim and Ausubel, 2005). Similar to higher organisms, the response to infection is characterized by the activation of a conserved ASK1-MAP kinase signaling cascade, which regulates the production of anti-microbial peptides (AMPs) (Pujol et al., 2008). AMPs are produced at high levels and are likely secreted by the exocytic pathway. Another arm of the C. elegans innate immune system involves the canonical cell death machinery. It has been shown that the execution machinery is required in the germline to protect against a subset of microbes. However, there does not seem to be a general requirement for apoptosis per se during immune challenge in C. elegans (Kim and Ausubel, 2005). Importantly, the upstream pattern recognition receptors that recognize non-self or their microbial patterns have not yet been identified. Moreover, how C. elegans distinguishes between pathogenic challenge, dead bacteria, and commensals has also not yet been established.
In this study, the authors identify Ced-1, classically identified as the apoptotic corpse receptor required for the uptake of dead cells, as required for the innate immune response to S. enterica (Haskins et al., 2008). While Ced-1 has homology to scavenger receptors and binds to dying cells, its ligand remains unknown (Yu et al., 2008; Zhou et al., 2001). Loss of function analysis demonstrated that Ced-1 is required for immunity to S. enterica, but not as a component of the classical apoptotic cascade (Haskins et al., 2008). Transcriptional profiling revealed a new role for this receptor in host defense, through the activation of a set of genes previously established to play a role in the unfolded protein response (Haskins et al., 2008).
The unfolded protein response (UPR) is a highly conserved cellular response that matches endoplasmic reticulum secretory load with protein folding capacity. The cellular response to ER misfolded proteins includes increased synthesis of ER chaperones, as well as inhibition of ER protein load via decreased ER protein synthesis (Ron and Walter, 2007). Recent work in C. elegans has shown that the IRE1/XBP1 branch, which activates the transcriptional upregulation of ER chaperones is sufficient for most inducible responses to ER protein misfolding (Shen et al., 2005). However, in the absence of a functional IRE1 response, acute ER stress induces the expression of a related set of genes referred to as Abu genes (activated in a blocked unfolded protein response) (Urano et al., 2002). Like ced-1, Abu genes are homologous to mammalian scavenger receptors, suggesting that ced-1 might be a distantly related abu gene. Interestingly, abu genes are likely to be present in the ER and their disruption causes constitutive UPR activation as well as enhancement of ER-associated degradation (ERAD) pathway mutants. Together, these data suggest that the abu genes may function to promote the folding of a subset of ER client proteins.
How might the membrane protein ced-1 link innate immunity to the unfolded protein response? Given the known function of ced-1 in apoptotic cell engulfment and its homology to mammalian scavenger receptors, one possibility is that cellular ‘debris’ may be bound by ced-1 thereby promoting their engulfment and delivery to the ER via ced-1 endocytosis. Ced-1 may therefore represent a DAMP receptor. Alternatively, Ced-1 may act directly as a PRR binding to non-self MAMPs. Once bound, and trafficked to the ER, these molecules could present as non-native peptides and activate the unfolded protein response. Such pathogen-induced UPR activation may be necessary to induce specialized branches of the UPR that increase ER folding capacity to accommodate the specialized synthesis, folding, and/or secretion of antimicrobial peptides (AMPs), which are rapidly induced in response to infection. Therefore, Ced-1 might be critical for activation specific folding pathways needed to coordinate the response to infection and the Abu genes represent one set of Ced-1 targets necessary for this response. While infection with a number of gram positive and negative pathogenic bacteria is not known to stimulate Abu gene expression, it should still be interesting to determine how UPR signaling pathways and target genes respond to different models of infection and whether individual branches of the UPR are specifically required for immunity.
Alternatively, ced-1 could also affect immunity responses through less direct mechanisms, all of which might impact AMP synthesis, folding, secretion, or expression or other unknown innate immune mechanisms. Clearly the repertoire of cellular innate immune response is intricate and likely functions as a network of interconnected system. Ced-1 provides an exciting model for how seemingly distinct pathways may be linked and should provide an interesting paradigm for understanding the complex interactions that make up host immune responses in other systems.
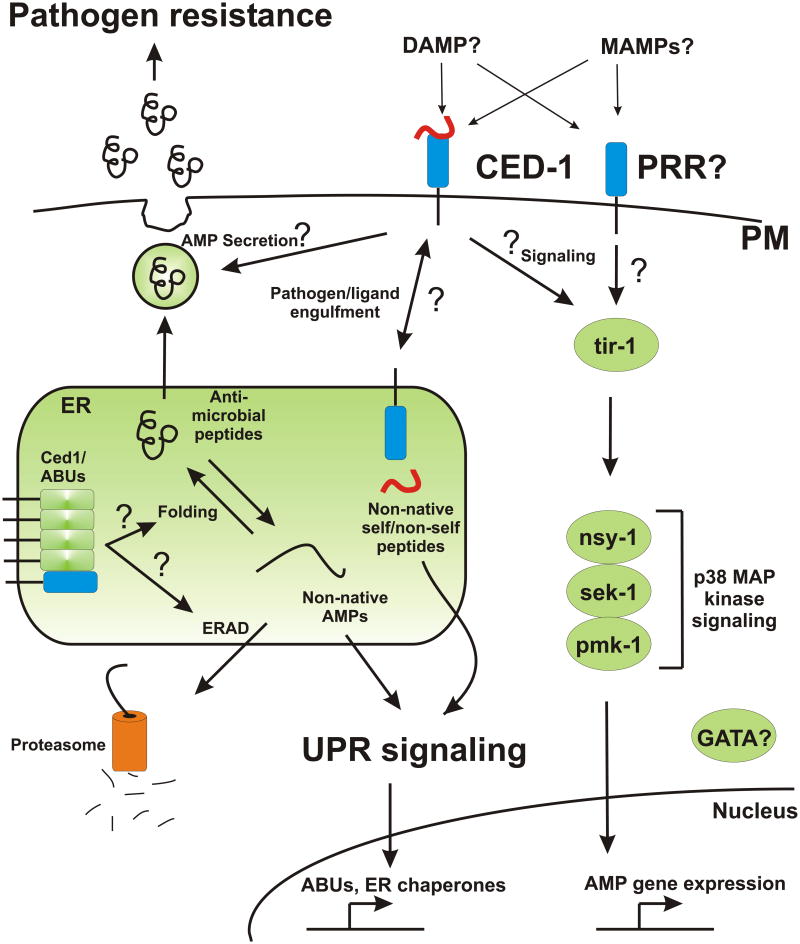
Figure 1.
Ced-1 may coordinate multiple mechanisms to link innate immunity to the Unfolded Protein Response. During pathogen infection, Ced-1 mediated-engulfment and ER delivery of self or non-self ‘damage signals’ might trigger the Unfolded Protein Response, possibly through acute elevation of ER protein load. Increased expression of UPR target genes, such as ER chaperones and Abu genes might, in turn, might be required to facilitate the folding and/or maturation of secreted antimicrobial peptides, which are also induced by pathogen infection via a conserved p38 MAP kinase signaling pathway. Ced-1 might also regulate immune responses through interactions with other cellular mechanisms, such as ERAD, secretory pathways, or direct regulation of MAP kinase-controlled AMP production.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Haskins KA, Russell RF, Gaddis N, Dressman HK, Agallay A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Developmental Cell. 2008 doi: 10.1016/j.devcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct Innate Immune Responses to Infection and Wounding in the C. elegans Epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]