Abstract
Malaria is a devastating parasitic disease that afflicts one-third of the world’s population. Commonly used malaria drugs address few targets and their efficacy is being undermined by parasite resistance. Most therapeutics target blood stage malaria, while only few compounds are active against malaria’s liver stage, the first stage of the Plasmodium parasite’s life cycle within the human host. The identification of inhibitors active against liver stage malaria would benefit both the development of chemical probes to elucidate the poorly understood biology of this phase of the parasite’s life cycle and potentially provide agents for preventing and eliminating the disease. Here, we report on the development of a live cell parasite traversal assay in 384-well format amenable to high-throughput screening that exploits the wounding of liver cells by the parasite. This method identifies small molecules that may inhibit the parasites actin-myosin motor system. The traversal assay, in addition to established methods, was used to evaluate the activity of halofuginone, a synthetic halogenated derivative of the natural alkaloid febrifugine, against liver stage Plasmodium berghei parasites. Halofuginone was found to inhibit P. berghei sporozoite load in HepG2 cells with an IC50 of 17 nM. While the compound does not affect parasite traversal through human liver cells, an inhibition time course assay indicates that it affects essential processes in both early and late stage parasite development.
Keywords: halofuginone, high-throughput screening, malaria, sporozoite, structure-activity relationships
Introduction
Malaria’s burden of mortality and morbidity continues to rise in several developing countries in Africa, South America and Asia.[1] The current suite of antimalarial agents targets only a handful of metabolic processes, primarily in the parasite’s blood stage, and drugs that inhibit other essential parasite pathways are needed to address prophylaxis and eradication.[2]
Parasites from the genus Plasmodium cause malaria[3] and they enter humans with the bite of an infected Anopheles mosquito. Sporozoites, the developmental form of the parasite transferred to the human host, travel from the dermis through the blood stream to the liver. On their route to the liver, motile sporozoites traverse, or migrate through several cells before infecting hepatocytes.[3] When parasite traversal is complete the sporozoites propagate, yielding tens of thousands of merozoites, the developmental form that infects red blood cells, within a few days.[3] The liver stage is asymptomatic, but once released from the liver the merozoites begin the cyclic blood stage that causes malaria’s characteristic symptoms. Some Plasmodium species, such as P. vivax and P. ovale, also have a dormant stage in the human liver termed hypnozoites that is difficult to target.[4] Activation of hypnozoites leads to relapses, which contribute significantly to malaria’s burden.
Drug development has traditionally focused on the blood stage of the parasite,[5] while only few chemical and genetic tools exist to investigate liver stage processes. Unlike liver stage malaria parasites, blood stage parasites can be maintained in cell culture, which greatly facilitates work with this Plasmodium form. To study liver stage parasites, viable Plasmodium sporozoites must be acquired through the dissection of live infected mosquitoes. Once these sporozoites infect liver cells, they develop into a form that can no longer invade liver cells. Few compounds with activity against liver stage sporozoites have been identified and many blood stage inhibitors, like artemisinin, are inactive against liver stage infection.
Primaquine remains the clinically used drug to clear P. vivax hypnozoites[2b] despite its many liabilities and a relatively low potency (IC50 ~10 μM) in vitro.[6] Additionally, primaquine can cause hemolytic anemia in people with glucose-6-phosphate dehydrogenase deficiency (G6PD), which is the most common enzyme deficiency in malarious regions of Africa, South America and Asia (reviewed in[7]). Another malaria drug, atovaquone, is a nanomolar inhibitor of liver stage malaria but is not effective against the dormant hepatic stage of P. vivax.[4]
Systematic searches for liver stage inhibitors could focus on compounds that prevent sporozoite traversal through cells, invasion of hepatocytes, or compounds that inhibit the development of sporozoites in their final host cell.[8] To date, methods have been developed to evaluate parasite propagation in 96-well and 384-well microtiter plates,[8-9] but there is no technique for the efficient analysis of parasite traversal applicable to a high-throughput screening platform. Here, we report the development of a miniaturized assay platform to profile inhibitors of Plasmodium sporozoite liver cell traversal in a 384-well microtiter plate format. Additionally, we have identified halofuginone (Fig. 1) as a potent inhibitor of sporozoite propagation within liver cells. Halofuginone is a synthetic derivative of febrifugine, a natural product isolated from the Chinese herb Dichroa febrifuga.[10] Both compounds have been widely recognized for their exceptional activity against blood stage malaria parasites and their unique mode of action that appears distinct form other antimalarials, however, neither compound has been evaluated for activity against liver stage parasites.[11] In this work we discovered that halofuginone has low nanomolar potency, similar to atovaquone, at reducing P. berghei sporozoite load in HepG2 cells. Halofuginone, like primaquine and atovaquone, does not affect sporozoite traversal. While the molecular target of halofuginone inhibition remains to be determined, it is likely common to both blood and liver stage parasites.
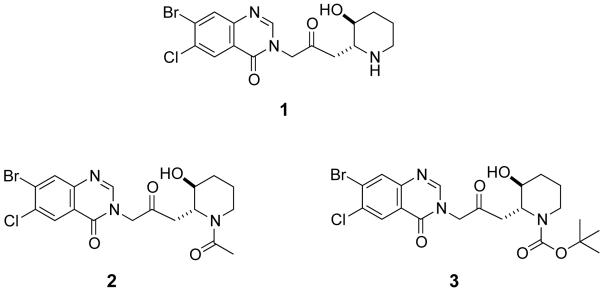
Figure 1.
Structure of halofuginone (1) and N-acetyl (2) and N-Boc (3) derivatives of halofuginone.
Results and Discussion
A limited number of compounds have been identified as inhibitors of the liver stage of the Plasmodium parasite. Current antimalarial drug discovery is focused on the parasite’s blood stages, and most mainstay malaria drugs, such as artemisinin and chloroquine,[2b] are inactive against the Plasmodium liver stages (Table 1), which represents the preferred life cycle stage for malaria prevention and prophylaxis. Differential gene expression and proteomic analysis during various life stages of the parasite indicates that some processes are essential only during specific life stages while others are important for every form of the parasite.[12] Unfortunately, the high percentage of genes with unknown function (~50%) makes it difficult to predict which genes are selective drug targets.[13] Halofuginone is one of the most potent known inhibitors of the malaria parasite’s blood stage,[11] however, activity against other Plasmodium life stages has not been investigated. Here, we utilized an in vitro infection system to evaluate if the compound targets a process that is essential for the Plasmodium parasite’s liver stage.
Table 1.
Activity of compounds against blood and liver stage Plasmodium parasites.
| Compound[[a]] | Blood stage P. falciparum 3D7 IC50 (nM) |
Liver stage P. berghei IC50 (nM) |
|---|---|---|
| Primaquine | 794[[a]] | 7500 |
| Atovaquone | 0.66[[b]] | 3 |
| Artemisinin | 46[[c]] | NO[[d]] |
| 1 | 0.7 | 17 |
| 2 | NO | NO |
| 3 | NO | NO |
Determined with P. falciparum Dd2.
Baniecki et al (2006) Antimicrob. Agent Chemother. 51 716-723.
Walsh et al (2007) Bio. Med. Chem. Lett. 17 3599-3602.
NO, not observed.
Halofuginone inhibits liver stage malaria
HepG2 cells, a human hepatoma cell line, were infected with P. berghei ANKA sporozoites (mouse strain) in the absence and presence of halofuginone (1 μM). The infection proceeded for 45 hrs and then cells were fixed, stained with an anti-P. berghei antibody,[14] and imaged on a fluorescence microscope. Visual inspection of the cells as well as quantitative analysis using high content imaging analysis software (Velos version 5.3.1.1, Molecular Devices) reveals that the parasite count is significantly lower in halofuginone-treated cells when compared to the DMSO control (Fig. S1). To rule out that halofuginone inhibited HepG2 growth, liver cell viability was assessed by microscopic imaging of the monolayer in bright field and quantified by a cell viability assay (CellTiter-Glo, Promega), which measures ATP. Both methods to evaluate cytotoxicity indicate that halofuginone (≤ 1 μM) does not inhibit liver cell viability.
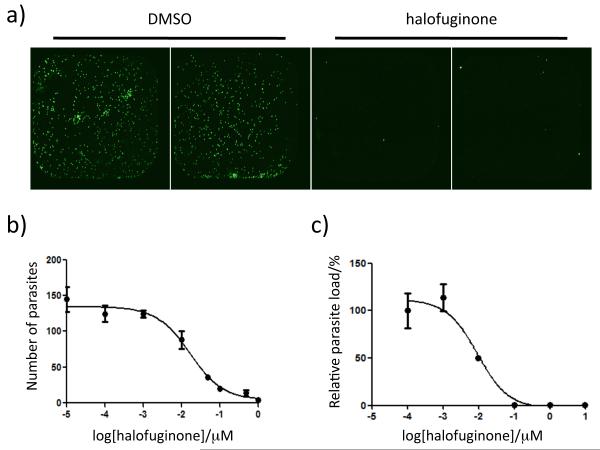
The inhibition of halofuginone was measured as a function of drug concentration. Dose-response curves were generated by quantifying the number of parasites with antibody staining at 45 hrs post infection in the presence of varying concentrations of the compound (Fig. 2B). The total number of parasites and potential morphological changes as a function of drug concentration was then determined by quantitative high content image analysis using Velos software. The parasite numbers determined by the software analysis were consistent with the numbers obtained by manually counting parasites. While halofuginone did inhibit P. berghei parasite load with an IC50 of 17 ± 8 nM, (validating it as one of the most potent inhibitors of liver stage Plasmodium reported), compound treatment did not affect parasite size as determined by measuring the signal area in the presence and absence of halofuginone (Fig. S2).
Figure 2.
Halofuginone inhibition of P. berghei parasite load in liver cells. Visualization of P. berghei sporozoite load in HepG2 cells by staining with an antimalarial antibody (A). Cells were infected in the presence of DMSO or 1 μM halofuginone and fixed 36 – 48 hours after sporozoite addition. Dose-response curves of parasite load assessed by quantification of parasite numbers with antibody staining (B) and by relative luminescence signal after infection with a transgenic luciferase reporter strain of P. berghei sporozoites (C) yield IC50 values of 17 ± 8 nM and 7.8 ± 3 nM, respectively. Data are shown as the mean ± the standard deviation.
As another means to evaluate parasite load in liver cells, HepG2 cells were infected with a luciferase-expressing P. berghei sporozoite strain.[15] Using this transgenic parasite strain the luciferase signal is directly proportional to the parasite load. A dose-response curve of the relative P. berghei luciferase signal in the presence of varying concentrations of halofuginone yielded an IC50 of 7.8 ± 3 nM (Fig. 2C), which is in good agreement with the IC50 determined by microscopy after parasite staining.
Halofuginone inhibits blood stage P. falciparum Dd2 with an IC50 that is 24-fold lower than its IC50 for liver stage P. berghei. It is possible that the metabolically active liver cells are reducing the effective concentration of the drug, or perhaps halofuginone’s target is less critical to the liver stage when compared to the blood stage. Both assays to quantitate parasite load indicate that halofuginone is significantly more potent than primaquine, but is 2 to 6-fold less effective than atovaquone (Table 1), the clinically used antimalarials that target liver stage Plasmodium.[7a]
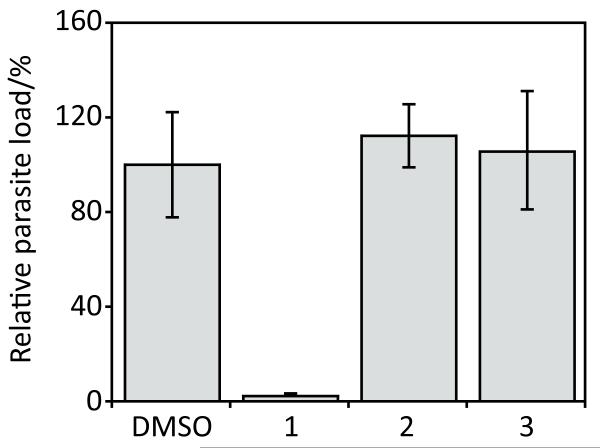
There are limited reports on structure-activity analysis of halofuginone inhibition of malaria. However, it is known that the piperidine ring (see Fig. 1) is important for antimalarial activity[16] but the substituent can be replaced with a pyrrolidine ring without a decrease in efficacy.[17] In this work, two N-acylated halofuginone derivatives (Fig. 1) were tested for the ability to reduce P. berghei sporozoite load in HepG2 cells. Figure 3 shows that these halofuginone derivatives were inactive at 1 μM in the assay, indicating that the basic nitrogen on halofuginone is essential for its mode of inhibition of the liver stage malaria parasite or that the steric hindrance introduced by these modifications prevents the molecule from binding its target. These halofuginone derivatives were also inactive against the blood stages of P. falciparum (Table 1). The agreement between both the blood and liver stage malaria assays with the halofuginone analogs suggests that halofuginone inhibits a common target or pathway in both stages of the parasite.
Figure 3.
Evaluation of halofuginone derivatives. Halofuginone (1 μM) inhibits P. berghei sporozoite load in HepG2 cells but N-acylated halofuginone derivatives (1 μM) are inactive. P. berghei parasite load in HepG2 cells was assessed by the relative luminescence signal 45 hours post-infection with a transgenic luciferase parasite reporter strain. Luminescence values were normalized to the DMSO control and reported as the average percent parasite load ± the standard deviation.
Plasmodium sporozoite traversal assay
Sporozoites have to traverse or migrate through several cells before they finally infect a hepatocyte and start propagating.[18] These traversal events ensure that the motile sporozoites progress from the blood stream to the liver[19] and are important for the parasites to cross the liver sinusoid barrier.[20] Traversal is maintained until the parasites encounter highly sulfated heparan proteoglycans in the liver that activate the sporozoites for invasion.[21]
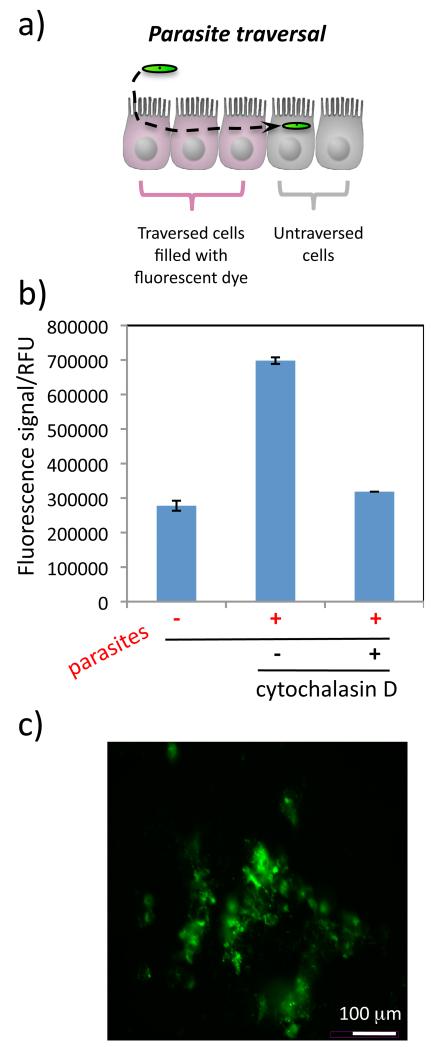
To our knowledge, there is no high-throughput screening method to rapidly evaluate inhibitors of sporozoite traversal. When a sporozoite traverses liver cells, it disrupts the host cell membrane, which then quickly reseals. Cell traversal can be measured by fluorescence microscopy with a cell-wounding assay. In this assay, rhodamine-labeled dextran is supplemented to the liver cell medium prior to sporozoite addition. Dextran can not diffuse through cell membranes, however, when the plasma membrane is disrupted by the traversing parasite the dye-labeled dextran enters the cytosol and gets trapped in the liver cells after resealing (Fig. 4A). Generally, analysis of this assay is time consuming, and involves trypsinization of the cells followed by fluorescence-activated cell sorting (FACS) for quantification.[22] Here, we used a fluorescently labeled dextran dye (Rhodamine Green) and optimized an assay that enables the sensitive detection of traversed cells in a 384-well microtiter plate using a fluorometer. The fluorescence signal in this assay is directly proportional to the number of sporozoites used to infect the HepG2 cells (Fig. S3) and is inhibited by cytochalasin D (Fig. 4B), an actin polymerization inhibitor that is known to prevent sporozoite traversal.[23] Heating the sporozoites for 30 min at 65°C also led to a reduction in the fluorescence signal and served as an independent control. Background fluorescence in this assay is observed due to autofluorescence of the HepG2 cells. When the data is normalized to the hepatocyte autofluorescence there is a 2.5 - 3.5-fold change in the fluorescence signal of cells that have been traversed by Plasmodium sporozoites (4,000 sporozoites added).
Figure 4.
Model of sporozoite traversal through cells (A). Within 2 hrs HepG2 cells that have been traversed by P. berghei sporozoites (in green) fill with Rhodamine Green dextran that was added to the cell medium (cells in pink). The relative numbers of traversed HepG2 cells can be measured using a fluorescence plate reader (B). Data are shown as the average fluorescence reading of 4 measurements on the same 384-well plate and error bars show the standard deviation. Sporozoites were incubated with cytochalasin D before addition to the cells as a control. Visualization of samples with a fluorescence microscope confirms that the fluorescently labeled dextran has entered a population of HepG2 cells (C).
To confirm the results obtained with the fluorescence plate reader, the dextran-containing cells were visualized with a fluorescence microscope (Fig. 4C). Visual inspection of the cells indicates that the fluorescent dye entered liver cells that were disrupted by the sporozoites in the DMSO control, but not in cells incubated with cytochalasin D-treated or heat-inactivated sporozoites. This is the first live cell traversal assay that has been optimized for 384-well format and it presents a promising method for identifying inhibitors of parasite traversal.
Halofuginone inhibits early and late liver stage parasite processes
Using the Plasmodium traversal assay it was found that halofuginone does not inhibit parasite traversal (percent change in fluorescence was not statistically different). Details concerning parasite traversal are active areas of investigation, but it is known that the Plasmodium parasite utilizes an actin-myosin motor system for motility. Clearly this process is not targeted by halofuginone and therefore the compound must inhibit another essential process that is important to parasite development in both liver and red blood cells.
It has been shown that halofuginone inhibits specifically the differentiation of naïve T-cells into TH17-cells by induction of the amino acid starvation response (AAR).[24] It is possible that a similar effect on the host could reduce Plasmodium infection of hepatocytes or that the drug acts on genes that are homologous to the host halofuginone-inducible genes. To test if a host halofuginone target exists, HepG2 cells were incubated with 1 μM halofuginone for 2 hours at 37 °C. After 2 hours the cells were washed 4 times with media before infection with P. berghei sporozoites. This treatment resulted in a 60% reduction in the P. berghei luciferase signal relative to the DMSO-treated control cells (45 hours post sporozoite addition). A similar magnitude of inhibition was also seen when cells were washed 8 times with media before infection with P. berghei sporozoites. Other liver stage Plasmodium inhibitors, like primaquine, are not effective at reducing parasite load if they are removed from the assay before sporozoite addition. This suggests that halofuginone has affinity to a host factor. Conceivably the compound’s ability to influence the amino acid starvation response is affecting liver stage Plasmodium development. It is also possible that halofuginone is trapped in a vesicle and its slow release after sporozoite addition is reducing the parasite load in liver cells.
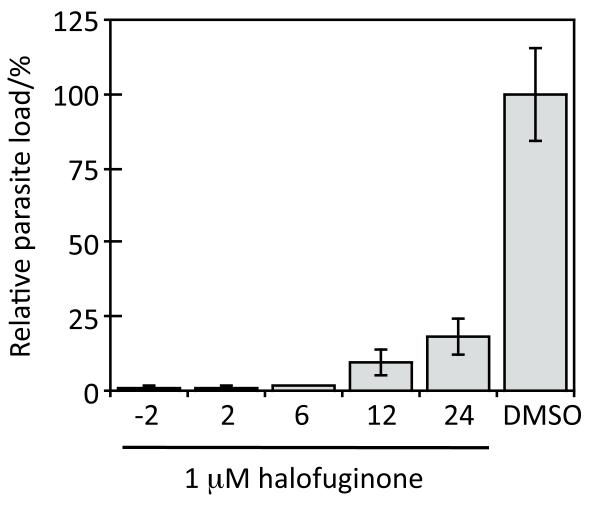
A time course assay of varying halofuginone addition to P. berghei sporozoite-treated liver cells was conducted next. The time course experiments show that halofuginone addition at 2, 6, 12, and 24 hours post sporozoite addition reduces the parasite load in HepG2 cells (Fig. 5). Thus halofuginone inhibits both early and late stage parasite development. Halofuginone’s target, which has remained elusive despite it’s progression to cancer clinical trials[10], is essential to several stages of P. berghei development within liver cells in addition to blood stage parasite development. Importantly, influencing this target has the ability to clear liver stage Plasmodium parasites after the infection has been established. The identification of the underlying mode of action and the molecular target of halofuginone is currently under investigation in our group.
Figure 5.
Time course assay of halofuginone inhibition. Halofuginone (1 μM) was added to HepG2 cells at the indicated times after P. berghei sporozoite addition (−2, 2, 6 12, and 24 hours). Parasite load in HepG2 cells was assessed by the relative luminescence signal 45 hours post-infection. Luminescence values were normalized to the DMSO control and reported as the average percent parasite load ± the standard deviation.
Conclusion
In summary, we optimized a live cell fluorescence assay to examine inhibitors of Plasmodium parasite traversal through liver cells in a high-throughput screening platform. This assay will facilitate systematic investigations for compounds that affect Plasmodium sporozoite traversal. Additionally, halofuginone was found to be a potent inhibitor of liver stage sporozoite development within liver cells, but does not interfere with parasite traversal of host cells. Although cancer clinical trials have revealed some side effects associated with the drug,[10] animal studies have shown halofuginone exhibits few observable side effects in the antimalarial dose range.[25] The high potency of inhibition of parasite development suggests that halofuginone efficiently targets a process essential to the parasite. This inhibition likely involves a target that is involved in both the liver and blood stages of the malaria parasite. While the precise nature of these targets remain to be determined, they remain candidates for future drug design with the hope to address the need for liver stage malaria inhibitors.
Experimental Section
Plasmodium sporozoite infection of liver cells
HepG2 cells (ATCC) were maintained in DMEM (Invitrogen), 10% FBS (Sigma) and 1% antibiotic-antimycotic (Invitrogen) in a standard tissue culture incubator (37°C, 5% CO2). Plasmodium-infected Anopheles stephensi mosquitoes were purchased from the New York University Langone Medical Center Insectary. Live P. berghei ANKA-infected mosquitoes were dissected to isolate sporozoites. Plasmodium sporozoites were then used to infect a monolayer of HepG2 cells in a 384-well plate by following a previously published procedure.[15]
Quantification of P. berghei sporozoite load in HepG2 cells
Parasite inhibition was assessed by immunofluorescence and luminescence. Liver stage Plasmodium assays were performed in the presence of varying concentrations of halofuginone (0 - 1 μM) and halofuginone analogs (1 μM). Halofuginone and halofuginone analogs were synthesized in house. The final concentration of DMSO was 0.3%. After 45 hours post-infection cells were washed 3 times with phosphate buffered saline (PBS) and then fixed with formaldehyde. The primary antibody was 2E6 against P. berghei heat-shock protein 70[14] and the secondary antibody was conjugated to Alexa Fluor 488 (Invitrogen). During the incubation with the secondary antibody the cells were also treated with Alexa Fluor 568 phalloidin (Invitrogen) to stain actin in the HepG2 cells. Parasites and cells were visualized with an ImageXpress Velos (Molecular Devices). Quantification of parasite numbers and size, and of liver cells was completed with Velos 5.3.1.1 software. Alexa Fluor 488 was measured with Ex 488 nm/Em 510-540 nm and Alexa Fluor 568 was measured with Ex 532 nm/Em 560-610 nm. Data analysis was carried out using GraphPad Prism software. Representative dose-response curves are shown where each point represents the average parasite count from triplicate measurements and error bars show the standard deviation. The curve was generated on three separate days and the reported IC50 is the average ± the standard deviation of the three independent measurements.
Liver stage Plasmodium assays were also completed using a luciferase-expressing sporozoite strain of P. berghei ANKA to infect HepG2 cells similar to a published procedure.[15] Parasite load was determined 45 hours post-infection with ONE-Glo (Promega) according to the manufacturer’s instructions. HepG2 viability in the presence of halofuginone was determined using CellTiter-Glo (Promega). Luminescence was quantified using an EnVision plate reader (Perkin-Elmer). Representative dose-response curves are shown where each point represents the average luminescence reading from triplicate measurements that were normalized to the DMSO control. Error bars show the standard deviation of the mean. The relative parasite load in HepG2 cells is proportional to the relative luminescence units. The curve was generated on three separate days and the reported IC50 is the average ± the standard deviation of the three independent measurements.
Probing stage specificity of halofuginone inhibition
P. falciparum 3D7 and Dd2 blood stage assays were performed following a previously published procedure.[26] The experiments were repeated 2-3 times to ensure reproducibility.
P. berghei sporozoites traverse, invade, and then develop within HepG2 cells. Traversal and invasion occurs within 2 hours post-infection and then parasite development continues for another ~55 hours.[27] Time course experiments were completed by adding halofuginone (1 μM) 2 hours before or 2, 6, 12 and 24 hours after sporozoite addition to HepG2 cells to probe which process the compound inhibits. To evaluate the possibility that halofuginone inhibits infection by modulating a host factor, HepG2 cells were incubated with 1 μM halofuginone for 2 hours at 37 °C. After 2 hours the cells were washed 4 - 8 times with media before addition of P. berghei sporozoites. The maximum predicted concentration of halofuginone at the time of sporozoite addition is 7 pM. P. berghei parasite load was measured 45 hours post-infection with ONE-Glo. Data are shown as the mean ± standard deviation.
Plasmodium sporozoite traversal assay
A parasite traversal assay was optimized in a 384-well plate to facilitate high-throughput analysis of liver stage Plasmodium inhibitors. A monolayer of 12,000 HepG2 cells in 25 μL in a black 384-well plate (Corning) was washed with cell medium (DMEM, 10% FBS, 1% antibiotic-antimycotic) and then treated with 4,000 P. berghei ANKA sporozoites. At the time of sporozoite addition 1 mg/mL of Rhodamine Green dextran (Invitrogen) was added to each well and the plate was spun for 10 min at 1,000 rpm. The plate was incubated at 37 °C for 1.5 hours. Cells were then washed 5 times with PBS and the fluorescence intensity was quantitated with an EnVision fluorescence plate reader (Perkin-Elmer). The assay was performed in the presence of increasing sporozoites (0 - 8,000) as a control. The negative control was DMSO and the positive controls were cytochalasin D, a known inhibitor of parasite traversal,[23] and sporozoites that were heat-inactivated for 30 min at 65 °C. Cells were also visualized after the assays with a confocal fluorescence microscope (Nikon Imaging Center, HMS) to evaluate the monolayer with phase contrast and to measure Rhodamine Green dextran inside the cells.
Supplementary Material
Battling a silent killer.
Before malaria symptoms appear several parasites have silently invaded and propagated within the liver. We have developed cell-based assays amenable to high-throughput screening to evaluate compounds for the ability to reduce parasite load in liver cells and to inhibit parasite traversal. These assays helped to identify halofuginone (shown) as a low nanomolar inhibitor of liver stage malaria parasites.
Acknowledgements
This work was supported by the Harvard Medical School-Portugal collaborative grant to J.C. and the Ruth L. Kirschstein National Research Service Award (F32GM093510) to E.R.D. We thank Maria M. Mota for critical input and providing the Plasmodium antibody, and Joseph F. Cortese for blood stage assays. We also thank the Nikon Imaging Center and the ICCB-Longwood Screening Facility at Harvard Medical School.
References
- [1]a.Sachs J, Malaney P. Nature. 2002;415(6872):680–685. doi: 10.1038/415680a. [DOI] [PubMed] [Google Scholar]; b Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. Nature. 2005;434(7030):214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]a.Kappe SH, Vaughan AM, Boddey JA, Cowman AF. Science. 2010;328(5980):862–866. doi: 10.1126/science.1184785. [DOI] [PubMed] [Google Scholar]; b Mazier D, Renia L, Snounou G. Nat Rev Drug Discov. 2009;8(11):854–864. doi: 10.1038/nrd2960. [DOI] [PubMed] [Google Scholar]
- [3]a.Miller LH, Baruch DI, Marsh K, Doumbo OK. Nature. 2002;415(6872):673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]; b Prudencio M, Rodriguez A, Mota MM. Nat Rev Microbiol. 2006;4(11):849–856. doi: 10.1038/nrmicro1529. [DOI] [PubMed] [Google Scholar]
- [4].Bassat Q, Alonso PL. Nat Med. 2011;17(1):48–49. doi: 10.1038/nm0111-48. [DOI] [PubMed] [Google Scholar]
- [5]a.Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, Vanderwall DE, Green DV, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Nature. 2010;465(7296):305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]; b Guiguemde WA, Shelat AA, Bouck D, Duffy S, Crowther GJ, Davis PH, Smithson DC, Connelly M, Clark J, Zhu F, Jimenez-Diaz MB, Martinez MS, Wilson EB, Tripathi AK, Gut J, Sharlow ER, Bathurst I, El Mazouni F, Fowble JW, Forquer I, McGinley PL, Castro S, Angulo-Barturen I, Ferrer S, Rosenthal PJ, Derisi JL, Sullivan DJ, Lazo JS, Roos DS, Riscoe MK, Phillips MA, Rathod PK, Van Voorhis WC, Avery VM, Guy RK. Nature. 2010;465(7296):311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Carraz M, Jossang A, Franetich JF, Siau A, Ciceron L, Hannoun L, Sauerwein R, Frappier F, Rasoanaivo P, Snounou G, Mazier D. PLoS Med. 2006;3(12):e513. doi: 10.1371/journal.pmed.0030513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]a.Burgoine KL, Bancone G, Nosten F. Malar J. 2010;9:376. doi: 10.1186/1475-2875-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Cappellini MD, Fiorelli G. Lancet. 2008;371(9606):64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- [8].Meister S, Plouffe DM, Kuhen KL, Bonamy GM, Wu T, Barnes SW, Bopp SE, Borboa R, Bright AT, Che J, Cohen S, Dharia NV, Gagaring K, Gettayacamin M, Gordon P, Groessl T, Kato N, Lee MC, McNamara CW, Fidock DA, Nagle A, Nam TG, Richmond W, Roland J, Rottmann M, Zhou B, Froissard P, Glynne RJ, Mazier D, Sattabongkot J, Schultz PG, Tuntland T, Walker JR, Zhou Y, Chatterjee A, Diagana TT, Winzeler EA. Science. 2011;334(6061):1372–1377. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]a.Gego A, Silvie O, Franetich JF, Farhati K, Hannoun L, Luty AJ, Sauerwein RW, Boucheix C, Rubinstein E, Mazier D. Antimicrob Agents Chemother. 2006;50(4):1586–1589. doi: 10.1128/AAC.50.4.1586-1589.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Prudencio M, Rodrigues CD, Hannus M, Martin C, Real E, Goncalves LA, Carret C, Dorkin R, Rohl I, Jahn-Hoffmann K, Luty AJ, Sauerwein R, Echeverri CJ, Mota MM. PLoS Pathog. 2008;4(11):e1000201. doi: 10.1371/journal.ppat.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Rodrigues CD, Hannus M, Prudencio M, Martin C, Goncalves LA, Portugal S, Epiphanio S, Akinc A, Hadwiger P, Jahn-Hofmann K, Rohl I, van Gemert GJ, Franetich JF, Luty AJ, Sauerwein R, Mazier D, Koteliansky V, Vornlocher HP, Echeverri CJ, Mota MM. Cell Host Microbe. 2008;4(3):271–282. doi: 10.1016/j.chom.2008.07.012. [DOI] [PubMed] [Google Scholar]
- [10].de Jonge MJ, Dumez H, Verweij J, Yarkoni S, Snyder D, Lacombe D, Marreaud S, Yamaguchi T, Punt CJ, van Oosterom A. Eur J Cancer. 2006;42(12):1768–1774. doi: 10.1016/j.ejca.2005.12.027. [DOI] [PubMed] [Google Scholar]
- [11].Samant BS, Sukhthankar MG. Med Chem. 2009;5(3):293–300. doi: 10.2174/157340609788185846. [DOI] [PubMed] [Google Scholar]
- [12]a.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. Nature. 2002;419(6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]; b Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Science. 2003;301(5639):1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]; c Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, Daly TM, Bergman LW, Kappe SH. Proc Natl Acad Sci U S A. 2008;105(1):305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Westenberger SJ, McClean CM, Chattopadhyay R, Dharia NV, Carlton JM, Barnwell JW, Collins WE, Hoffman SL, Zhou Y, Vinetz JM, Winzeler EA. PLoS Negl Trop Dis. 2010;4(4):e653. doi: 10.1371/journal.pntd.0000653. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Williams CT, Azad AF. PLoS One. 2010;5(4):e10267. doi: 10.1371/journal.pone.0010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tsuji M, Mattei D, Nussenzweig RS, Eichinger D, Zavala F. Parasitol Res. 1994;80(1):16–21. doi: 10.1007/BF00932618. [DOI] [PubMed] [Google Scholar]
- [15].Ploemen IH, Prudencio M, Douradinha BG, Ramesar J, Fonager J, van Gemert GJ, Luty AJ, Hermsen CC, Sauerwein RW, Baptista FG, Mota MM, Waters AP, Que I, Lowik CW, Khan SM, Janse CJ, Franke-Fayard BM. PLoS One. 2009;4(11):e7881. doi: 10.1371/journal.pone.0007881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kikuchi H, Yamamoto K, Horoiwa S, Hirai S, Kasahara R, Hariguchi N, Matsumoto M, Oshima Y. J Med Chem. 2006;49(15):4698–4706. doi: 10.1021/jm0601809. [DOI] [PubMed] [Google Scholar]
- [17].Zhu S, Zhang Q, Gudise C, Wei L, Smith E, Zeng Y. Bioorg Med Chem. 2009;17(13):4496–4502. doi: 10.1016/j.bmc.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mota MM, Pradel G, Vanderberg JP, Hafalla JC, Frevert U, Nussenzweig RS, Nussenzweig V, Rodriguez A. Science. 2001;291(5501):141–144. doi: 10.1126/science.291.5501.141. [DOI] [PubMed] [Google Scholar]
- [19].Amino R, Thiberge S, Blazquez S, Baldacci P, Renaud O, Shorte S, Menard R. Nat Protoc. 2007;2(7):1705–1712. doi: 10.1038/nprot.2007.120. [DOI] [PubMed] [Google Scholar]
- [20].Ishino T, Yano K, Chinzei Y, Yuda M. PLoS Biol. 2004;2(1):E4. doi: 10.1371/journal.pbio.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Coppi A, Tewari R, Bishop JR, Bennett BL, Lawrence R, Esko JD, Billker O, Sinnis P. Cell Host Microbe. 2007;2(5):316–327. doi: 10.1016/j.chom.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Prudencio M, Rodrigues CD, Ataide R, Mota MM. Cell Microbiol. 2008;10(1):218–224. doi: 10.1111/j.1462-5822.2007.01032.x. [DOI] [PubMed] [Google Scholar]
- [23].Kumar KA, Oliveira GA, Edelman R, Nardin E, Nussenzweig V. J Immunol Methods. 2004;292(1-2):157–164. doi: 10.1016/j.jim.2004.06.017. [DOI] [PubMed] [Google Scholar]
- [24].Sundrud MS, Koralov SB, Feuerer M, Calado DP, Kozhaya AE, Rhule-Smith A, Lefebvre RE, Unutmaz D, Mazitschek R, Waldner H, Whitman M, Keller T, Rao A. Science. 2009;324(5932):1334–1338. doi: 10.1126/science.1172638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jiang S, Zeng Q, Gettayacamin M, Tungtaeng A, Wannaying S, Lim A, Hansukjariya P, Okunji CO, Zhu S, Fang D. Antimicrob Agents Chemother. 2005;49(3):1169–1176. doi: 10.1128/AAC.49.3.1169-1176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baniecki ML, Wirth DF, Clardy J. Antimicrob. Agents. Chemother. 2007;51(2):716–723. doi: 10.1128/AAC.01144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stanway RR, Mueller N, Zobiak B, Graewe S, Froehlke U, Zessin PJ, Aepfelbacher M, Heussler VT. Cell Microbiol. 2011;13(11):1768–1782. doi: 10.1111/j.1462-5822.2011.01657.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.