Abstract
Autophagy is a membrane-trafficking process that delivers cytoplasmic constituents to lysosomes for degradation. It contributes to energy and organelle homeostasis and the preservation of proteome and genome integrity. Although a role in cancer is unquestionable, there are conflicting reports that autophagy can be both oncogenic and tumor suppressive, perhaps indicating that autophagy has different roles at different stages of tumor development. In this report, we address the role of autophagy in a critical stage of cancer progression—tumor cell invasion. Using a glioma cell line containing an inducible shRNA that targets the essential autophagy gene Atg12, we show that autophagy inhibition does not affect cell viability, proliferation or migration but significantly reduces cellular invasion in a 3D organotypic model. These data indicate that autophagy may play a critical role in the benign to malignant transition that is also central to the initiation of metastasis.
Keywords: autophagy, cancer, invasion, migration, organotypic model
Introduction
Autophagy is required for cellular homeostasis, and perturbations in the process can lead to a variety of diseases, including neurodegeneration, impaired immunity and cancer.1-5 The molecular mechanism of autophagy is conserved from yeast to humans and has been well characterized.6 During autophagy, double-membraned vesicles termed autophagosomes engulf and transport cytoplasmic contents such as protein aggregates, pathogens and damaged mitochondria to lysosomes for degradation. Despite our relatively good understanding of the molecular mechanism of autophagy, we are only starting to learn how autophagy contributes to cellular responses in different contexts. For example, autophagy is essential for the cellular response to a plethora of different stresses that regulate cell viability, but in certain contexts this can promote cell survival, whereas in others it can contribute to cell death.7,8 This presents a potential difficulty for using inhibitors of autophagy to treat complex diseases such as cancer, as the effect of inhibiting autophagy in different settings is difficult to predict.
Clinical trials are nonetheless underway to assess the effect of autophagy inhibition on cancer progression in a variety of tumors types, and there is evidence to show that inhibition of autophagy can increase survival outcomes for patients with glioblastoma multiforme.9 However, similar to the bipolar reports on the role of autophagy in cell death and cell survival, studies in vitro and in mouse models of cancer suggest that inhibition of autophagy may not be beneficial in all tumor types. While some studies show that certain tumors depend on autophagy for survival,10,11 deletion of autophagy genes has also revealed tumor suppressive effects.12-15 These seemingly contradictory reports may indicate that autophagy is pro- or anti-tumorogenic depending on the context or the stage of tumor development.
The development of cancer is a multi-stage process.16 In the initial stages of tumor development, the capacity to replicate beyond normal constraints and the ability to evade programmed cell death pathways are key events.16 As tumors develop and grow, tumor cells need to invade local tissue, survive upon tissue detachment and acquire the characteristics needed to form a secondary mass at a distant site.16 Although the role of autophagy in controlling programmed cell death has been heavily investigated, the role of autophagy in these other tumor cell attributes is poorly understood. We report here a study to analyze the role played by autophagy in tumor cell invasion using a 3D organotypic model designed to mimic in vivo interactions between invasive cells and the surrounding stroma.17 Using cells containing a doxycycline-regulated shRNA against a key component of the autophagy machinery, Atg12, we show that inhibition of autophagy impairs tumor cell invasion in an organotypic model. We consider, therefore, that these findings indicate that autophagy may be required for this key characteristic of tumor cells as they progress toward malignant disease.
Results
RNAi-mediated knockdown of Atg12 in glioma cells impedes autophagy.
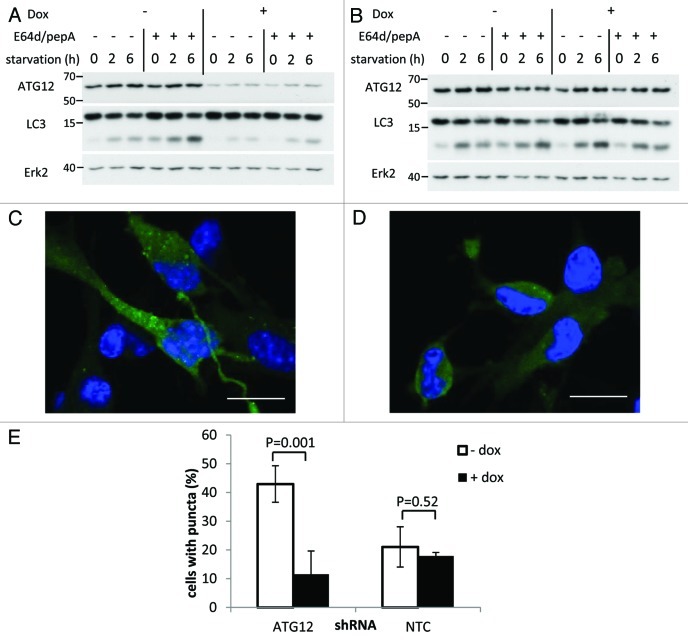
To investigate the role of autophagy in key characteristics of tumor cell behavior, we utilized a glioma cell line that expresses a shRNA under the control of the tetracycline-inducible promoter that targets the essential autophagy gene Atg12 (shAtg12 cells). Treatment of this cell line with the tetracycline analog doxycycline (Dox) induced expression of the shRNA and decreased levels of Atg12 expression (Fig. 1A). In contrast, no decrease in Atg12 levels was observed following Dox treatment of a control line containing a non-targeting shRNA (NTC cells) (Fig. 1B).
Figure 1. Atg12 knockdown inhibits basal autophagy and starvation induced autophagy. Where indicated GL261 shAtg12 cells (A) and NTC cells (B) were cultured with doxycycline for 72 h to induce shRNA expression. Cells were then starved for indicated time periods and the cell lysates analyzed by western blot. Where indicated, cells were incubated with lysosomal protease inhibitors E64d and pepstatin A for a total of 7 h including any starvation period. (C–E) Cells were infected with adenovirus containing GFP-LC3 and then starved for 6 h to induce autophagy. Representative images (cropped 60 × images) showing GFP-LC3 localization in shAtg12 cells cultured without (C) and with (D) doxycycline are shown. (E) Quantification of the number of cells containing GFP-LC3 puncta. Microscope images (60 × objective) were taken at random positions and the percentage of cells containing more than five GFP-LC3 puncta were counted. Scale bars are 10 μm. The graph shows the mean and standard deviations from four uncropped 60 × images per condition.
To monitor the impact of decreased levels of Atg12 on autophagy, we analyzed the levels of a form of the LC3 protein termed LC3-II18 after Dox treatment of shAtg12 cells. LC3 is encoded by the essential autophagy gene MAP1LC3B and, once expressed, is immediately cleaved into a form termed LC3-I. Upon initiation of autophagy, LC3-I is conjugated to the lipid phosphatidylethanolamine, and this form of LC3, termed LC3-II, integrates into the autophagosome membrane.18 The ratio of LC3-I to LC3-II is a measure of autophagic activity and can be assessed based on differences in electrophoretic mobility, which can be discerned by western blotting.18,19 Treatment of shAtg12 cells with Dox increased the ratio of LC3-I to LC3-II compared with untreated shAtg12 cells (Fig. 1A), especially when cells were incubated in Earle’s balanced salt solution (EBSS), a medium lacking amino acids that induces autophagy. In contrast, Dox treatment of NTC cells did not affect the LC3-I/LC3-II ratio (Fig. 1B). These results suggest that ATG12 knockdown could impair the initiation of autophagy. However, LC3-II is also degraded by autophagy, and a reduction in the amount of LC3-II could potentially indicate an enhancement in autophagic degradation. The use of lysosomal inhibitors E64d and pepstatin A blocks the degradation stage of autophagy and therefore enhances accumulation of LC3-II following induction of autophagy. As expected, incubation of shAtg12 cells in EBSS in the absence of Dox caused accumulation of LC3-II that was enhanced by treatment with E64d and pepstatin A (Fig. 1A). Conversely, the smaller accumulation of LC3-II seen in Dox-treated shAtg12 cells following incubation in EBSS was not enhanced by treatment with E64d and pepstatin A. This clearly indicates that ATG12 knockdown inhibited the initiation of autophagy. Treatment of NTC cells with Dox had no effect on EBSS-induced accumulation of LC3-II either in the absence or presence of E64d and pepstatin A (Fig. 1B).
We also assessed if the decreased levels of LC3-II seen in shAtg12 cells following treatment with Dox genuinely represented a decrease in autophagosome number. To do this, we infected cells with a previously described adenovirus that expresses LC3 fused to GFP.20 Using this virus, autophagosomes can be detected as distinct puncta via fluorescent microscopy. These studies revealed that knockdown of Atg12 by Dox treatment of shAtg12 cells greatly impaired the accumulation of autophagosomes following incubation in EBSS (Fig. 1C–E). By contrast, no effect on autophagosome number was observed in NTC cells following treatment with Dox (Fig. 1E). Taken together, these data indicate that shAtg12 cells constitute a robust system in which to investigate tumor-related cell behavior in both autophagy-competent and autophagy-deficient states.
Loss of autophagy does not impair cell proliferation or cell viability in glioma cells.
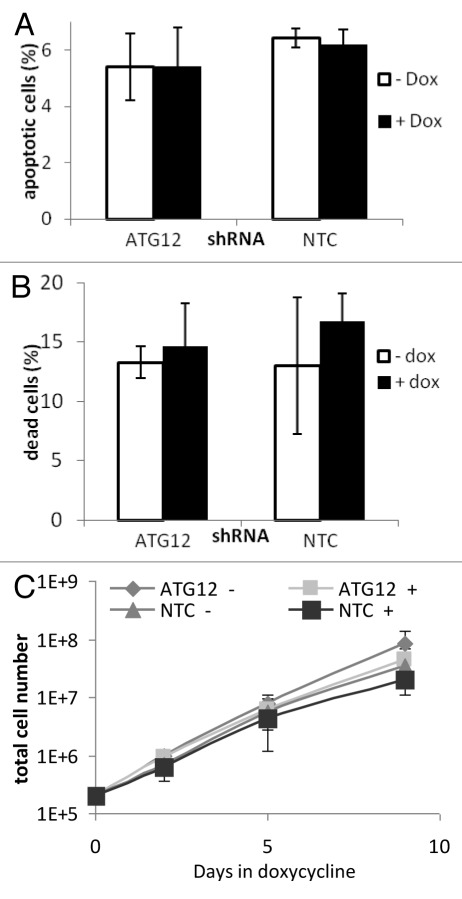
Using shAtg12 cells, we were interested to know if autophagy was required for several tumor-associated phenotypes. First, we tested if loss of autophagy following depletion of Atg12 has any effect on cell viability. Cells that had been incubated in the absence or presence of Dox for 9 d were analyzed by flow cytometry for the appearance of cells with sub-G1 DNA content, a reliable marker of apoptotic death. As can be seen in Figure 2A, loss of Atg12 had no impact on the appearance of apoptotic cells, as the percentage of cells with a sub-G1 DNA content was equal to cells that had been incubated in the absence of Dox. Similarly, treatment of NTC cells with Dox also had no effect on the appearance of apoptotic cells (Fig. 2A).
Figure 2. Cell viability and growth are unaffected by Atg12 knockdown. Cells were cultured in doxycycline for 9 d. (A) After 9 d the percentage of apoptotic cells (cells containing less than a G1 compliment of DNA) was measured by flow cytometry of permeablized cells stained with propidium iodide. (B) The total numbers of dead/necrotic cells were determined by incubating unfixed cells in propidium iodide without permeablisation and counting the stained (permeable/dead) cells by flow cytometry. (C) Total cell numbers over 9 d were monitored by counting trypsinized cells at 2, 5 and 9 d.
In addition to apoptotic cell death, cells can also die by necrosis, and it has been shown that autophagy can act to repress this form of death. To test if loss of Atg12 has an impact on necrosis, cells were incubated in propidium iodide (PI), which enters permeabilized cells (a characteristic of necrosis), where it fluoresces following intercalation with DNA. Similar to what was observed following analysis of apoptosis, no increase in PI positivity was observed following Dox treatment of either shAtg12 cells or NTC cells, indicating that inhibition of autophagy does not lead to necrotic cell death (Fig. 2B).
The growth and proliferation of cells is connected to availability of nutrients, and since autophagy is intricately connected to nutrient availability, we next considered whether autophagy may be required for another characteristic of cancer cells’ sustained continual replication. shAtg12 cells and NTC cells were incubated in the absence or presence of Dox for a period of 9 d, while each cell population was assessed periodically by measuring cell number. This revealed that under normal growth conditions, the loss of Atg12, and by association the loss of autophagy, had no impact on proliferation as cells numbers did not significantly differ after 9 d of Dox treatment when compared with controls (Fig. 2C).
Loss of autophagy does not affect glioma cell migration, but impairs invasion in a 3D organotypic model.
The ability of tumor cells to migrate and invade local tissue is a key characteristic of cancer and a defining transition that distinguishes benign from malignant growth.16 We questioned, therefore, using the shAtg12 cell model, whether autophagy was required for these processes.
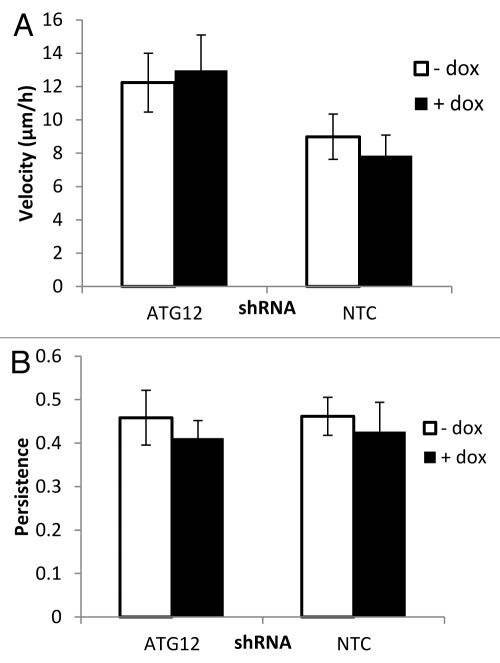
As in previous assays, shATG12 cells (and NTC cells) were incubated in the absence or presence of Dox for 9 d to allow for generation of autophagy-competent and autophagy-deficient cells. These cells were then plated and when confluent were subjected to a “scratch assay” to measure cell migration. Cell populations were scratched with a plastic implement to create a wound in the cell monolayer. The ability of cells to fill this wound was monitored by time-lapse microscopy, and parameters of cell migration were measured over time. As can be seen in Figure 3, the ability of glioma cells to migrate in response to a scratch wound was not affected by loss of autophagy. More detailed analysis of the data from this assay indicates that loss of autophagy has no impact on two specific parameters of migration, velocity or persistence, in this glioma cell model (Fig. 3A and B).
Figure 3. Cell migration velocity and persistence are unaffected by Atg12 knockdown. Migration of cells across a scratch wound was monitored by time-lapse microscopy and individual cells were tracked using Image J software. Average velocity (A) and persistence (B) are shown. Persistence was calculated as the Euclidean distance (distance in a straight line from start to finish) divided by the total accumulated distance. Error bars are the standard deviation from two independent experiments with three images per time point for each experiment.
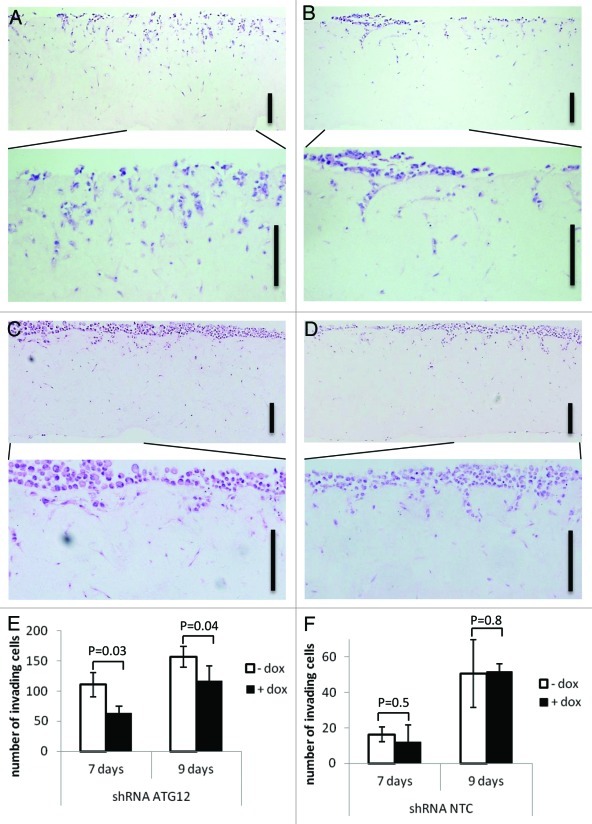
Cell migration is only one facet of invasive and metastatic tumor cells. In an in vivo setting, tumor cells must be able to invade adjacent tissue to reach local blood and lymphatic vessels in order to metastasize. To examine the role of autophagy in this process, we utilized a previously characterized organotypic model of invasion involving fibroblast-contracted collagen matrices.17,21,22 Cells were seeded onto matrices in the absence or presence of Dox and incubated for 72 h to induce Atg12 knockdown under conditions that did not promote invasion. Cells were then allowed to invade toward a chemoattractant (FBS) for up to 9 d. Treatment of shRNA Atg12 cells with Dox reduced invasion by ~45% and ~25% after 7 and 9 d, respectively (Fig. 4A, B and E). By contrast, no significant difference in cellular invasion was detected in NTC cells following treatment with Dox (Fig. 4C, D and F). When taken together, these results indicate that autophagy is required for glioma cell invasion in a manner independent of cell viability, cell proliferation and cell migration. Since autophagy is known to be deregulated in multiple tumor types, these findings reveal an additional function of autophagy that may be relevant to the benign-to-malignant conversion and for the establishment of metastatic disease.
Figure 4. Atg12 knockdown inhibits tumor cell invasion. (A–D) Hematoxylin and eosin-stained sections of GL261 cells on organotypic matrix. GL261 shAtg12 cells (A and B) and NTC cells (C and D) were seeded on to an organotypic matrix in the absence (A and C) or presence (B and D) of doxycycline. Images are shown of GL261 shAtg12 cells and NTC cells after 7 or 9 d of invasion respectively. (E and F) Quantification of invasion. The average number of invading shAtg12 cells (E) and NTC cells (F) in four representative Hematoxylin and eosin-stained images (10 × objective lens) were counted for 7 and 9 d of invasion. Scale bars are 200 μm and error bars are standard deviations from four different microscope images.
Discussion
The ability of tumor cells to invade local tissue is a key characteristic of the malignant phenotype and is a characteristic required for the initiation of metastasis.16 Numerous studies have indicated that autophagy has roles during tumor development, and to the best of our knowledge, we report here for the first time that autophagy is important for cell invasion. We show, in an organotypic model involving 3D fibroblast-contracted collagen I matrices, that inhibition of autophagy via RNAi-mediated knockdown of the essential autophagy gene Atg12 greatly impairs the invasive nature of glioma cells. Perhaps the most interesting aspect of our discovery is that inhibition of autophagy made no difference to several other tumor cell phenotypes in our model when measured in 2D. Cell death and cell growth have both been connected to autophagy,23-25 but inhibition of autophagy had no impact on cell viability or cell proliferation under normal growth conditions. Most strikingly, analysis of cell migration, a characteristic important for tumor cell invasion, was also unaffected by inhibition of autophagy when assayed in 2D. This naturally raises the question of how cell invasion in our organotypic model is affected by loss of autophagy. Ultimately, this may simply reflect differences in cell behavior in 2D vs. 3D. For example, it has been reported that epithelial-mesenchymal transitions of cells can be different upon transfer from 2D to 3D, and that their motility, and therefore migratory capacity, can be affected as a result.26-28 Clearly, culture in 2D has its limitation and analysis of invasion in the organotypic model more accurately reflects the environment in which migration and invasion would occur in vivo. In this context, tumor cells have a more complex interaction with the microenvironment that may reveal and/or invoke the development of additional tumor cell characteristics. It may be the case that autophagy-competent cells are better at digesting extracellular matrix components either externally, via secretion of proteins such matrix metalloproteases, or internally, via digestion of extracellular matrix components in lysosomes, and this may aid invasion through the matrix. Equally, autophagy-competent cells may respond differently to the availability of extracellular matrix-derived ligands such as fibronectin or collagen. Finally, it must be remembered that culture in our organotypic model involves an intricate relationship between tumor cells and fibroblasts. These cells in the organotypic model secrete factors to which tumor cells can respond in an invasive manner, and it may be the case that autophagy-deficient cells are impaired in their response to these agents. Moreover, studies have shown that migrating fibroblasts can act as “path leaders” for tumor cell invasion, and again, autophagy in tumor cells may be required for this process.29
Although the ability to tease out the mechanistic details underlying the role of autophagy in invasion is beyond the scope of this current study, we feel it is without question that the findings we present here have important implications for our understanding of the role of autophagy in cancer. Reports have shown that autophagy is required for other tumor-associated traits, and we consider that we add to this list by showing that autophagy is required for tumor cell invasion. As a result of our data, it is natural to speculate, as has previously been suggested,30 that autophagy may be required for metastasis, although it not clear whether inhibition of autophagy would affect established metastases or just the ability to initiate metastasis. Further studies are therefore required to address these intriguing issues by analysis of the role of autophagy in in vivo models of human cancer.
Materials and Methods
Cell lines and cell culture.
Mouse glioblastoma cell line GL261 was cultured at 37°C under 5% CO2 in a high-humidity atmosphere in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin and 100 ∝g/ml streptomycin. GL261 cells expressing pTRIPZ shRNA NTC or ATG12 were supplemented with puromycin (0.5 ∝g/ml). Where indicated, doxycycline (Sigma Aldrich) was used at a concentration of 2 ∝g/ml. E64d and pepstatin A (both from Calbiochem) were both used at a concentration of 10 ∝g/ml.
Construction of inducible shRNA cell line.
Mouse pGIPZ-shATG12mir individual clone (ID V2LMM_72549) was purchased from Open Biosystems, and gene knockdown was verified. The shRNAmir clone was moved from pGIPZ to the pTRIPZ lentiviral inducible system (Open Biosystems) according to the manufacturers instructions. Briefly, pGIPZ-shATG12mir and pTRIPZ vectors were prepared for ligation by performing a MluI/XhoI double digestion. Clones were verified by Sal1 digestion, and the sequence was verified by using the pTRIPZ sequencing primer GGA AAG AAT CAA GGA GG. The pTRIPZ empty vector and the non-silencing pTRIPZ lentiviral shRNAmir control were also purchased from Open Biosystems. The Translentiviral shRNA Packaging System with Arrestin-In transfection reagent (Open Biosystems) was used to generate lentivirus particles in HEK293FT cells. GL261, murine glioma cell line was obtained from NCI-Frederick and were transduced with lentivirus using 8 ug/ml polybrene and selected with 1 ug/ ml puromycin. Doxyclycine was used at a concentration of 2 ug/ ml to induce expression of TurboRFP and ATG12 gene knockdown; single clones demonstrating efficient TurboRFP expression upon doxycycline treatment were isolated.
Western blotting.
Cell lysates were prepared in lysis buffer (1% triton-X, 0.1% SDS, 50 mM HEPES pH 7.4, 150 mM NaCl, 100 mM NaF, 10 mM EDTA, 10 mM Na4P2O7 containing protease inhibitor cocktail from Roche) and transferred to nitrocellulose or immobulin-P membranes as previously described in reference 31. Membranes were probed using standard immunoblotting techniques. ATG12 and LC3B antibodies were from Cell Signaling, and the Erk2 antibody was a kind gift from Prof. Chris Marshall (Ab122, CJ Marshall, Institute of Cancer Research).
Starvation-induced autophagic flux assay.
GL261 cells were cultured with doxycycline for 72 h to induce expression of shRNA. Cells were then washed three times in warm PBS and once in warm Earle’s balanced salt solution (EBSS, Sigma Aldrich) and incubated in EBSS for 2 or 6 h to induce autophagy. Autophagic degradation of LC3II was inhibited by addition of lysosomal protease inhibitors E64d and pepstatin A. The inhibitors were added prior to starvation and were included in the starvation medium, such that cells were incubated with the inhibitors for a total of 7 h. Cell lysates were then analyzed by western blot.
LC3 puncta assay.
GL261 cells were seeded on microscope coverslips and incubated overnight. Cells were then infected with an adenovirus containing LC3 fused to GFP. After 16 h incubation with the virus, the medium was changed, and the cells were incubated for a further 24 h. Autophagy was induced by starving the cells in EBSS for 6 h. Cells were then fixed (4% paraformaldehyde for 20 min at room temperature) and the coverslips mounted on slides using immunofluorescence mounting solution (Dako) containing 0.5 ∝g/ml DAPI. Cells were viewed under a 60x objective with a 488 nm excitation laser to view GFP-LC3 positive puncta. The number of cells containing puncta was counted for four different microscope views.
Cell proliferation and death assays.
GL261 cells were cultured with or without doxycycline for 9 d, and the total cell numbers were counted periodically as a measure of cell proliferation. As an assessment of necrosis, cells (including floating and adherent cells) that had been incubated in the absence or presence of Dox for 9 d were harvested with trypsin and incubated with propidium iodide to label the permeabilized (necrotic) cells. The proportion of PI-positive cells was determined by flow cytometry. As an assessment of apoptosis, total cell populations including floating and adherent cells were harvested fixed in methanol for 24 h. After this time, cells were stained with propidium iodide, and the population analyzed by flow cytometry for the appearance of cell with sub-G1 DNA content, a reliable measure of apoptotic cells.32
Wound-healing assay.
Cells were cultured with or without doxycycline for 7 d and then seeded into a 6-well plate and incubated overnight. A scratch wound was then made, and the wound area was photographed by time lapse microscopy over the following 24 h at 20 min intervals. Individual cells were then tracked over time using ImageJ software, and the average velocity and persistence were calculated (17 cells per 10x objective view, 3 views per wound).
Organotypic invasion assay.
Organotypic matrices comprising primary human fibroblasts embedded in polymerized collagen I were constructed, and invasion experiments were performed as described previously in references 17, 21 and 22. Briefly, rat tail tendon collagen solution (final protein concentration ~2 mg/ml) was extracted from tendons with 0.5 M acetic acid. Subsequently ~7.5 x 104/ml primary human fibroblasts were mixed with collagen solution (2.5 ml) immediately before it was allowed to set in a 35 mm dish. Polymerized matrices were detached from the edges of the dishes and were allowed to contract for approximately 6 d in complete media (DMEM, 10% FCS), until the fibroblasts had contracted the matrix to ~1.5 cm diameter. GL261 cells were seeded (4 x 104 cells in complete medium) on to the matrix, and doxycycline was added where indicated. Cells were allowed to attach and grow to confluence for 72 h before the matrix was transferred to a raised grid in a 6 cm dish containing complete medium. The level of the grid was set to the air/liquid interface, resulting in the matrix being fed from below with complete media that was changed every day. Cells were then allowed to invade toward the culture medium for 7 or 9 d. The organotypic matrix was then fixed using 4% paraformaldehyde and processed by standard methods for hematoxylin and eosin staining. Four representative images (10x objective) were taken for each organotypic matrix, and the number of GL261 cells that invaded past the surface of the matrix were counted as invading cells. Representative 20x images were also taken.
Statistical analyses.
Statistical significance was determined by the Student t-test, and p-values of less than 0.05 were considered significant.
Glossary
Abbreviations:
- dox
doxycycline
- shRNA
short-hairpin RNA
- Atg
autophagy-related gene
- NTC
non-targeting control
- EBSS
Earle’s balanced salt solution
- LC3
microtubule-associated protein 1 light chain 3 β
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Work in the Tumor Cell Death Laboratory is supported by Cancer Research UK and the Association for International Cancer Research. Paul Timpson and Kurt Anderson of the Tumor Cell Migration Group were also supported by Cancer Research UK. Work in the Thorburn laboratory is supported by NIH grants R01 CA111421 and R01 CA150925. We are grateful to members of the Tumor Cell Laboratory for critical reading of the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20424
References
- 1.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–8. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Mah LY, Ryan KM. Autophagy and cancer. Cold Spring Harb Perspect Biol. 2012;4:a008821. doi: 10.1101/cshperspect.a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenfeldt MT, Ryan KM. The multiple roles of autophagy in cancer. Carcinogenesis. 2011;32:955–63. doi: 10.1093/carcin/bgr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–9. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sotelo J, Briceño E, López-González MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–43. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 10.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Timpson P, McGhee EJ, Erami Z, Nobis M, Quinn JA, Edward M, et al. Organotypic collagen I assay: a malleable platform to assess cell behaviour in a 3-dimensional context. J Vis Exp. 2011;•••:e3089. doi: 10.3791/3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2007;3:4. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bampton ETW, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 21.Timpson P, McGhee EJ, Morton JP, von Kriegsheim A, Schwarz JP, Karim SA, et al. Spatial regulation of RhoA activity during pancreatic cancer cell invasion driven by mutant p53. Cancer Res. 2011;71:747–57. doi: 10.1158/0008-5472.CAN-10-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edward M, Gillan C, Micha D, Tammi RH. Tumour regulation of fibroblast hyaluronan expression: a mechanism to facilitate tumour growth and invasion. Carcinogenesis. 2005;26:1215–23. doi: 10.1093/carcin/bgi064. [DOI] [PubMed] [Google Scholar]
- 23.Long JS, Ryan KM. New frontiers in promoting tumour cell death: targeting apoptosis, necroptosis and autophagy. Oncogene. 2012 doi: 10.1038/onc.2012.7. [DOI] [PubMed] [Google Scholar]
- 24.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 25.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Töyli M, Rosberg-Kulha L, Capra J, Vuoristo J, Eskelinen S. Different responses in transformation of MDCK cells in 2D and 3D culture by v-Src as revealed by microarray techniques, RT-PCR and functional assays. Lab Invest. 2010;90:915–28. doi: 10.1038/labinvest.2010.63. [DOI] [PubMed] [Google Scholar]
- 27.Cukierman E, Bassi DE. Physico-mechanical aspects of extracellular matrix influences on tumorigenic behaviors. Semin Cancer Biol. 2010;20:139–45. doi: 10.1016/j.semcancer.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen CD, Sahai E. Cancer dissemination--lessons from leukocytes. Dev Cell. 2010;19:13–26. doi: 10.1016/j.devcel.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 30.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. 2010;22:241–5. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell HS, Dufes C, O’Prey J, Crighton D, Bergamaschi D, Lu X, et al. A p53-derived apoptotic peptide derepresses p73 to cause tumor regression in vivo. J Clin Invest. 2007;117:1008–18. doi: 10.1172/JCI28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellicciari C, Manfredi AA, Bottone MG, Schaack V, Barni S. A single-step staining procedure for the detection and sorting of unfixed apoptotic thymocytes. Eur J Histochem. 1993;37:381–90. [PubMed] [Google Scholar]