Abstract
E2F1 is a eukaryotic transcription factor that is known to regulate various cellular pathways such as cell cycle progression, DNA replication, DNA damage responses and induction of apoptosis. Given its versatile roles, a precise and tight regulation of E2F1 is very critical to maintain genomic stability. E2F1 is regulated both at transcriptional and posttranslational levels during cell cycle and upon DNA damage. After S phase, E2F1 is targeted for degradation and is kept at low levels or in an inactive state until the next G1/S phase transition. Our studies show that APC/C ubiquitin ligase in conjunction with its co-activator Cdh1 (APC/CCdh1) can downregulate E2F1. We also identify an APC/C subunit APC5 that binds to E2F1 and is essential for E2F1 ubiquitination. We confirm an interaction between E2F1 and Cdh1 as well as an interaction between E2F1 and APC5 both in vivo and in vitro. In vitro GST pull-down assays have mapped the C-terminal 79 a.a. of E2F1 as Cdh1 interacting residues. Ectopically expressed Cdh1 downregulates the expression of E2F1–4. Our studies have also shown for the first time that E2F1 can be modified by K11-linkage specific ubiquitin chain formation (Ub-K11). The formation of Ub-K11 chains on E2F1 is increased in the presence of Cdh1 and accumulated in the presence of proteasome inhibitor, suggesting that APC/CCdh1 targets E2F1 for degradation by forming Ub-K11 chains. We also show that the effect of Cdh1 on E2F1 degradation is blocked upon DNA damage. Interestingly, Ub-K11-linked E2F1 accumulates after treatment of DNA damaging agents. The data suggest that DNA damage signaling processes do not inhibit APC/CCdh1 to ubiquitinate E2F1. Instead, they block the proteasomal degradation of Ub-K11-linked E2F1, and therefore lead to its accumulation.
Keywords: APC/C, Cdh1, DNA damage, E2F1, K11-linked ubiquitin
Introduction
The E2F family of transcription factors are known to regulate various cellular processes, including cell cycle progression, cell differentiation and apoptosis. Among the E2F family proteins, E2F1 has the best-established role in regulating apoptosis in addition to cell proliferation.1 During cell cycle progression, E2F1 accumulates during G1/S transition and induces the transcription of several DNA replication and S-phase specific genes that are essential for cell growth and proliferation. In response to genotoxic stress, E2F1 is stabilized and triggers apoptosis.2-4 E2F1 undergoes several posttranslational modifications upon DNA damage. E2F1 is phosphorylated by ATM/ATR kinases at Ser-313 and subsequently stabilized by binding to a 14-3-3 family member 14-3-3τ 5. E2F1 can also be phosphorylated by Chk2 kinase upon DNA damage at Ser-364.6 In fact, both Chk1 and Chk2 have been shown to be important for E2F1 stabilization.7 In addition to phosphorylation, E2F1 is acetylated by p300/CREB-binding protein-associated factor (P/CAF) upon DNA damage.8 It is believed that these events together result in E2F1 stabilization and activation, which induces upregulation of various pro-apoptotic genes such as p19ARF 9, 10, p73, Apaf-1,11 caspase 3, 7, 8 and 9.12-14
Given the multifaceted roles of E2F1 in maintaining genomic integrity, E2F1 is precisely regulated by several transcriptional and posttranslational mechanisms. Transcriptionally, E2F1 levels are regulated by a feedback loop mechanism through the E2F responsive genes on its own promoter.15-17 At the translational level, expression of E2F1 is negatively regulated by two c-Myc-responsive microRNAs, miR-17–5p and miR-20a.18 Posttranslationally, the transcriptional activity of E2F1 is mainly regulated through binding to hypophosphorylated form of Rb.19 Another mechanism of controlling E2F1 is by targeting it to the proteasome-dependent degradation pathway. Several different E3 ligases have been identified to be able to ubiquitinate and degrade E2F1 during various cell cycle phases. SCFskp2 is known to ubiquitinate and degrade E2F1 during S/G2 phases.20 However, absence of Skp2 was not sufficient to stabilize E2F1 in Skp2−/− mouse embryonic fibroblasts,5,21 suggesting that E2F1 can be regulated by multiple E3 ligases. The Drosophila E2F1 is targeted for degradation in a PCNA-dependent manner by the CRL4-Cdt2 E3 Ligase;22 nevertheless, absence of canonical PIP box in mammalian E2F1 required for its binding to PCNA excludes the possibility of a role for CRL4-Cdt2 in mammalian E2F1 degradation.
Recently, E2F1 was shown to be targeted for degradation by the APC/CCdc20 E3 ligase during prometaphase.23 The anaphase-promoting complex/cyclosome (APC/C) is a multi-component E3 ligase that regulates the temporal progression of eukaryotic cells through the M-phase and subsequent transition into the G1 phase. APC/C is activated by one of its co-activators, Cdc20 and Cdh1, which facilitates the recruitment of substrates and confers substrate specificity through their WD40 domains. In addition to Cdc20 and Cdh1, the APC/C core subunits have been suggested to participate in substrate binding, since the budding yeast APC/C lacking its Doc1/Apc10 subunit cannot bind and ubiquitinate the substrates.24-26 In fact, recent structural studies of yeast APC/C confirmed that both Doc1/Apc10 and co-activators contribute recognition sites for substrates.27-29 After recruiting substrates, APC/C then specifically synthesizes K-11 linkage-specific ubiquitin chains on the substrates and target them for 26S proteasome-dependent degradation.
Low levels of E2F1 are detected during M phase and early G1 phase. The precise mechanisms by which E2F1 is maintained at low levels are yet to be fully understood. We have identified in a yeast two-hybrid experiment that APC5 interacts with E2F1. This prompted us to investigate if APC/C E3 ligase has a role in E2F1 degradation process. In the process of our study, Peart et al. reported that Cdc20 can target E2F1 for degradation during prometaphase.23 However, during M-phase exit and early G1 phase, Cdc20 is degraded by APC/CCdh1 30, leaving the question of how E2F1 is degraded at these phases. Therefore, we investigated if APC/C can target E2F1 via its early G1 phase co-activator Cdh1 through the formation of K11-specific ubiquitin chain synthesis. We also investigated the effect DNA damage on APC/CCdh1-mediated E2F1 degradation.
Results
Cdh1 and APC5 interact with E2F1 in vitro
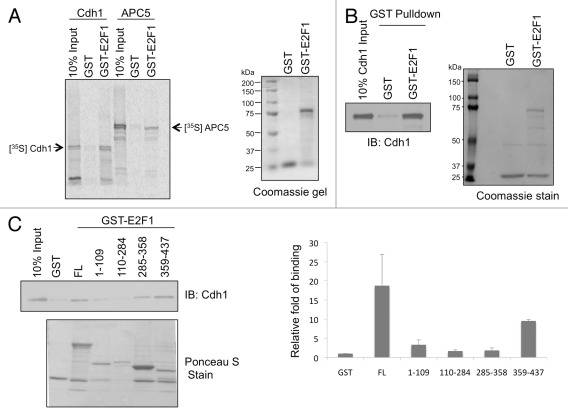
To identify the E2F1-interacting proteins that might regulate E2F1 protein stability, we performed a yeast two-hybrid screen using E2F1 N terminus as a bait.31 APC5, a subunit of APC/C complex, was identified as a potential E2F1-interacting protein in that screen. Since Cdh1 is an adaptor protein that brings in APC/C substrates, we therefore investigated the interaction between E2F1 and both APC5 and Cdh1. First, we tested an interaction between E2F1 and APC5 or Cdh1 in vitro by incubating [35S]-labeled in vitro-translated APC5 or Cdh1 with GST-E2F1 in a GST pull-down assay (Fig. 1A). Both APC5 and Cdh1 could be pulled down along with GST-E2F1 but not GST, suggesting an interaction between APC5 or Cdh1 and E2F1. A direct interaction between GST-E2F1 and Cdh1 was further confirmed using purified recombinant Cdh1 protein (rCdh1) and purified GST-E2F1 protein in a GST pull-down assay (Fig. 1B). We also mapped the domain of E2F1 that directly interacted with Cdh1. Incubation of rCdh1 with GST-E2F1 full-length and domain mutants (a.a. 1–109, 110–284, 285–358 and 359–437) immobilized on glutathione-Sepharose beads showed that rCdh1 bound to the C-terminal 359–437 a.a. residues of E2F1 with higher affinity compared with other domains (Fig. 1C), suggesting that Cdh1 binds to the C-terminal 79 a.a. of E2F1. We cannot rule out a possibility that Cdh1 might bind to multiple domains of E2F1.
Figure 1. E2F1 directly binds to Cdh1 via its C terminus. GST-E2F1 WT (A and B) or truncation mutants (C) were incubated with in vitro translated [S35]-labeled Cdh1 or APC5 (A) or bacterially purified Cdh1 (B). E2F1-bound Cdh1 was detected by autoradiography (A) or by immunoblotting (B and C). Right panel of (C): The intensity of Cdh1 signals bound to E2F1 WT or truncation mutants as well as the amounts of GST, GST-E2F1 and GST-E2F1 truncation mutants on the corresponding GST pull-down assay were quantified using Image J software. The bound Cdh1 signals were then normalized by the abundance of each corresponding GST fusion protein in that assay. The results averaged from three independent experiments are plotted here. FL: full-length; the numbers indicate the amino acid residues of hE2F1.
E2F1 interacts with APC/C components Cdh1 and APC5 in vivo
To further determine if E2F1 can interact with Cdh1 and APC5 in vivo, HEK293 cells were transfected with HA-E2F1 and APC5 or GFP-Cdh1 and were left untreated or treated with proteasome inhibitors. Then, co-immunoprecipitation assays were performed. Presence of ectopically expressed APC5 was readily detected in lysates co-immunoprecipitated with HA-E2F1 only in the presence of proteasome inhibitor-MG132 compared with lysates without treatment or lysates expressing HA-E2F1 or APC5 alone (Fig. 2A). Similarly, immunoprecipitation of GFP-Cdh1 co-precipitated higher amounts of E2F1 in the presence of MG132 corresponding to the increased accumulation of E2F1 in the presence of MG132 within the input lysates (Fig. 2B). Requirement of proteasome inhibition to capture E2F1 and APC5 or Cdh1 interaction suggested a very transient interaction between E2F1 and APC/C components in the cells, or that the complex of E2F1 with APC/CCdh1 is rapidly degraded.
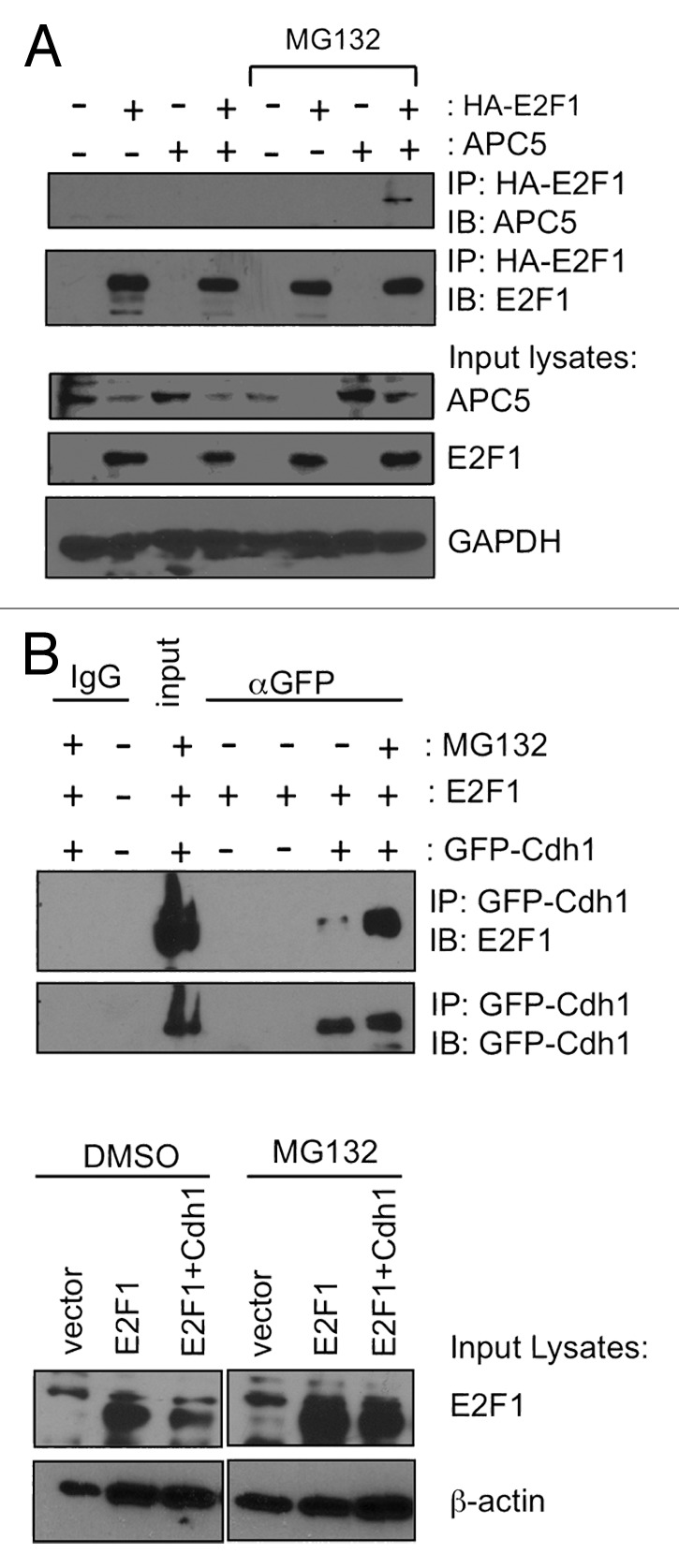
Figure 2. E2F1 interacts with APC/C components Cdh1 and APC5 in vivo. HEK293 cells were transfected with plasmids expressing HA-E2F1 and APC5 (A) or GFP-Cdh1 (B), and then treated with MG132 as indicated. (A) HA-tagged E2F1 was immunoprecipitated from cell lysates with HA beads and the co-immunoprecipitated APC5 was detected by immunoblotting. (B) GFP-Cdh1 was immunoprecipitated from cell lysates using GFP antibody and the co-immunoprecipitated E2F1 was detected by immunoblotting.
Cdh1 downregulates E2F1 protein levels
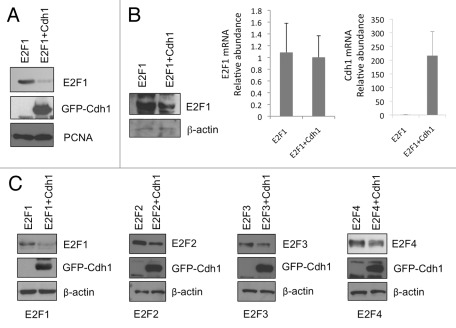
We then investigated whether expression of Cdh1 could downregulate E2F1 levels. Indeed, co-expression of GFP-Cdh1 led to a significantly lower level of E2F1 when compared with that in the GFP control sample (Fig. 3A). Downregulation of E2F1 by Cdh1 was mediated through a post-transcriptional mechanism, as the E2F1 mRNA levels were not significantly affected (Fig. 3B). Analysis of HEK293T lysates with ectopically expressed HA-E2F1 and GFP-Cdh1 in the presence of cycloheximide at different time points showed that the presence of Cdh1 significantly shortened the half-life of E2F1 from ~3-4 h to 1 h (Fig. 5C, top left and bottom panels).
Figure 3. Cdh1 downregulates E2F1 protein levels. (A) HEK293T cells were transfected with HA-E2F1 along with GFP-Cdh1 or GFP vector control. Cell lysates were prepared and subject to SDS-PAGE and immunoblot analysis as indicated. (B) To determine whether downregulation of E2F1 by Cdh1 is mediated through a post-transcriptional mechanism, HEK293T cells were transfected as in (A). Cells were harvested for mRNA isolation and E2F1 mRNA and Cdh1 mRNA were measured by real-time RT-PCR. The p value for a difference in E2F1 mRNA levels between GFP and GFP-Cdh1 samples is 0.70 (two-tailed t-test). A fraction of cells were used to prepare lysates and the lysates were analyzed by SDS-PAGE and immunoblot analysis as indicated. (C) HEK293T cells were transfected with HA-E2F1 or E2F2 or E2F3 or E2F4 along with GFP-Cdh1 or GFP control. Two days later, cells were harvested and analyzed by immunoblot analysis as indicated.
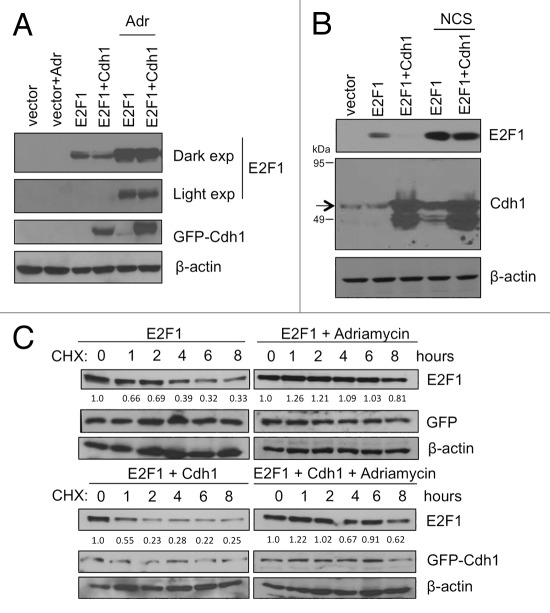
Figure 5. Cdh1-mediated degradation of E2F1 is inhibited upon DNA damage. (A) HEK293T cells were transfected with HA-E2F1 with GFP-Cdh1 (or with GFP control) and cells were either left untreated or treated with adriamycin (5 μM) for 5 h. E2F1 and GFP-Cdh1 in total lysates were analyzed by SDS-PAGE and immunoblotting. (B) HEK293T cells were transfected with HA-E2F1 with pcDNA3-Cdh1 (or with an empty vector control) and cells were untreated or treated with neocarzinostatin (NCS, 300 ng/ml). E2F1 and Cdh1 in total lysates were analyzed by SDS-PAGE and immunoblotting. The arrow indicates endogenous and overexpressed Cdh1. The lower molecular-weight band recognized by Cdh1 antibody was seen sometimes in cells transfected with untagged Cdh1 and could be a degradation product of Cdh1. (C) HEK293T cells were transfected with HA-E2F1 with GFP control (upper panels) or HA-E2F1 with GFP-Cdh1 (lower panels). The cells were then either left untreated or treated with adriamycin (5 μM) for 16 h. Next, the cells were treated with cycloheximide (20 μg/ml) for various time points. HA-tagged E2F1 and GFP-Cdh1 proteins were detected by western blot. E2F1 signals were measured by densitometry and the relative intensities compared with the respective 0 h sample are shown below each panel.
Since the C terminus of E2F1 binds to Cdh1, given the conservation of the C terminus among E2F1–4 proteins, it is not surprising that overexpression of GFP-Cdh1 also led to various degrees of downregulation in the levels of E2F2, 3 and 4 (Fig. 3C). Like E2F1, E2F2 and E2F4 interacted well with Cdh1 in vitro (Fig. S1). An interaction between E2F3 and Cdh1 can also be demonstrated by Daniele Guardavaccaro and colleagues (personal communication; Ping et al., in this issue). These data suggest that APC/CCdh1 also regulates other E2Fs. We had focused on E2F1 in the subsequent studies.
Cdh1 promotes K11 linkage-specific ubiquitination of E2F1
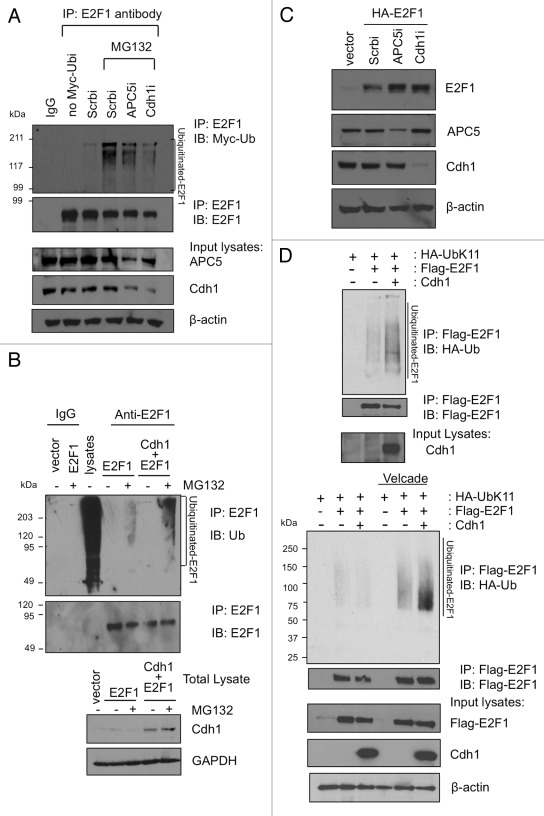
Next we wanted to test if E2F1 ubiquitination is altered in the presence or absence of Cdh1 and APC5. To this end, we first knocked down Cdh1 or APC5 expression in HEK293 cells that were transfected with Myc-ubiquitin and looked at the ubiquitination of endogenous E2F1. Indeed, knockdown of APC5 or Cdh1 decreased the ubiquitination of E2F1 compared with a scrambled RNAi control (Fig. 4A). When we ectopically overexpressed Cdh1 along with HA-E2F1, we detected increased accumulation of ubiquitinated E2F1 upon immunoprecipitation of cell lysates for E2F1 followed by immunoblotting for endogenous ubiquitin (Fig. 4B). Consistent with a role for Cdh1 and APC5 for E2F1 degradation, depletion of either Cdh1 or APC5 by shRNA-expressing constructs led to an increased level of coexpressed HA-E2F1 (Fig. 4C). These results strongly support a role for APC/CCdh1 in the ubiquitination of E2F1.
Figure 4. Cdh1 promotes K11-linkage specific ubiquitination of E2F1. (A) HEK293 cells were transfected with Myc-ubiquitin along with vectors expressing siRNA for APC5, Cdh1 or a scrambled (Scrb) sequence control. The cells were left untreated or treated with MG132. Lysates were immunoprecipitated with normal mouse IgG or a monoclonal E2F1 antibody, followed by immunoblotting to detect ubiquitinated E2F1. An aliquot of total lysates were immunoblotted as indicated. (B) HEK293 cells were transfected either with a control empty vector, E2F1 alone, or E2F1 with Cdh1. The cells were left untreated or treated with MG132. Lysates were immunoprecipitated with normal mouse IgG or a monoclonal E2F1 antibody, followed by immunoblotting with ubiquitin antibody to detect ubiquitinated E2F1. “Lysates” lane is from total cellular lysates without immunoprecipitation and serves as a positive control for ubiquitin immunoblotting. Aliquots of each input lysate were also immunoblotted as indicated (below). (C) HEK293 cells were transfected with an empty vector or HA-E2F1 along with a vector expressing siRNA for APC5, Cdh1 or a scrambled (Scrb) sequence control. Two days later, cells were harvested and lysates were immunoblotted with indicated antibodies. (D) HA-Ub-K11-specific ubiquitin was overexpressed in HEK293T cells along with GFP-Cdh1 (or GFP vector control) and Flag-E2F1 as indicated in the presence (lower panel) or absence (upper panel) of proteasome inhibitor Velcade. Lysates were processed as described in Materials and Methods, and then immunoprecipitated with anti-Flag beads. The HA-Ub-K11-conjugated E2F1 were detected by HA immunoblotting.
APC/C E3 ligase has been shown to form specific K11-linked ubiquitin chains on its substrates.32 Therefore, we wanted to test if presence of Cdh1 could induce K11-specific ubiquitin chain linkages on E2F1. HEK293T cells were transfected with Flag-E2F1 and GFP-Cdh1 along with HA-Ub-K11, which can form only K11 Ub chains, as all other lysines except K11 are mutated in this ubiquitin. Cell lysates were immunoprecipitated for Flag-E2F1 and blotted with HA antibody to detect Ub-K11 chains conjugated with E2F1 in the presence or absence of Cdh1. Increased accumulation of Ub-K11 chains on E2F1 was noticed in the presence of Cdh1 (Fig. 4D, top panel). Also, the Ub-K11 chains were accumulated in the presence of proteasome inhibitor Velcade (Fig. 4D, bottom panel), suggesting that APC/CCdh1 can, indeed, ubiquitinate E2F1 via K11-specific Ub chain synthesis and target it to proteasome-mediated degradation.
Cdh1-mediated degradation of E2F1 is inhibited upon DNA damage
We then tested if the stabilization of E2F1 after DNA damage was due to inhibition of APC/CCdh1-mediated degradation. Cell lysates were prepared and analyzed from HEK293T cells transfected with E2F1 along with or without Cdh1 and treated with two different DNA damaging agents, adriamycin (Adr) and neocarzinostatin (NCS). Interestingly, presence of DNA damage did inhibit the Cdh1 effect on E2F1. In spite of presence of ectopically overexpressed Cdh1, E2F1 protein levels remained accumulated after the treatment of cells with DNA damaging agents (Fig. 5A and B). We also performed a time course experiment in the presence of cycloheximide to measure the half-life of E2F1. The HEK293T cells were transfected with Flag-E2F1 along with GFP-Cdh1 or with control GFP and cells were harvested at various time points after the treatment of cycloheximide. Analysis of these lysates indicated that while the half-life of E2F1 decreased from 3 ~4 h to 1 h in the presence of Cdh1 before DNA damage (Fig. 5C, left, top and bottom panels), the half-life of E2F1 remained at ≥ 8 h in the presence of adriamycin regardless whether Cdh1 was overexpressed or not (Fig. 5C, right, top and bottom panels). This data suggests that E2F1 accumulation after DNA damage may result from perturbation of APC/CCdh1-mediated ubiquitin proteasome degradation pathway.
Next, we wanted to investigate the mechanism by which APC/CCdh1-mediated E2F1 degradation is inhibited after DNA damage. We transfected HEK293T cells with HA-Ub-K11, Flag-E2F1 with or without Cdh1 and then left the cell untreated or treated the cells with adriamycin. Ubiquitination assay analysis was performed as described above (Fig. 4 and Methods section). Surprisingly, we detected increased accumulation of K11-specific ubiquitin chains on E2F1 after DNA damage (Fig. 6). This data suggests that DNA damage signaling directly inhibits the proteasome-dependent degradation of K11-specific, Ub-linked E2F1 but does not block the process of Cdh1-dependent E2F1 ubiquitination. The data also shows a large accumulation of Ub-K11-E2F1 after adriamycin treatment. The accumulation of Ub-K11-E2F1 is probably due to a block at the proteasomal degradation.
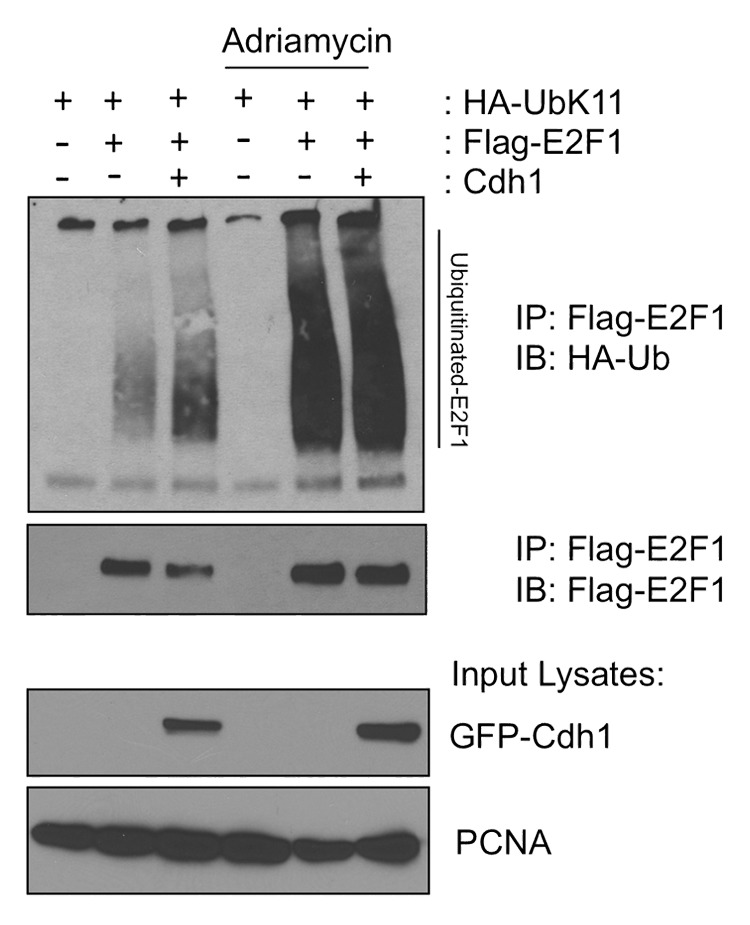
Figure 6. Cdh1-promoted K11-linkage-specific ubiquitination of E2F1 is enhanced and accumulated after DNA damage. HEK293T cells were transfected with Flag-E2F1, HA-Ub-K11 and GFP-Cdh1 (or GFP vector control) as indicated. The cells were either left untreated or treated with 5 μM adriamycin for 5 h before harvesting. The lysates were prepared and analyzed for E2F1 ubiquitination as described in methods section. Fractions of the total lysates were analyzed as well (lower panels).
Discussion
E2F1, a critical modulator of cell proliferation and apoptosis, is known to be precisely and tightly regulated at multiple levels, starting from its transcription control to its post-translation modifications and degradation. Multiple E3 ligases have been shown to target E2F1 for ubiquitin-mediated degradation in various cellular contexts. Our data demonstrates a novel role for one of the master E3 ligases of cell cycle, APC/CCdh1, in ubiquitinating and degrading E2F1 by catalyzing its characteristic K11-specific ubiquitin chain formation. Our current studies provide evidence for an interaction between E2F1 and an APC/C subunit APC5, for the first time, and with Cdh1 as shown earlier by Peart and colleagues23 both in vivo and in vitro (Figs. 1 and 2). Cdh1 binds to the C-terminal 79 a.a. residues of E2F1 (Fig. 1C). We demonstrate that E2F1 is targeted for degradation by ectopically expressed Cdh1 (Figs. 3 and 5C). Further, we show that the endogenous E2F1 ubiquitination requires Cdh1 and APC5 (Fig. 4A). Depletion of either APC5 or Cdh1 led to accumulation of E2F1 (Fig. 4C). Overexpression of Cdh1 also induces E2F1 ubiquitination with K11-specific Ub linkage (Fig. 4B and D). Our data establishes that E2F1 transcription factor is a substrate of APC/CCdh1 E3 ligase. APC/C is the only E3 ligase known to catalyze K11-specific chains on its substrates, and importantly, for the first time, we show that E2F1 is ubiquitinated by K11-specific ubiquitin chain formation.
The fact that the effect of Cdh1 on E2F1 stability is abolished in the presence of DNA damage (Fig. 5) suggests that blocking of the APC/CCdh1-mediated degradation of E2F1 might be responsible for E2F1 stabilization upon DNA damage. We anticipated that Cdh1-mediated E2F1 ubiquitination would be inhibited by adriamycin treatment. To our surprise, Ub-K11-linked E2F1 greatly accumulates after DNA damage (Fig. 6). Given that APC/C is the only E3 ligase known to form K11-specific ubiquitin chains on the substrates, these data suggest that DNA damage treatment does not inhibit the process of Cdh1-mediated ubiquitination; rather, it might inhibit the proteasomal degradation of the ubiquitinated E2F1. Thus, we observed a very significant accumulation of Ub-K11-linked E2F1 after adriamycin treatment. The mechanism and significance for the accumulation of Ub-K11-E2F1 upon DNA damage is to be investigated. It is worth noting that accumulation of ubiquitinated E2F1 was also observed after camptothecin treatment along with stabilization and acetylation of E2F1.33 Together, these data suggest a regulatory mechanism on the proteasomal degradation of ubiquitinated E2F1. Whether some or all of phosphorylation, acetylation and 14–3-3τ binding events on E2F1 operate directly to prevent proteasome from degrading ubiquitinated E2F1 deserves further investigation. In this regard, 14–3-3τ can directly bind the C8 subunit of the 20S proteasome.34 Although it promotes ubiquitin-independent proteasomal degradation of p21 via C8 interaction, it would be interesting to test whether 14–3-3τ/C8 interaction could, instead, perturb the process of ubiquitin-dependent proteasomal degradation of E2F1. On the other hand, the potential of alteration in the transcriptional activity or target gene specificity of E2F1 upon K11-specific ubiquitination poses another interesting venue to explore and could provide another layer of regulation of E2F1 transcriptional activity.
The identification of APC5 as an APC/C core subunit that binds to E2F1 is quite interesting. The 3D structure of the APC/C shows an asymmetric triangular morphology with a large inner cavity surrounded by an outer protein wall.27-29,35-38 The complexity suggests that certain subunits may guide substrates into the inner cavity where substrates are ubiquitinated. While it is well established that the substrate recognition of APC/C involves a bipartite degron receptor between co-activator Cdh1 and an APC/C core subunit, Apc10/Doc1,27,28 whether other APC/C subunits contribute to substrate recognition and specificity remains to be explored. Our study suggests that Cdh1 and APC5 can form another bipartite degron receptor. Although Apc1, Apc4 and Apc5 form a scaffolding platform to connect a catalytic subcomplex (Apc2 and Apc11) and the other tetratricopeptide repeat (TPR)-containing proteins subcomplex (for substrate/co-activator recruitment), Cdh1 and Apc5 are juxtaposed in space according to a high-resolution structure of yeast APC/C complex;29 therefore, it is possible that E2F1 might sit in the in-between space and interact with both Cdh1 and APC5 through different domains. Mutation of Drosophila IDA, the human APC5 homolog, blocks the degradation of cyclin B, but does not affect Securins turnover,39 also supporting a role for substrate recognition by APC5. In fact, like other subunits in the substrate recognition TPR subcomplex, APC5 also contains TPR motifs.
The proteasomal degradation of E2F1 can be regulated at both its N terminus and C terminus. Deletion of an N-terminal 4140 or 853 amino acid fragment stabilizes E2F1. This N terminus coincides with the Skp2 binding domain; therefore, Skp2 was proposed to be responsible for the N terminus-mediated E2F1 degradation.40 Hofmann et al. also mapped another destabilizing element of E2F1 to a.a. 363–378.41 Interestingly, we show that E2F1 a.a. 359–437 can bind Cdh1. Thus, APC/CCdh1 may be responsible for the C terminus-mediated degradation of E2F1. The a.a. 363–378 region is very close to pocket protein binding domain, and pRb can protect E2F1 from degradation.41 However, overexpression of pRb did not protect E2F1 from Cdc20- and Cdh1-induced degradation in HeLa cells.23 Whether pRb plays a role in regulating APC/CCdh1-mediated E2F1 degradation is yet to be determined.
Although E2F1 contains two RXXL motifs in the cyclin A binding domain, mutations of both motifs did not affect E2F1 degradation by Cdc20 or Cdh1.23 The C-terminal Cdh1 binding domain of E2F1 does not contain consensus sequences for known APC/C degrons.42 The degradation recognition region of yeast Iqg1p, a direct APC/C target, also does not contain a known APC/C-recognition sequence.43 It is possible that E2F1 is recognized by both Cdh1 and APC5 through its spatial structure or through yet-to-be-determined recognition sequences. Future study is warranted to further define the molecular details of how E2F1 is recognized by Cdh1 and APC5.
Materials and Methods
Cell culture, transfection and treatment
HEK293 or 293T cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (50 IU/ml), and streptomycin (50 μg/ml). Cells were grown in a humidified incubator at 37°C with 5% CO2 and 95% air. Transfection was performed with a standard calcium phosphate method or Lipofectamine 2000 (Invitrogen). After transfection, cells were incubated for 48 h before analysis. Cells were treated with 20 μg/ml of cycloheximide (Calbiochem) or 5 μM of adriamycin (Pfizer) or 20 ng/ml of Bortezomib (VELCADE, Millenium Pharmaceuticals, Inc.) or 20 μM MG132 (Calbiochem) for indicated time points as described in each experiment.
Plasmids construction
An hCdh1 cDNA in pOTB7 vector was purchased from ATCC (Image Clone ID: 3160334). To construct pcDNA3-hCdh1, the hCdh1 cDNA was excised with SmaI and subcloned to the EcoRV-digested pcDNA3. To construct pEGFP-C1-hCdh1 and pGEX6P3-hCdh1, hCdh1 cDNA was PCR amplified using a pair of primers: 5′-CGCGAATTCCATGGACCAGGACTATGAG-3′ and 5′-CAAGTCGACTTACCGGATCCTGGTGAA-5′. The PCR product was digested by EcoRI/SalI and then subcloned into EcoRI/SalI-digested pBluescript vector. The hCdh1 sequence was verified by DNA sequencing. The hCdh1 cDNA was then swapped from pBluescript into pEGFP-C1 and pGEX-6P3 vectors by EcoRI/SalI digestion. The hAPC5 cDNA in pOTB7 vector was also purchased from ATCC (Image Clone ID: 2823401). The hAPC5 cDNA was excised with EcoRI/XhoI and ligated with EcoRI/XhoI digested pcDNA3 to construct pcDNA3-hAPC5. To construct Flag-E2F1, hE2F1 cDNA was excised from pcDNA3-HA-E2F13 to pCMV-Tag2B vector by BamHI/EcoRI digestion. The hE2F4 cDNA was swapped from pcDNA3-HA-E2F4 to pGEX-6P1 by BamHI/EcoRI digestion for production of GST-E2F4 fusion protein in E. coli. The vectors for GST-E2F1 domain mutants have been described.44
The 19-nt target sequences for siCdh1 is: 5′-TGAGAAGTCTCCCAGTCAG-3′; for siAPC5 is: 5′-CCTCCGTGTCCAAGATGTTTT-3′; for a random scrambled sequence (siScr) is 5′-GCGCGCTTTGTAGGATTCG-3′. The target sequences were constructed to pSUPERIOR.puro vector (OligoEngine).
Immunoprecipitation, western blot analysis and immunofluoresence studies
Cells were harvested 48 h after transfection in TNN buffer, and immunoprecipitation was performed as described.31 The specific signals were detected with appropriate antibodies. The antibodies specific to GFP (B-2), Cdh1 (Dh-01), HA (Y11), PCNA (PC10) and E2F1 (C-20 or KH-95) were purchased from Santa Cruz. The β-actin and FLAG antibody were purchased from Sigma. GAPDH antibody was purchased from Alexis. The β-actin and FLAG antibody were purchased from Sigma. GAPDH antibody was purchased from Alexis.
GST pull-down assay
The GST fusion proteins were induced by 0.5 mM IPTG (isopropyl-β-D-thiogalactopyranoside) in E. coli strain BL21 or DH5α and purified. The GST portion on GST-Cdh1 was excised by PreScission Protease (Pharmacia). Approximately, 5 μg of GST-E2F1 full-length or truncation mutants were immobilized on glutathione-Sepharose beads and incubated with 0.5 -1 μg of rCdh1 or 2 μl of [35S]-labeled in vitro translated products and rotated at 4°C for 3 h in NETN-A Buffer (250 mM NaCl, 5 mM EDTA, 50 mM TRIS pH 7.5 and 0.1% NP-40). The beads were washed five times with NETN-B buffer (150 mM NaCl, 5 mM EDTA, 50 mM TRIS pH 7.5 and 0.5% NP-40) and then subject to SDS-PAGE and analyzed by western blot with anti-Cdh1 antibody or by Storm Phosphorimager.
In vitro translation of proteins
TNT rabbit reticulolysate kit from Promega was used to transcribe and translate Cdh1 or APC5 using pcDNA3-Cdh1 and pcDNA3-APC5 plasmids in the presence of [S35]-labeled methionine.
RNA extraction and real-time reverse transcription-PCR (RT-PCR)
RNA was extracted using TRIzol reagent (Invitrogen). Quantitative PCR was performed in triplicate on an MX3005P thermal cycler using SYBR green dye method to track the progress of the reactions with ROX dye added as reference. GAPDH was run in parallel with test genes. PCR condition is: 95°C denaturation step for 30 sec and 55°C annealing for 1 min and 72°C for 2 min. Results were analyzed with MxPro 4.0 QPCR software (Stratagene). The PCR primers GAPDH promoters have been described.45 The other PCR primers were as follows. For Cdh1, the forward primer was 5′-tggagcgtgaacttccacag-3′, and the reverse primer was 5′-CGTGAACAGACCCTTCTTCT-3′. For E2F1, the forward primer was 5′-CCGCCATCCAGGAAAAGGT-3′, and the reverse primer was 5′-GCCCTCAAGAGACGTTGGTG-3′.
In vivo ubiquitination assay
HEK293 or 293T cells were transfected with Myc-ubiquitin (a gift from Gen Wang) or HA-ubiquitin containing only K11 lysine (HA-Ub-K11)46 (obtained from Sandra Weller through Addgene Plasmid 22901), Flag-E2F1 or untagged E2F1 and GFP-Cdh1 or a control GFP vector as indicated. Two days later, cells were treated with 5 μM adriamycin or 60 ng/ml of Velcade for 6 h. Cells were then lysed in SDS lysis buffer (1% SDS and 60 mM TRIS pH 6.8) followed by boiling for 5 min at 95°C. The cell lysates were reconstituted to 0.1% SDS by 1:10 dilution in NETN A buffer (described above) supplemented with a cocktail of protease inhibitors. The lysates were then sonicated and clarified by 10 min centrifugation at 14,000 rpm in a microfuge. Equivalent amounts of lysates were incubated overnight with Flag-agarose beads (Sigma) at 4°C. Beads were washed four times with RIPA buffer (50 mM TRIS pH 8.0, 150 mM NaCl, 0.5% Sodium Deoxycholate, 1% NP-40 and 0.1% SDS), once with 0.5 M LiCl buffer and one final wash with RIPA buffer. The beads were then boiled in Laemmli buffer and analyzed by SDS-PAGE followed by western blot analysis using HA antibody.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (RO1CA100857, RO1CA138641 and ARRA 3 P30CA125123–03S5) and Department of Defense Breast Cancer Research Program (W81XWH-09–1-0338). E.D.W. was supported by NIH “Research Supplements to Promote Diversity in Health-Related Research” Program. W.C.L. is a Leukemia and Lymphoma Society Scholar. All authors disclose that they have no financial interests that will pose a conflict of interest regarding the submitted article.
Glossary
Abbreviation:
- APC/C
anaphase-promoting complex/cyclosome
- Ub
ubiquitination
- K11
Lysine 11 residue of ubiquitin
- a.a.
amino acid
- GST
glutathione-S transferase
- P/CAF
p300/CREB-binding protein-associated factor
- TPR
tetratricopeptide repeat
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20643
References
- 1.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 2.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–13. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–44. [PMC free article] [PubMed] [Google Scholar]
- 4.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–8. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Liu K, Lin FT, Lin WC. A role for 14-3-3 tau in E2F1 stabilization and DNA damage-induced apoptosis. J Biol Chem. 2004;279:54140–52. doi: 10.1074/jbc.M410493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5:401–9. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- 7.Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–54. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ianari A, Gallo R, Palma M, Alesse E, Gulino A. Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage. J Biol Chem. 2004;279:30830–5. doi: 10.1074/jbc.M402403200. [DOI] [PubMed] [Google Scholar]
- 9.Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, et al. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–5. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 10.Hiebert SW, Packham G, Strom DK, Haffner R, Oren M, Zambetti G, et al. E2F-1:DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol Cell Biol. 1995;15:6864–74. doi: 10.1128/mcb.15.12.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–8. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 12.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4:859–64. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh JK, Fredersdorf S, Kouzarides T, Martin K, Lu X. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 1997;11:1840–52. doi: 10.1101/gad.11.14.1840. [DOI] [PubMed] [Google Scholar]
- 14.Phillips AC, Bates S, Ryan KM, Helin K, Vousden KH. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes Dev. 1997;11:1853–63. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- 15.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–25. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 16.Hsiao KM, McMahon SL, Farnham PJ. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–37. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 17.Neuman E, Flemington EK, Sellers WR, Kaelin WG., Jr. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–15. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 19.Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–93. [PubMed] [Google Scholar]
- 20.Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–9. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–81. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ, 3rd, Reis T, Edgar BA, et al. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell. 2008;15:890–900. doi: 10.1016/j.devcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peart MJ, Poyurovsky MV, Kass EM, Urist M, Verschuren EW, Summers MK, et al. APC/C(Cdc20) targets E2F1 for degradation in prometaphase. Cell Cycle. 2010;9:3956–64. doi: 10.4161/cc.9.19.13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–8. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 25.Passmore LA, Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–8. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, et al. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 2003;22:786–96. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, et al. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–8. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–32. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 30.Peart MJ, Poyurovsky MV, Kass EM, Urist M, Verschuren EW, Summers MK, et al. APC/C(Cdc20) targets E2F1 for degradation in prometaphase. Cell Cycle. 2010;9:3956–64. doi: 10.4161/cc.9.19.13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol. 2003;23:3287–304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–65. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galbiati L, Mendoza-Maldonado R, Gutierrez MI, Giacca M. Regulation of E2F-1 after DNA damage by p300-mediated acetylation and ubiquitination. Cell Cycle. 2005;4:930–9. doi: 10.4161/cc.4.7.1784. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Liu K, Lin HY, Bellam N, Ling S, Lin WC. 14-3-3Tau regulates ubiquitin-independent proteasomal degradation of p21, a novel mechanism of p21 downregulation in breast cancer. Mol Cell Biol. 2010;30:1508–27. doi: 10.1128/MCB.01335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gieffers C, Dube P, Harris JR, Stark H, Peters JM. Three-dimensional structure of the anaphase-promoting complex. Mol Cell. 2001;7:907–13. doi: 10.1016/S1097-2765(01)00234-9. [DOI] [PubMed] [Google Scholar]
- 36.Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–81. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohi MD, Feoktistova A, Ren L, Yip C, Cheng Y, Chen JS, et al. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol Cell. 2007;28:871–85. doi: 10.1016/j.molcel.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Passmore LA, Booth CR, Vénien-Bryan C, Ludtke SJ, Fioretto C, Johnson LN, et al. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol Cell. 2005;20:855–66. doi: 10.1016/j.molcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Bentley AM, Williams BC, Goldberg ML, Andres AJ. Phenotypic characterization of Drosophila ida mutants: defining the role of APC5 in cell cycle progression. J Cell Sci. 2002;115:949–61. doi: 10.1242/jcs.115.5.949. [DOI] [PubMed] [Google Scholar]
- 40.Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–9. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- 41.Hofmann F, Martelli F, Livingston DM, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–59. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 42.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–38. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 43.Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, et al. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol Biol Cell. 2007;18:5139–53. doi: 10.1091/mbc.E07-05-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paik JC, Wang B, Liu K, Lue JK, Lin WC. Regulation of E2F1-induced apoptosis by the nucleolar protein RRP1B. J Biol Chem. 2010;285:6348–63. doi: 10.1074/jbc.M109.072074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu K, Bellam N, Lin HY, Wang B, Stockard CR, Grizzle WE, et al. Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol Cell Biol. 2009;29:2673–93. doi: 10.1128/MCB.01140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livingston CM, Ifrim MF, Cowan AE, Weller SK. Virus-Induced Chaperone-Enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 2009;5:e1000619. doi: 10.1371/journal.ppat.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.