Abstract
Combinations of nucleoside and nonnucleoside inhibitors (NNRTIs) of HIV-1 reverse transcriptase (RT) are widely used in anti-AIDS therapies. Five NNRTIs including nevirapine are clinical drugs; however, the molecular mechanism of inhibition by NNRTIs is not clear. We determined the crystal structures of RT–DNA–nevirapine, RT–DNA, and RT–DNA–AZT-triphosphate complexes at 2.85, 2.70, and 2.80 Å, respectively. The RT–DNA complex in the crystal could bind nevirapine or AZT-triphosphate; however, not both. Binding of nevirapine led to opening of the NNRTI-binding pocket. The pocket formation caused shifting of the 3’-end of DNA primer by ~5.5 Å away from its polymerase active site position. Nucleic acid interactions with fingers and palm subdomains were reduced, the dNTP-binding pocket was distorted, and the thumb opened up. The structures elucidate complementary roles of nucleoside and nonnucleoside inhibitors in inhibiting RT.
The enzyme reverse transcriptase (RT) of HIV-1 is responsible for copying the viral single-stranded RNA genome to double-stranded (ds) DNA in the cytoplasm of infected cells. This 117 kDa heterodimeric (p66 and p51) protein performs three catalytic steps: (1) RNA-dependent DNA polymerization to synthesize a (−) strand DNA complementing the viral (+) strand RNA genome, (2) RNase H cleavage of the RNA strand, and (3) DNA-dependent DNA polymerization to synthesize dsDNA using the (−) strand DNA as the template. The dsDNA is transported into the nucleus as a pre-integration complex and integrated into the chromosome of the infected cell. HIV-1 infection is chronic and requires life-long treatment. Emergence of drug-resistant HIV-1 strains and side effects impede the long-term use of drugs; therefore, new drugs against existing and new targets are required and constantly being developed. HIV-1 infection, in general, is treated with combinations of three or more antiviral agents. Twenty-six individual drugs are approved of which thirteen inhibit RT1. RT drugs are either (1) nucleoside or nucleotide inhibitors (NRTIs) that are incorporated into the growing DNA strand and act as chain terminators because NRTIs lack a 3’-OH group, or (2) nonnucleoside RT inhibitors (hereafter called NNRTIs or nonnucleosides) that are allosteric inhibitors of DNA polymerization. Several anti-retroviral therapy regimens use nonnucleosides in combinations with NRTIs; nevirapine, delavirdine, efavirenz, etravirine, and rilpivirine (TMC278, Edurant) are nonnucleoside drugs.
Structures of RT have been known for almost two decades when binary complexes of RT with nevirapine2 and with DNA3 were reported. An innovative protein-nucleic acid cross-linking technique helped obtain an RT–DNA–dTTP ternary complex structure4. Subsequently, a large number of RT structures have been studied that help in understanding the enzymatic activities, inhibition and mechanisms of drug resistance5,6, and have aided design of new drugs7. RT has a hand-like structure8 (Fig. 1). The palm contains the polymerase active site and nonnucleoside-binding pocket located ~10 Å apart. The major conformational changes in RT9 characterized by structural studies are: (1) the thumb lifts up to bind nucleic acid10,11, (2) the fingers fold down to capture dNTP substrates in the presence of nucleic acid4, and (3) nonnucleoside binding leads to thumb hyperextension. Pre-steady and steady state kinetics data suggested that the binding of a nonnucleoside inhibits the chemical step of DNA polymerization12,13; however, precise effects on nucleic acid and dNTP are unclear14, and RT–nonnucleoside association and dissociation are complex processes15, which are not yet conclusively explained by kinetics experiments. Binding of a nonnucleoside can enhance p66/p51 dimerization16. Recent single-molecule FRET studies17,18 revealed that RT frequently flips and slides over nucleic acid substrates in the process of copying the viral RNA into dsDNA. An RT–nucleic acid complex is stabilized in a polymerization-competent conformation when dNTP is present. In contrast, nevirapine has a destabilizing effect that was interpreted as the consequence of loss of thumb and fingers interactions with nucleic acid18. Binding of an incoming dNTP at the polymerase active site decreased the efficiency of cross-linking, whereas, NNRTI binding increased cross-linking19; site-directed photocrosslinking of the fingers subdomain of HIV-1 RT to an extended template using photolinkers of different length to monitor changes in the distance between particular positions on the surface of the protein and a nucleic acid substrate. Pre-steady state kinetics analyses12,13,20 reported no decrease in binding of DNA or dNTP upon binding of an NNRTI; in fact, dNTP-binding was enhanced at saturating concentrations. Potential mechanisms of inhibition by nonnucleosides postulated include: (1) restriction of thumb mobility2, (2) distortion of the catalytic triad21, (3) repositioning of the primer grip22, and (4) loosening of the thumb and fingers clamp18.
Figure 1. Polymerase domain of HIV-1 RT in complex with DNA.

Nevirapine and AZTTP are placed based on superposition of the palm subdomain of nevirapine- and AZTTP-ternary structures, respectively, on the RT–DNA structure. The 3’-azido group of AZT-terminated primer in the current RT–DNA and AZTTP-ternary structures occupies the metal A position, whereas metal B is present in the AZTTP-ternary structure; metal ion A is positioned based on its location in the dTTP-ternary structure4. RT binds dNTP and catalytically incorporates nucleotides by a cation-dependent nucleotidyltransferase reaction. Incorporation of an NRTI, like AZT, or binding of a nonnucleoside, like nevirapine, inhibits DNA polymerization by RT.
Nonnucleosides indirectly interfere with DNA polymerization. Therefore structures of RT–nucleic acid–NNRTI (± dNTP or analog) complexes are essential for understanding inhibition of polymerization and excision23,24 by a nonnucleoside and to visualize how both types of RT drugs synergistically inhibit DNA polymerization25. Here we report a RT–DNA binary crystal form in which RT–DNA could undergo conformational changes to accommodate an AZT (zidovudine, 3’-azido-3’-deoxythymidine26)-triphosphate (or AZTTP) at the dNTP-binding site or nevirapine at the nonnucleoside-binding pocket. The differences between the structures directly reflect the impact of nevirapine binding on RT–DNA complex and help illustrate the structural basis of nonnucleoside inhibition.
RESULTS
Polymerase competent state of RT–DNA binary complex
In completing synthesis of a dsDNA from viral RNA genome, RT makes several jumps and binds different nucleic acid sequences27. Recent single-molecule studies17,18 revealed that RT binds a nucleic acid in two distinct modes: polymerase-competent (mode-1), analogous to that observed in crystal structures, and a switched mode (mode-2). Binding of dNTP stabilizes mode-1 for RT–nucleic acid complexes, and only mode-1 complex can bind dNTP and incorporate nucleotide. Previous structures of RT in complexes with non-cross-linked nucleic acids3,28 were naturally formed by mixing DNA or with RT in solution; however, RT cross-linking to DNA4 stabilized the complex for studying the polymerase-relevant states of the enzyme. In our current study, RT–DNA cross-linking was used for obtaining a stable polymerase-relevant structure in the presence of a nonnucleoside drug at high resolution, in light of the fact that nevirapine binding increases dynamics of an RT–nucleic acid complex, as revealed by the single-molecule FRET study18.
AZTTP or nevirapine binding to RT–DNA complex in crystal
Recent successful experiments were designed based on the facts that (1) a typical protein crystal has >50% disordered solvent which may permit repositioning of a flexible domain if the domain is not constrained by interactions with neighboring symmetry-related molecules in the crystal, (2) in an earlier experiment we replaced tenofovir diphosphate with its natural counterpart dATP in crystal by soaking29, and (3) dNTP analogs were successfully soaked into RT–DNA crystals30. Therefore, a crystal form of RT–DNA complex in which the thumb and fingers subdomains have enough room to flex may permit binding of dNTPs and nonnucleosides by soaking. We discovered such a crystal form for D498N mutant RT cross-linked with DNA containing AZT-terminated primer (see online Methods), and soaked AZTTP as the incoming dTTP analog and nevirapine (Fig. 1). An attempt to crystallize RT cross-linked with a ddTTP (2’,3’-dideoxythymidine triphosphate) primer-terminated DNA did not produce diffraction quality crystals; change in DNA sequence and incoming dNTP in the past had led to new crystal forms31 or were unsuccessful in yielding crystals. Our attempts enabled us to obtain the crystal structures of RT–DNA, RT–DNA–AZTTP (AZTTP-ternary), and RT–DNA–nevirapine (nevirapine-ternary) complexes at 2.70, 2.80, and 2.85 Å resolution, respectively (Table 1). The crystals soaked with AZTTP together with nevirapine in the presence or absence of MgCl2 (or MnCl2) formed nevirapine-ternary complex only, despite this crystal form allowing RT–DNA to bind either individually. In all three structures, the primer 3’-end AZT was designed to occupy the P site and blocked further incorporation of nucleotides; incoming AZTTP in AZTTP-ternary structure occupied the N site (Fig. 2a) and the thymine was paired with the template adenine base.
Table 1.
Data collection and refinement statistics.
| RT:DNA binary complex | AZTTP-ternary complex | Nevirapine-ternary complex | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 89.48, 133.17, 139.90 | 90.05, 132.65, 138.01 | 89.82, 132.05, 142.73 |
| α, β, γ (°) | 90, 98.67, 90 | 90, 98.12, 90 | 90, 100.84, 90 |
| Resolution (Å) | 50.0–2.7 (2.75–2.70)* | 50.0–2.8 (2.9–2.8) | 40–2.85 (2.9–2.85) |
| Rmerge | 0.089 (0.625) | 0.083 (0.523) | 0.090 (0.618) |
| I / σI | 11.6 (1.9) | 11.7 (1.9) | 10.1 (1.7) |
| Completeness (%) | 98.4 (97.2) | 92.8 (85.1) | 99.0 (97.7) |
| Redundancy | 3.9 (3.2) | 3.2 (2.4) | 3.6 (2.9) |
| Refinement | |||
| Resolution (Å) | 44.0–2.7 | 45.0–2.8 | 38.4–2.85 |
| No. reflections (Rfree set) | 86,138 (2,606) | 73,170 (2,206) | 75,395 (2,287) |
| Rwork / Rfree | 0.238/0.274 | 0.229/0.265 | 0.250/0.299 |
| No. atoms | |||
| Protein+DNA | 17,635 | 17,640 | 17,478 |
| Ligand/ion | 46 | 110 | 40 |
| Water | - | - | - |
| B-factors (Å2) | |||
| Protein | 83 | 65 | 82 |
| Ligand/ion | 87 | 76 | 72 |
| Water | - | - | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.011 | 0.010 |
| Bond angles (°) | 1.26 | 1.38 | 1.33 |
One crystal was used for each data set.
Values in parentheses are for highest-resolution shell.
Figure 2. Effects of nevirapine on polymerase active site conformation and dNTP-binding.
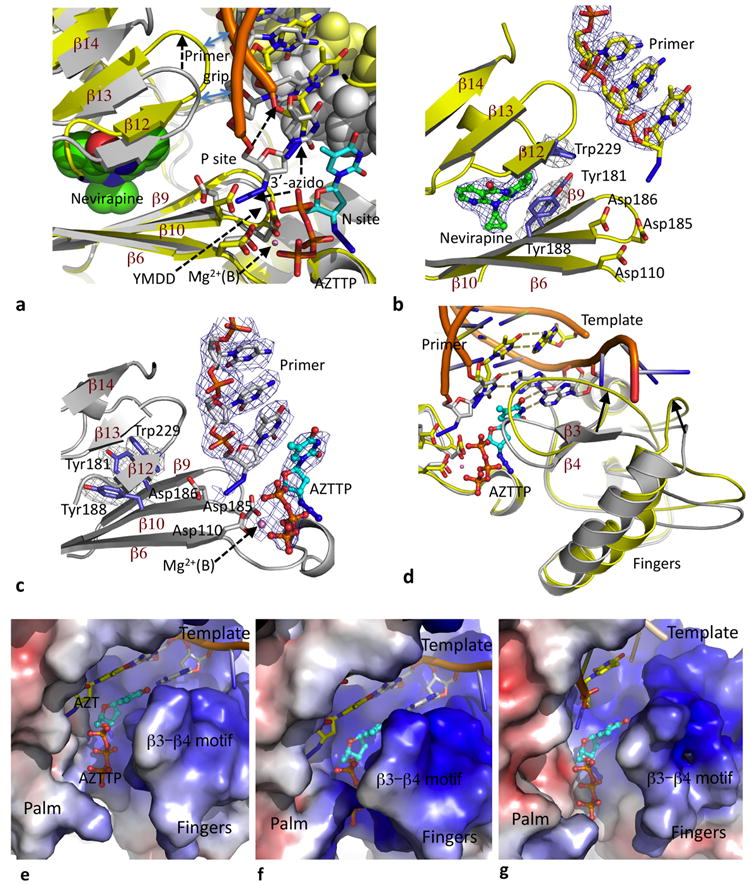
a. Palm superposition of nevirapine-ternary (yellow protein and DNA) on AZTTP-ternary (gray protein and DNA; cyan AZTTP) structures; the template strands are shown as space-filled models. Binding of nevirapine (green space filling) shifts the “primer grip” by ~4 Å which pulls the AZT at the primer end away from the P site and repositions the template strand. b & c. Electron density maps calculated using six-fold symmetry averaged phases (see online methods) show the binding of nevirapine and AZTTP in respective ternary structures; comparison of the two structures show rotamer switching for Y181 and Y188, shearing of the β12–β13–β14 sheet, and the shift of DNA primer. d. Repositioning of fingers subdomain based on the superposition and color code described in panel a. In nevirapine-ternary structure, the template (yellow) overhang is disordered because portions of the repositioned fingers subdomain in the structure would interfere with the template-binding track in AZTTP-ternary structure (gray). Electrostatic potential surfaces of (e) RT–DNA, (f) AZTTP-ternary, and (g) nevirapine-ternary complexes show the conformational variations of the dNTP-binding region upon binding of AZTTP or nevirapine. AZTTP is present only in AZTTP-ternary (f) structure and is placed in RT–DNA (e) and nevirapine-ternary (g) structures based on superposition of palm subdomains.
Impact of nevirapine or AZTTP binding on RT–DNA complex
Binding of AZTTP shrunk the unit cell volume of RT–DNA crystals by 1.2%, and binding of nevirapine expanded the volume by 0.67%; however, individual cell parameters changed more upon binding of nevirapine (Table 1). The AZTTP-ternary structure represents an RT-polymerase complex having architecture similar to that in all RT-ternary structures despite differences in RT or DNA sequences, incoming dNTP or analogs, and crystallization conditions or parameters (Supplementary Fig. 1 and supplementary Table 1) in different studies; the base-pairing, metal chelation, and other interactions of AZTTP with RT resemble that of dTTP4 substrate and AZTppppA (ATP-mediated excision product of AZT)6,23,24 in respective co-crystal ternary structures. AZTTP binding causes the tip of the fingers to close and wrap around the dNTP analog (Supplementary Fig. 2a). Upon AZTTP binding, the conserved β3–β4 region folds in by ~2 Å and side chains rearrange to form a closed dNTP-binding pocket in the ternary structure, analogous to that observed upon soaking of GS-914830. Unlike in the binary complex structure determined in the presence of a monoclonal antibody3, the fingers subdomain in the current binary structure is not wide open (Supplementary Fig. 2b); however, it is highly flexible. Apart from the fingers rearrangement, AZTTP binding has no major discernible impact on RT–DNA conformation. The 3’-azido group of the primer terminal AZT occupies the position of cofactor metal ion A (Fig. 1).
The mode of binding and interactions of nevirapine in nevirapine-ternary and nevirapine-binary2,21 structures are similar; however, the binding of nevirapine had a profound effect on RT–DNA conformation compared to the binding of AZTTP. The opening of the nonnucleoside-binding pocket upon nevirapine binding is accompanied by switching of the Tyr181 and Tyr188 rotamer conformations and shearing of the β12–β13–β14 sheet away from the β6–β10–β9 sheet (Figs. 2a-c; supplementary Movie 1); the β6–β10–β19 sheet contains the polymerase “catalytic triad” (Asp110, Asp185, and Asp186), and the β12–β13–β14 sheet contains the “primer grip” that positions the primer strand appropriately for nucleotide incorporation2,21. Upon binding of nevirapine, the primer grip is shifted by ~4 Å, the shifted primer grip lifts the DNA primer terminus away from the P site (Fig. 2a), and key interactions of the conserved catalytic Y183MDD motif with the primer terminus are lost. The nucleotide complementary to AZT and upstream template overhang are displaced and are partially or completely disordered in the nevirapine-ternary structure. The loss of contacts between the template strand and fingers has a profound impact on the positional stability of both, and hence, the fingers subdomain is also weakly ordered. The binding and incorporation of a dNTP requires interactions with fingers, in particular the β3–β4 motif, base pairing with the first template overhang, and chelation with the metal ion B4. The β3–β4 motif is deformed in the nevirapine-ternary structure, and parts of the fingers are shifted by 5-7 Å into regions that otherwise accommodate the template overhang in catalytically competent RT–DNA structures (Fig. 2d). The open dNTP-binding site (Fig. 2e) in RT–DNA is closed when AZTTP is bound (Fig. 2f). The open and rearranged dNTP-binding site (Fig. 2g) in nevirapine-ternary complex could provide easy access for dNTPs to reach the site; however, this structure did not provide evidence of formation of a closed dNTP-bound quaternary complex when soaked with AZTTP.
The thumb subdomain in nevirapine-ternary structure has an extended conformation similar to that in the first nevirapine-binary structure (PDB ID 3HTV)2, and the inward movement of the thumb is restricted by the displaced β12–β13–β14 sheet (Supplementary Fig. 3). The thumb in the higher-resolution nevirapine-binary structure (PDB ID 1VRT)21 is further extended (Fig. 3a) and involved in crystal contacts. The catalytic triad in nevirapine-ternary structure has an intermediate conformation (Fig. 3b) between that in AZTTP-ternary and nevirapine-binary21 structures, suggesting that the Y183MDD motif may not adopt a rigid conformation in the presence of a nonnucleoside. The fingers subdomain has a semi-open or open conformation in nevirapine-ternary or nevirapine-binary structure (Fig. 3c), respectively. The interactions of DNA with palm and fingers subdomains are substantially reduced with nevirapine present (Fig. 3d); however, the remaining DNA-protein interactions are less perturbed despite all subdomains being rearranged. A Y183MDD sheet (β6–β10–β9) superposition (Supplementary Fig. 4) between nevirapine-ternary and RT–DNA binary structures showed that RNase H active site has moved ~6.5 Å away upon nevirapine binding, and DNA is rearranged while retaining its interactions with RNase H domain (Fig. 3d).
Figure 3. Impact of nevirapine binding on thumb, fingers and DNA.
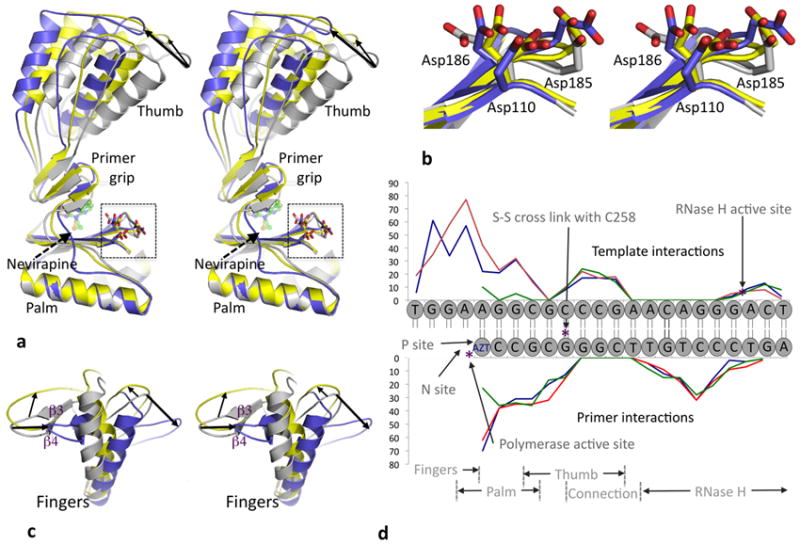
a. Stereo view of a palm superposition of nevirapine-ternary (yellow), AZTTP-ternary (gray), and nevirapine-binary21 (blue) structures shows that the thumb subdomain is hyper extended upon nevirapine binding; the tip of the thumb is ~7.5 and 11 Å away in nevirapine-ternary and nevirapine-binary21 structures, respectively, from the tip in AZTTP-ternary structure based on described superposition. The thumb of nevirapine-ternary and the first nevirapine-binary structures2 however are in close proximity based on palm superposition (Supplementary Fig. 3). In the higher resolution nevirapine-binary structure21, the thumb was further extended as a result of crystal contacts. b. Zoomed stereo view of the polymerase active site region of superimposed structures in a. The YMDD loop in nevirapine-ternary (yellow) has an intermediate state between AZTTP-ternary (gray) and nevirapine-binary21 (blue) structures indicating active site flexibility even when nevirapine is bound; the Cα atom of D185 in the nevirapine-ternary structure is displaced ~1 Å from its counterpart in the other two structures. c. Repositioning of fingers subdomain based on described superposition and color scheme in panel a. The thick arrows represent fingers repositioning to nevirapine-binary and thin arrow represents repositioning to nevirapine-ternary from AZTTP-ternary structure. d. Comparison of interactions of DNA template-primer with RT in RT–DNA (blue), AZTTP-ternary (red) and nevirapine-ternary (green) structures; the extent of interactions (y-axis) are represented by the number of interatomic distances (<4.5 Å) between a nucleotide and RT. The interactions of DNA with fingers and palm are decreased upon nevirapine binding; the interactions with thumb, connection, and RNase H are not altered.
DISCUSSION
Primer repositioning obstructs polymerization and excision
DNA polymerization by RT is carried out by cyclic repetitions of three structurally distinct steps (Fig. 4a) representing (1) binding of dNTP, (2) nucleotide incorporation with concomitant release of pyrophosphate, and (3) translocation of the incorporated nucleotide from N to P site. Incorporation of an NRTI stalls the cycle by blocking further incorporation. An incorporated AZT can readily shuttle between P and N sites, and a pyrophosphate donor such as ATP, with the assistance of AZT-resistance mutations in RT, excises the AZT molecule from the N site. Excision is carried out by completing one reverse cycle of polymerization, structurally represented by RT–DNA–AZT (P-complex) → RT–DNA–AZT (N-complex)6 → RT–DNA–AZTppppA (excision product complex)6. The P’-complex, represented by the nevirapine-ternary structure, is unlikely to accommodate ordered binding of dNTP in a catalytically competent mode19. Even if a dNTP were to bind, the primer 3’-end at the P’ site would not allow nucleotide incorporation.
Figure 4. A molecular mechanism of nonnucleoside inhibition and impact of DNA binding on resistance to nonnucleoside drugs.

a. Scheme representing effect of nonnucleoside inhibitor binding on DNA polymerization by RT. For incorporating a nucleotide, RT completes three structurally distinct steps in a clockwise cycle. Incorporation of an NRTI drug blocks the cyclic process, leading to a P-complex structure lacking 3’-OH, like the structure of RT–DNA–AZT-terminated complex. RT removes AZT by following the three steps in a reverse cycle and using a pyrophosphate donor (ATP or PPi). Binding of a nonnucleoside inhibitor shifts the primer end from P site to P’ site. This P’-complex is catalytically incompetent because the 3’-end is positioned away from the polymerase active site. Release of nonnucleoside inhibitor would shift the P’-complex to P-complex and restore DNA polymerization by RT. b. Positions of three structural elements: β6–β10–β9 sheet (yellow), β12–β13–β14 sheet (gray), and loop 95-103 (cyan) + Glu138 loop (blue) of p51 in nevirapine-ternary structure. These elements are responsible for and rearranged upon the binding of nevirapine (green). Nonnucleoside inhibitor binding locks the primer grip away from the active site, whereas binding of dsDNA requires the primer grip to be positioned near the active site as in RT–DNA and AZTTP-ternary structures. Red arrows indicate the structural movements (Supplementary Movie 1) between the polymerase-competent and nonnucleoside-bound states.
dNTP binding in the presence of nevirapine
Earlier reported kinetics data suggested that nevirapine improves binding of dNTPs and inhibits the chemical step of nucleotide incorporation12,13 that directly correlates with the proposed “catalytic distortion” model21 of nonnucleoside inhibition. A later pre-steady kinetics study20 using elemental effect32 (dTTPαS vs. dTTP) revealed that a conformational state and not the chemical step leading to nucleotide incorporation is blocked by a nonnucleoside inhibitor and favored the “primer-grip distortion” model22 as a possible molecular mechanism for nonnucleoside inhibition. The nevirapine-ternary structure presented in this study defines the impact of nevirapine binding on DNA positioning and RT–DNA interactions. Also, the structure shows that the primer grip repositioning locked the thumb at a hyper-extended position2, and results in a loss of interactions between DNA and the polymerase domain of RT18. While the RT–DNA complex in crystal could bind nevirapine or AZTTP, the complex did not bind both simultaneously and thereby did not provide direct evidence of dNTP binding. In the nevirapine-ternary structure, the flexible fingers (Fig. 3c), repositioned template–primer, and disordered template-overhang would reduce ordered dNTP binding at the N site. However, the flexibility of the fingers may allow alternative modes of dNTP binding to RT in presence of nevirapine. Structures of DNA polymerases (such as Klenow fragment)33,34 have shown that transition from open to closed fingers is responsible for binding and incorporation of correct nucleotides complementing the template overhang, and a recent single-molecule FRET experiment on Klenow fragment demonstrated a high degree of flexibility of the fingers in the absence of substrates35.
According to pre-steady state kinetics20 dNTPs are expected to bind nevirapine-ternary complex in a metal-dependent but not in a catalytically relevant mode. Structures of DNA polymerases before and after misincorporation of a dATP opposite to an abasic nucleotide have revealed rearrangements of DNA and protein at the polymerase active sites. An open complex of KlenTaq DNA polymerase is formed upon binding of ddATP in a catalytically relevant mode opposite to a template furan36; the amino acid residue Y671 on O-helix interacts with the ddATP base and partly compensates for the loss of canonical base-pairing. A bacteriophage RB69 DNA polymerase open complex structure37 (PDB ID 2P5G) was determined with a dAMP incorporated into the primer opposite to a template furan; apparently, lack of stable interactions (base pairing and stacking) of the terminal dAMP led to multiple conformations of the DNA in the four copies of the complex in asymmetric unit. HIV-1 RT appears to follow the “A-rule” for misaligned primer extension opposite to an abasic moiety in vitro38, which suggests that the polymerase active site conformation of RT could rearrange to accommodate and incorporate a dNTP without canonical base-pairing at the N site. In contrast to the rearrangements of the DNA polymerases discussed, a nevirapine-ternary structure of RT is expected to bind dNTP in a polymerase incompetent mode. Although DNA polymerases share remarkably similar domain architecture and chemistry of nucleotide incorporation, characteristics such as rate of nucleotide incorporation, mode and extent of protein nucleic acid interactions, presence or absence of a proof-reading mechanism, fidelity, and rate-limiting steps that select correct vs. incorrect nucleotide incorporation39 are highly enzyme specific.
It is likely that dNTPs enter the N-site of nevirapine-ternary complex (Fig. 2g) and bind in multiple orientations. Alternatively, the primer 3’-end of a nucleic acid duplex may slide past the polymerase active site18, and a rearranged dNTP-binding region with the duplex resting over it may define a ordered yet non-productive binding of dNTP. Further biochemical, mutational, and structural studies are needed to clarify if there is ordered dNTP-binding to RT, and to map the RT-dNTP interactions in presence of nonnucleosides and nucleic acid.
Conformational states of RT in presence of NNRTI and DNA
Taking the biochemical data, clinical observations, and structural states of RT into consideration, it appears that RT with nonnucleoside and nucleic acid in solution would exist in three states: (I) nevirapine-ternary, (II) nevirapine-binary or switched mode-2 complex17, and (III) RT–DNA. Among the three, state III is active, and states I and II are inactive for nucleotide incorporation. The relative proportion of the three states would depend upon binding characteristics and effective concentrations of nevirapine and nucleic acid17. The fraction of RT in state III would incorporate nucleotides unless DNA primers are terminated by NRTIs, which may be a reason why (1) a combination of nonnucleoside and NRTI is more effective in decreasing the viral load in treatment-naive patients than when treated with individual components and (2) both NRTI and nonnucleoside resistance mutations co-emerge in response to a drug combination. For example, a recent phase III trial40 of a triple combination of two NRTIs (tenofovir-disoproxil-fumarate and emtricitabine) and a nonnucleoside (rilpivirine) reported that M184V/I and E138K mutations are predominantly associated with treatment failure. An arm of the trial that used efavirenz instead of rilpivirine showed M184I/V and K103N as the predominant resistance mutations, however at a lower frequency. Also, additive effects of nevirapine and AZT have been observed biochemically25 and clinically41.
NNRTI inhibition and resistance mutations
Excision of AZT is diminished in the presence of a nonnucleoside42,43. AZT at the primer 3’-terminus is locked at the P’ position (Fig. 4a) in nevirapine-bound RT and not permitted to translocate back to the N site for excision. Additionally, the distorted dNTP-binding site (Fig. 2g) may decrease the binding of excision substrate ATP (or PPi). Nevirapine blocks both DNA polymerization and NRTI excision.
Binding of nevirapine is accompanied by repositioning of the primer grip away from the active site whereas binding of DNA in a catalytically competent mode requires the primer grip to be near the active site (Fig. 4b; supplementary Movie 1). A bound nonnucleoside is surrounded by three walls: (1) β6–β10–β9 sheet, (2) β12–β13–β14 sheet, and (3) 100-105 loop of p66 + Glu138 loop of p51. Rearrangements of the loops would permit entry and exit of most nonnucleosides. In addition to the pocket mutations that directly influence inhibitor binding, the loop mutations such as K103N and E138K (of p51) do not have extensive interactions with a bound nonnucleoside. The loop mutations facilitate exit of nonnucleosides from the pocket, permitting the primer grip to position nucleic acid in a catalytically relevant mode and shifting the equilibrium towards state III. The mutation K103N confers relatively uniform resistance to chemically diverse nonnucleosides, and as discussed above, two loop mutations E138K and K103N are predominantly associated with treatment failure of rilpivirine and efavirenz arms, respectively.
Nonnucleoside resistance mutations impact NRTI susceptibility, implying communication between the two sites even in absence of a nonnucleoside. A common nonnucleoside resistance mutation Y181C enhances AZT sensitivity in a thymidine analog mutant background44. A recent study has shown that E138K mutation is compensatory for M184I/V and vice versa45. The individual mutations E138K or M184V/I reduce the virus fitness by 2-3 fold; however E138K+M184I/V in combination restores the fitness equivalent to that of wild-type HIV-1.
Conclusions
The current structures of three RT complexes, earlier RT structures, and abundant available clinical and biochemical data enabled us to analyze the specific effects of nonnucleosides on RT structure and function. Nevirapine binding revealed several impacts on RT–DNA complex. It has a direct impact by displacing the primer grip, and consequently, the primer terminus moves out, the thumb is locked at a hyper-extended position2, interactions between DNA and the polymerase domain is decreased18, dNTP-binding site is distorted, and the RNase H active site is repositioned with respect to the polymerase active site. Enhanced understanding of mechanistic details of nonnucleoside inhibition may also aid development of allosteric small molecules that inhibit by blocking conformational mobility of molecular machines such as hepatitis C NS5B polymerase46, bacterial RNA polymerase47, and other macromolecular assemblies associated with various diseases.
Methods and any associated references are available in the online version of the paper at www.nature.com/nsmb.
METHODS (online)
RT expression, RT–DNA cross-linking, and purification
HIV-1 reverse transcriptase (RT) construct RT127A used for the current structural studies was derived from a previously reported construct RT13A48. The mutation D498N in the RT127A construct was introduced using the methods described therein. The amino acid residue D498 is a part of the RNase H active site. The D498N mutant RT blocks RNase H activity; however, the mutant RT has polymerase activity comparable to wild-type RT49. The D498N mutant RT was expressed and purified as previously reported48. Briefly, the RT was expressed in BL21-CodonPlus-RIL cells, induced with 1 mM IPTG at an OD600 of 0.9, followed by expression at 37°C for 3 hours. The cells were sonicated at a power output of ~45 watts with a Misonix 3000 sonicator. The samples were purified using a Ni-NTA column according to manufacturer’s recommendations (Qiagen, Valencia, CA USA). The final purification step was carried out using a Mono Q column and the purified RT samples were buffer exchanged into 10 mM Tris pH 8.0 and 75 mM NaCl.
The 27-mer DNA template (5’ ATGGAAGGCGCCCGAACAGGGACTGTG 3’) was synthesized by Integrated DNA Technologies (Coralville, IA). The 20-mer primer (5’ ACAGTCCCTGTTCGGGCGCC 3’) bearing a cross-linkable thioalkyl tether (on G in the primer strand)50 was custom synthesized by Midland Certified Reagent Company (Midland, TX) using phosphoramidite that was custom synthesized by Chemgenes (Wilmington, MA), and the primer was annealed to the template. The 27:20-mer dsDNA was cross-linked to RT127A at the mutated Q258C site of p66, and the cross-linked primer was extended with an AZT at the 3’-end through RT polymerization51. The cross-linked RT127A–DNA (AZT-terminated primer) complex was purified using Ni-NTA and heparin columns in tandem as previously described51. The His-tag was removed by adding human rhinovirus 14 3C protease at a 1:10 ratio to RT–DNA complex at 4°C for 48 hours. Ni-NTA beads were added to remove the cleaved His-tag and His-tagged protease. An Amicon Ultra-4 Ultracel unit (30 kDa cutoff) was used to exchange the buffer into 75 mM NaCl, 10 mM Tris-HCl pH 8.0. The protein was concentrated to 15 mg.ml-1.
Crystallization
Sitting drops of 0.5 μl protein plus 0.5 μl of well solution were set up at 4°C on Combiclover Jr. plates from Emerald BioStructures. The well solution contained 10-12% PEG 8000 (w/v), 50 mM Bis-Tris propane pH 6.8–7.2, 100 mM (NH4)2SO4, 5% sucrose (w/v), 5% glycerol (v/v), and 20 mM MgCl2. Crystals appeared in a few days, and continued growing slowly for an additional 2-3 weeks to a full size of ~200 μm long and 50-150 μm thick. For the nevirapine soak below, the crystals were grown with 2 mM AZTTP added to the drop.
Crystal soaking and freezing
Binary complex: An RT–DNA (AZT-terminated primer) binary complex crystal of size 200 × 200 × 60 μm3 was transferred to 150 μl of a stabilization solution containing 12% PEG 8000, 10% glycerol at pH 7 for 30 minutes. The crystal was dipped in 50 μl cryoprotective solution containing 20% glycerol (v/v) for 1 minute and flash cooled in liquid N2.
AZTTP soak
An RT–DNA (AZT-terminated primer) binary complex crystal of size 180 × 120 × 40 μm3 was transferred to 50 μl of a stabilization solution containing 12% PEG 8K (w/v), 50 mM Bis-Tris propane pH 7.2, 100 mM (NH4)2SO4, 5% sucrose (w/v), 5% glycerol (v/v), 20 mM MgCl2, 2 mM AZTTP for five minutes. The crystal was transferred to 50 μl of a cryoprotective solution raised to 20% glycerol (v/v) for one minute then flash cooled in liquid N2.
AZTTP+nevirapine or nevirapine soaks
In an effort to obtain a structure of RT–DNA in complex with both AZTTP at the dNTP-binding site and nevirapine at the nonnucleoside inhibitor-binding site, RT–DNA (AZT-terminated primer) ternary complex crystals, grown in 2 mM AZTTP were transferred to 150 μl of the stabilization solution (same as for the binary complex data set above) to remove free AZTTP and Mg2+ ions for 10 - 15 minutes. The crystals were then transferred to 50 μl of the stabilization solution plus 2 mM nevirapine for one hour, then 50 μl with 2 mM each nevirapine and AZTTP and 20 mM MgCl2 for 15 minutes. Finally the crystals were stabilized for one minute in 50 μl this solution raised to 20% glycerol (v/v) for cryo-protection. This experiment produced only nevirapine-ternary complex; no Mg2+ ion or AZTTP binding was detected at the dNTP-binding site when nevirapine was bound to HIV-1 RT in datasets from multiple crystals, or when Mn2+ was substituted for Mg2+. Crystals soaked with nevirapine after washing out Mg2+ ions and without adding AZTTP also produced nevirapine-ternary complex.
X-ray crystallography
Multiple X-ray diffraction datasets were collected from the above-described crystals and duplicate soaks using synchrotron sources: the F1 beamline at Cornell High Energy Synchrotron Source (CHESS) and the X25 beamline at Brookhaven National Laboratory (BNL). The data were processed and scaled using HKL200052 (Table 1). The diffraction data from nevirapine-ternary complex crystals were highly anisotropic, and observed structure factors were corrected using UCLA MBI – Diffraction Anisotropy Server53. The structures were solved by molecular replacement using protein atoms in the crystal structure of RT–DNA– tenofovir-diphosphate29, and the subdomains were positioned by rigid-body refinements in which each RT molecule was divided into thirteen segments. Each crystal structure had two RT complexes per asymmetric unit. The three datasets corresponding to three RT complexes were non-isomorphous with each other, which allowed us to conduct 6-fold real space averaging among the three crystal forms and two-fold non-crystallography symmetry (NCS) within each crystal form. DMMULTI54, as implemented in CCP4, was used for averaging and phase extension. The electron density maps calculated from averaged phases and figures of merit (FOM) helped position the individual amino acid residues, DNA, nevirapine, and AZTTP. The fingers subdomain is the least ordered region in all three structures. The degree of disorder of the fingers varies from higher to lower in the sequence nevirapine-ternary > RT–DNA-binary > AZTTP-ternary; the binding of AZTTP increased the positional stability of the fingers subdomain in the AZTTP-ternary structure. Individual structures were refined using PHENIX55 (m) and the model building was carried out using COOT56. The figures showing structural information were generated using PyMOL (http://www.pymol.org/).
Supplementary Material
Acknowledgments
We acknowledge personnel at the Cornell High Energy Synchrotron Source (CHESS) and Brookhaven National Laboratory (BNL) for generous allocation of data collection time. We thank Art Clark, Steve Hughes, and Steve Tuske for advice, and Stefan Sarafianos and Aaron Shatkin for helpful comments on the manuscript. We are grateful to the U.S. National Institutes of Health (NIH) for R21 Award AI 087201 to K.D. and R37 MERIT Award AI 27690 to E.A.
Footnotes
Author Contributions: K.D. and E.A. designed the project, S.E.M. performed the experiments, K.D. carried out structural studies and analyses, J.D.B. cloned and expressed the protein. K.D., S.E.M., and E.A. wrote the paper.
Author Information: The authors declare no competing financial interests.
Accession codes The atomic coordinates and structure factors for the structures of RT–dsDNA, AZTTP-ternary, and nevirapine-ternary complexes have been deposited in the Protein Data Bank (PDB) under accession codes xxxx, yyyy, and zzzz respectively.
Supplementary Information is linked to the online version of the paper at www.nature.com/nsmb.
References
- 1.U. S. F. & D. Administrtion. http://www.fda.gov/ForConsumers/byAudience/ForPatientAdvocates/HIVandAIDSActivities/ucm118915.htm.
- 2.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 3.Jacobo-Molina A, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 5.Ren J, Stammers DK. Structural basis for drug resistance mechanisms for non-nucleoside inhibitors of HIV reverse transcriptase. Virus Res. 2008;134:157–170. doi: 10.1016/j.virusres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Tu X, et al. Structural basis of HIV-1 resistance to AZT by excision. Nat Struct Mol Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das K, et al. Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants. J Med Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 8.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 9.Sarafianos SG, et al. Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem Biol. 1999;6:R137–R146. doi: 10.1016/s1074-5521(99)80071-4. [DOI] [PubMed] [Google Scholar]
- 10.Rodgers DW, et al. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiou Y, et al. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 12.Rittinger K, Divita G, Goody RS. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc Natl Acad Sci U S A. 1995;92:8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sluis-Cremer N, Temiz NA, Bahar I. Conformational changes in HIV-1 reverse transcriptase induced by nonnucleoside reverse transcriptase inhibitor binding. Curr HIV Res. 2004;2:323–332. doi: 10.2174/1570162043351093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geitmann M, Unge T, Danielson UH. Biosensor-based kinetic characterization of the interaction between HIV-1 reverse transcriptase and non-nucleoside inhibitors. J Med Chem. 2006;49:2367–2374. doi: 10.1021/jm0504048. [DOI] [PubMed] [Google Scholar]
- 16.Tachedjian G, Orlova M, Sarafianos SG, Arnold E, Goff SP. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 2001;98:7188–7193. doi: 10.1073/pnas.121055998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbondanzieri EA, et al. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peletskaya EN, Kogon AA, Tuske S, Arnold E, Hughes SH. Nonnucleoside inhibitor binding affects the interactions of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase with DNA. J Virol. 2004;78:3387–3397. doi: 10.1128/JVI.78.7.3387-3397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Q, Radzio J, Anderson KS, Sluis-Cremer N. Probing nonnucleoside inhibitor-induced active-site distortion in HIV-1 reverse transcriptase by transient kinetic analyses. Protein Sci. 2007;16:1728–1737. doi: 10.1110/ps.072829007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J, et al. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 22.Das K, et al. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J Mol Biol. 1996;264:1085–1100. doi: 10.1006/jmbi.1996.0698. [DOI] [PubMed] [Google Scholar]
- 23.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 24.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3’-azido-3’-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 25.Gu Z, Quan Y, Li Z, Arts EJ, Wainberg MA. Effects of non-nucleoside inhibitors of human immunodeficiency virus type 1 in cell-free recombinant reverse transcriptase assays. J Biol Chem. 1995;270:31046–31051. doi: 10.1074/jbc.270.52.31046. [DOI] [PubMed] [Google Scholar]
- 26.Mitsuya H, et al. 3’-Azido-3’-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc Natl Acad Sci U S A. 1985;82:7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz RA, Skalka AM. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 28.Sarafianos SG, et al. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das K, et al. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J Biol Chem. 2009;284:35092–35100. doi: 10.1074/jbc.M109.022525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lansdon EB, et al. Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex. J Mol Biol. 2010;397:967–978. doi: 10.1016/j.jmb.2010.02.019. S0022-2836. [DOI] [PubMed] [Google Scholar]
- 31.Tuske S, et al. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat Struct Mol Biol. 2004;11:469–474. doi: 10.1038/nsmb760. [DOI] [PubMed] [Google Scholar]
- 32.Mizrahi V, Henrie RN, Marlier JF, Johnson KA, Benkovic SJ. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985;24:4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- 33.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santoso Y, et al. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc Natl Acad Sci U S A. 2010;107:715–720. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obeid S, et al. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 2010;29:1738–1747. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahn KE, Belrhali H, Wallace SS, Doublie S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 38.Cai H, Bloom LB, Eritja R, Goodman MF. Kinetics of deoxyribonucleotide insertion and extension at abasic template lesions in different sequence contexts using HIV-1 reverse transcriptase. J Biol Chem. 1993;268:23567–23572. [PubMed] [Google Scholar]
- 39.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 40.Molina JM, et al. Rilpivirine versus efavirenz with tenofovir and emtricitabine in treatment-naive adults infected with HIV-1 (ECHO): a phase 3 randomised double-blind active-controlled trial. Lancet. 2011;378:238–246. doi: 10.1016/S0140-6736(11)60936-7. [DOI] [PubMed] [Google Scholar]
- 41.Carr A, et al. A controlled trial of nevirapine plus zidovudine versus zidovudine alone in p24 antigenaemic HIV-infected patients The Dutch-Italian-Australian Nevirapine Study Group. AIDS. 1996;10:635–641. doi: 10.1097/00002030-199606000-00009. [DOI] [PubMed] [Google Scholar]
- 42.Odriozola L, et al. Non-nucleoside inhibitors of HIV-1 reverse transcriptase inhibit phosphorolysis and resensitize the 3’-azido-3’-deoxythymidine (AZT)-resistant polymerase to AZT-5’-triphosphate. J Biol Chem. 2003;278:42710–42716. doi: 10.1074/jbc.M212673200. [DOI] [PubMed] [Google Scholar]
- 43.Basavapathruni A, Bailey CM, Anderson KS. Defining a molecular mechanism of synergy between nucleoside and nonnucleoside AIDS drugs. J Biol Chem. 2004;279:6221–6224. doi: 10.1074/jbc.C300523200. [DOI] [PubMed] [Google Scholar]
- 44.Selmi B, et al. The Y181C substitution in 3’-azido-3’-deoxythymidine-resistant human immunodeficiency virus, type 1, reverse transcriptase suppresses the ATP-mediated repair of the 3’-azido-3’-deoxythymidine 5’-monophosphate-terminated primer. J Biol Chem. 2003;278:40464–40472. doi: 10.1074/jbc.M302928200. [DOI] [PubMed] [Google Scholar]
- 45.Xu HT, et al. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J Virol. 2011;85:11300–11308. doi: 10.1128/JVI.05584-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswal BK, et al. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J Biol Chem. 2005;280:18202–18210. doi: 10.1074/jbc.M413410200. [DOI] [PubMed] [Google Scholar]
- 47.Mukhopadhyay J, et al. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauman JD, et al. Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design. Nucleic Acids Res. 2008;36:5083–5092. doi: 10.1093/nar/gkn464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeStefano JJ, et al. Characterization of an RNase H deficient mutant of human immunodeficiency virus-1 reverse transcriptase having an aspartate to asparagine change at position 498. Biochim Biophys Acta. 1994;1219:380–388. doi: 10.1016/0167-4781(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 50.Hou X, Wang G, Gaffney BL, Jones RA. Synthesis of guanosine and deoxyguanosine phosphoramidites with cross-linkable thioalkyl tethers for direct incorporation into RNA and DNA. Nucleosides Nucleotides Nucleic Acids. 2009;28:1076–1094. doi: 10.1080/15257770903368385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarafianos SG, et al. Trapping HIV-1 reverse transcriptase before and after translocation on DNA. J Biol Chem. 2003;278:16280–16288. doi: 10.1074/jbc.M212911200. [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. DENZO and SCALEPACK. Vol F. Crystallography of Biological Macromolecules. Kluwer Academic Publishers; Boston: 2001. [Google Scholar]
- 53.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


