Abstract
Abstracts
Background
The lipase subfamilies I.1 and I.2 show more than 33% homology in the amino acid sequences and most members share another common property that their genes are clustered with the secondary genes whose protein products are required for folding the lipase into an active conformation and secretion into the culture medium. In previous studies, the lipase (LipA) and its chaperone (LipB) from Ralstonia sp. M1 were overexpressed in E. coli and the lipase was successfully refolded in vitro. The purpose of this study was to enhance the production of the active lipase LipA from Ralstonia sp. M1 in the heterologous host E. coli without in vitro refolding process, using two-plasmid co-expression systems and dual expression cassette plasmid systems.
Results
To produce more active lipase from Ralstonia sp. M1 in E. coli without in vitro refolding process but with the help of overexpression of the chaperone (LipB1 and LipB3 corresponding to 56-aa truncated and 26-aa truncated chaperone LipB), six different expression systems including 2 two-plasmid co-expression systems (E. coli BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k) and 4 dual expression cassette plasmid systems (BL21/pELipAB-LipB1a, BL21/pELipAB-LipB3a, BL21/pELipA-LipB1a, and BL21/pELipA-LipB3a) were constructed. The two-plasmid co-expression systems (E. coli BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k) produced the active lipase at a level of 4 times as high as the single expression cassette plasmid system E. coli BL21/pELipABa did. For the first time, the dual expression cassette plasmid systems BL21/pELipAB-LipB1a and BL21/pELipAB-LipB3a yielded 29- and 19-fold production of the active lipase in comparison with the single expression cassette plasmid system E. coli BL21/pELipABa, respectively. Although the lipase amount was equally expressed in all these expression systems (40% of total cellular protein) and only a small fraction of the overexpressed lipase was folded in vivo into the functional lipase in soluble form whereas the main fraction was still inactive in the form of inclusion bodies. Another controversial finding was that the dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a and E. coli/pELipAB-LipB3a secreted the active lipase into the culture medium of 51 and 29 times as high as the single expression cassette plasmid system E. coli pELipABa did, respectively, which has never been reported before. Another interesting finding was that the lipase form LipA6xHis (mature lipase fused with 6× histidine tag) expressed in the dual expression cassette plasmid systems (BL21/pELipA-LipB1a and BL21/pELipA-LipB3a) showed no lipase activity although the expression level of the lipase and two chaperone forms LipB1 and LipB3 in these systems remained as high as that in E. coli BL21/pELipABa + pELipB1k, BL21/pELipABa + pELipB3k, BL21/pELipAB-LipB1a, and BL21/pELipAB-LipB3a. The addition of Neptune oil or detergents into the LB medium increased the lipase production and secretion by up to 94%.
Conclusions
Our findings demonstrated that a dual expression cassette plasmid system E. coli could overproduce and secrete the active chaperone-dependent lipase (subfamilies I.1 and I.2) in vivo and an improved dual expression cassette plasmid system E. coli could be potentially applied for industrial-scale production of subfamily I.1 and I.2 lipases.
Keywords: Ralstonia sp. M1, Lipase, Chaperone, Functional expression, Secretion
Background
Lipases (triacylglycerol ester hydrolases, EC 3.1.1.3) catalyze the hydrolysis of long-chain triacylglycerols in aqueous media to liberate diacyl-, monoacylglycerol, glycerol and fatty acids [1]. In non-aqueous media, they catalyze the esterification and transesterification reactions [2]. Unlike esterases, lipases prefer long chain fatty acids and are activated in the interface of lipid substrate and water. Owing to their substrate specificity, region specificity, chiral selectivity, thermostability, and alkaline stability, microbial lipases have been utilized widely in industrial fields for the production or modification of detergents, dairy products and diagnostics, as well as for oil processing, biotransformations, and chiral separation or synthesis [3].
Bacterial lipase family I has been divided into seven subfamilies on basis of amino acid sequence homology [3]. Subfamily I.1 contains the lipases from Pseudomonas aeruginosa [4,5], Pseudomonas fragi [6], and Acinetobacter sp. [7,8], and subfamily I.2 comprises the lipases from Pseudomonas glumae [9], Pseudomonas luteola [10], Burkholderia cepacia [11], Chromobacterium viscosum [12], and Ralstonia sp. M1 [13].
These two lipase subfamilies I.1 and I.2 show more than 33% homology in the amino acid sequences [14] and most members share another common property that their genes are clustered with the secondary genes located immediately downstream [4,5,8,11,13,15] or upstream of the lipase genes [7]. Protein products of the secondary genes are so-called lipase-specific foldases (modulator, activator, helper protein or chaperone) required for folding the lipase into an active conformation and secretion into the culture medium. During the secretion process of the lipase via the two-step type II secretion pathway, the lipase chaperones directly participate in the in vivo folding and secretion process.
In spite of various applications of Pseudomonas and Burkholderia lipases from subfamilies I.1 and I.2, respectively, as detergent additive and for variety of biotransformations, overproduction of the functional lipase in E. coli has been much difficult due to the requirement of the chaperone to fold the lipase into an active conformation [14]. To enhance the functional expression of the lipases in E. coli, two strategies have been developed. The first one was to produce and purify both the lipase and its chaperone separately in E. coli and the fully active lipase was subsequently achieved by in vitro refolding process using various approaches [11,16-20]. Recently, the second strategy was directed to produce in vivo functional lipases in heterologous host E. coli using two-plasmid co-expression system without the need of in vitro refolding [21-24]. To date, expression of in vivo functional lipase in heterologous host E. coli using a dual expression cassette plasmid system has not been achieved.
In the previous study [13], the lipase-encoding gene lipA and its chaperone-encoding gene lipB from the Ralstonia sp. strain M1 were cloned, sequenced, and overexpressed in E. coli BL21 separately. The recombinant lipase was an alkaline, thermophilic, highly organic-solvent-resistant and detergent-inducible lipase. The plasmid pELipABa carrying the gene cluster encoding LipA and LipB under the control of T7 promoter was expressed in E. coli strain BL21. The amount of LipA accounted for 40% total cellular protein, however the amount of LipB was just less than 1% in comparison with the total amount of proteins. On the other hand, for the optimum in vitro refolding of the lipase, both LipA and LipB are required in equal amounts to form the lipase:chaperone (1:1) complex [19]. That means the unequal expression level of the lipase and its chaperone in the heterologous host led to the low formation of the active lipase in vivo, only 1-3% of the expressed lipase were in vivo activated.
To enhance the amount of the functional lipase, we overexpressed LipA (70 mg per gram cell wet weight) and LipB1 (12 mg per gram cell wet weight) in E. coli BL21 and the in vitro refolding increased the lipase activity by 4.4 times. However, in these studies, in vitro refolding was required to fold the lipase into active conformation. The purpose of the present study was to enhance the production of the active lipase LipA from Ralstonia sp. M1 in the heterologous host E. coli without in vitro refolding process, using two-plasmid co-expression systems and dual expression cassette plasmid systems. For the first time, an active lipase was expressed in a dual expression cassette plasmid system E. coli.
Results and discussions
Functional expression of lipase using two-plasmid co-expression system
To express the functional lipase in E. coli without in vitro refolding process, the first strategy was developed to enhance the expression level of the chaperone in E. coli using two-plasmid co-expression systems E. coli BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k.
Expression level of LipA and LipB
E. coli BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k were cultivated and induced by IPTG for the production of the lipase and chaperone LipA + LipB1 and LipA + LipB3, respectively. In the previous study, the active lipase was negligible when the co-expressed chaperone LipB in the gene cluster was expressed at a negligible level in comparison to the lipase LipA using plasmid pELipABa [25]. Thus, in this study, we tried to enhance the expression level of the chaperone in the form of LipB1 or LipB3 to produce more active lipase molecules in vivo.
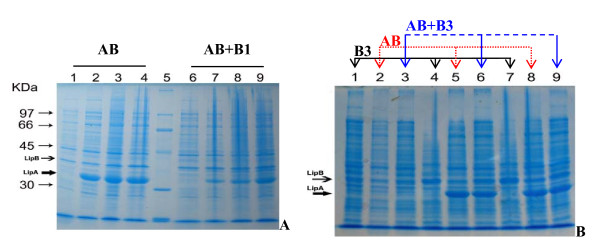
The expression level of LipA in BL21/pELipABa + pELipB1k (10-21% of total cellular protein, using densitometry analysis Dolphin 1-D software operation) (Figure 1A, lane 7-9) was quite lower than that in BL21/pELipABa (30-40%) (Figure 1A, lane 2-4). The expression level of LipA in BL21/pELipABa after induction by IPTG for 1 hour (40%) (Figure 1A, lane 2) was higher than that in BL21/pELipABa + pELipB1k after induction by IPTG for 4 hours (21%) (Figure 1A, lane 9). However, the amount of chaperone LipB (Figure 1A, lane 2-4) and LipB + LipB3 (Figure 1A, lane 7-9) expressed in two systems BL21/pELipABa and BL21/pELipABa + pELipB1k, respectively was also not observed by SDS-PAGE even after induction for 4 hours.
Figure 1.
SDS-PAGE of BL21 cell lysates using two-plasmid co-expression system. (A). pELipABa (1-4), pELipABa + pELipB1k (6-9). Lanes (1-4) and (6-9) indicated induction by IPTG for 0, 1, 2, and 4 hours, respectively; Lane 5: standard protein. (B). pELipB3k (1, 4, 7), pELipABa (2, 5, 8) and pELipABa + pELipB3k (3, 6, 9). Lanes (1, 2, 3), (4, 5, 6), (7, 8, 9) indicated induction by IPTG for 0, 2, and 4 hours, respectively.
The lipase LipA was expressed in BL21/pELipABa (Figure 1B, lane 2, 5, 8) and BL21/pELipABa + pELipB3k (Figure 1B, lane 3, 6, 9) in an equal level (25%) and obviously observed, whereas the expression level of the protein chaperones LipB3 (in BL21/pELipB3k, Figure 1B, lane 1, 4, 7), LipB (in BL21/pELipABa, Figure 1B, lane 2, 5, 8) and LipB + LipB3 (BL21/pELipABa + pELipB3k, Figure 1B, lane 3, 6, 9) was not observed by SDS-PAGE even after induction by IPTG for 4 hours.
Production of functional lipase
The lipase assay was performed with olive oil as substrate by a continuous titration method. The two-plasmid co-expression systems E. coli BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k produced the functional lipase of 493 U/g and 440 U/g cell wet weight corresponding to 4.6 and 4.1 times as high as the single-plasmid expression system BL21/pELipABa (107 U/g) did, respectively (Table 1) although the expression level of LipA in E. coli BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k (21-25%) was lower than that of LipA in BL21/pELipABa (25-40%). The only explanation was that the two-plasmid co-expression systems BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k expressed more chaperone molecules (LipB + LipB1 and LipB + LipB3, respectively) than the single plasmid expression system BL21/pELipABa (only LipB) and this led to approximately 4-5 times in vivo lipase folding in both the two-plasmid co-expression systems.
Table 1.
Production of the active lipase LipA by the two-plasmid co-expression systems BL21/pELipABa + pELipB1k and BL21/pELipABa + pELipB3k
| Expression system BL21/ | Whole cell lysate | |
|---|---|---|
| U/g | % | |
| pELipABa | 107.3 ± 3.6 | 100 ± 3.4 |
| pELipABa + pELipB1k | 492.7 ± 3.1 | 458.9 ± 2.9 |
| pELipABa + pELipB3k | 440.3 ± 5.7 | 410.2 ± 5.3 |
The result of in vivo folding was very similar to in vitro refolding in the previous study [13]. In vitro refolding of the lipase LipA expressed in the single plasmid expression system BL21/pELipABa using the optimal ratio of lipase to chaperone for refolding (1:10) led to the optimal formation of the functional lipase (650 U/mg lipase protein), 4 times as high as the functional lipase LipA without in vitro refolding (151 U/mg). Recently, in vivo expression of functional Pseudomonas and Burkholderia lipases of the subfamilies I.1 and I.2 were achieved by two-plasmid co-expression system E. coli BL21(DE3) [21-23]. However, no direct comparison between in vitro refolding and in vivo folding efficacy was reported so far in these studies.
Functional expression of lipase using dual expression cassette plasmid system
The second strategy was directed to enhance the expression level of the chaperone in E. coli and thus to enhance the production of the functional lipase without in vitro refolding process using the dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a and pELipAB-LipB3a.
Expression level of LipA and LipB
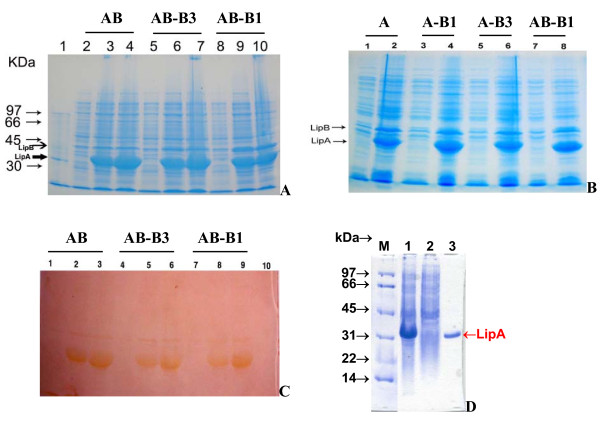
The expression level of the lipase LipA (Figure 2) was relatively equal (~40%) in all dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a (Figure 2A, lane 10, B lane 8), BL21/pELipAB-LipB3a (Figure 2A, lane 7), BL21/pELipA-LipB1a (Figure 2B, lane 4), BL21/pELipA-LipB3a (Figure 2B, lane 6) as well as in the single expression cassette plasmid systems BL21/pELipABa (Figure 2A, lane 4) and BL21/pELipAa (Figure 2B, lane 2). The lipase activity staining also confirmed that the amount of the lipase LipA was produced equally in the dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a (Figure 2C, lane 9), BL21/pELipAB-LipB3a (Figure 2C, lane 6), and BL21/pELipABa (Figure 2C, lane 3).
Figure 2.
SDS-PAGE of BL21 cell lysates using dual expression cassette plasmid system. (A). pELipABa (2-4), pELipAB-LipB3a (5-7), pELipAB-LipB1a (8-10). Lanes (2, 5, 8), (3, 6, 9), (4, 7, 10) indicated induction by IPTG for 0, 2, and 4 hours, respectively; Lane 1: standard protein. (B). pELipAa (1-2), pELipA-LipB1a (3-4), pELipA-LipB3a (5-6), pELipAB-LipB1a (7-8). Odd and even lanes indicated induction by IPTG for 0 and 4 hours, respectively. (C). Lipase-activity stained SDS-PAGE of BL21 cell lysate harboring the recombinant plasmids: pELipABa (1-3), pELipAB-LipB3a (4-6), pELipAB-LipB1a (7-9). Lanes (1, 4, 7), (2, 5, 8), and (3, 6, 9) indicated induction by IPTG for 0, 2, and 4 hours, respectively. (D). pELipAB-LipB1a whole cell lysate (1), cell lysate's supernatant (2), cell lysate precipitate (3).
Similar to the two-plasmid co-expression systems, the protein chaperone LipB, LipB1 and LipB3 expressed in the dual expression cassette systems E. coli BL21/pELipAB-LipB1a (Figure 2A, lane 10, B lane 8), BL21/pELipAB-LipB3a (Figure 2A, lane 7), BL21/pELipA-LipB1a (Figure 2B, lane 4), BL21/pELipA-LipB3a (Figure 2B, lane 6) and in the single expression cassette plasmid system BL21/pELipABa (Figure 2A, lane 4) was difficult to be detected by SDS-PAGE.
Production of functional lipase
After induction by 1 mM IPTG for 4 hours, the dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a and pELipAB-LipB3a produced the functional lipase at the level of 6473 U/g and 4282 U/g cell wet weight corresponding to 28.5 and 18.9 times as high as the single expression cassette plasmid system BL21/pELipABa (227 U/g cell wet weight) did, respectively (Table 2). The dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a and pELipAB-LipB3a secreted the functional lipase at the level of 5065 and 2930 U/g cell wet weight, corresponding to 50.7 and 29.3 times as high as the single expression cassette plasmid system BL21/pELipABa (100 U/g cell wet weight) did, respectively.
Table 2.
Production of the active lipase LipA by the dual expression cassette plasmid systems BL21/pELipAB-LipB1a and BL21/pELipAB-LipB3a
| Expression system BL21/ | Culture supernatant | Whole cell lysate | Whole culture | Cell lysate's supernatant | Cell lysate's precipitate | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| U/g | % | U/g | % | U/g | % | U/g | % | U/g | % | |
| pELipABa | 99.9 ± 1.5 | 100 ± 1.5 | 127.5 ± 1.1 | 100 ± 0.8 | 227.4 ± 0.9 | 100 ± 0.4 | 32.4 ± 1.6 | 21.3 ± 1.1 | 119.7 ± 8.6 | 78.7 ± 5.6 |
| pELipAB-LipB1a | 5064.7 ± 11.2 | 5068.4 ± 11.2 | 1408.3 ± 6.9 | 1104.5 ± 5.4 | 6473.0 ± 18.0 | 2846.1 ± 7.9 | 267.9 ± 9.6 | 17.2 ± 0.6 | 1287.9 ± 49.4 | 82.8 ± 3.2 |
| pELipAB-LipB3a | 2930.8 ± 4.5 | 2932.9 ± 4.5 | 1352.1 ± 4.2 | 1060.4 ± 3.3 | 4282.9 ± 8.5 | 1883.1 ± 3.7 | 192.0 ± 7.0 | 11.8 ± 0.4 | 1437.9 ± 47.1 | 88.2 ± 2.9 |
Although the expression level of the chaperone (LipB + LipB1 and LipB + LipB3) in the dual expression cassette plasmid systems was not able to be observed on SDS-PAGE and the expression level of the lipase LipA was relatively equal (~40% of total cellular protein), the production of the functional lipase was increased to 29 and 19 times, respectively. The explanation might be that the number of the chaperone molecules (LipB + LipB1 and LipB + LipB3) expressed in the dual expression cassette plasmid system was higher than that (only LipB) in the single expression cassette plasmid system. In the sequence, it led to enhance the number of in vivo folded lipase molecules. In our previous study [26], we also expressed the lipase from Burkholderia cepacia ATCC 21808 in heterologous host E. coli using a dual expression cassette plasmid with the pre-signal peptide ompA. The functional lipase expressed in the dual expression cassette plasmid system increased to 450 folds (18000 U/g) in comparison with that expressed in the single expression cassette plasmid system (40 U/g), although the lipase level decreased from 40% to 15% of the total proteins [26] due to simultaneous overexpression of the chaperone. To our knowledge, there has been no report on the expression of the functional (Pseudomonas and Burkholderia, subfamily I.1 and I.2) lipase using a dual expression cassette system.
The functional lipase in the culture supernatant of the dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a and pELipAB-LipB3a was 50.7 and 29.3 times as high as the functional lipase in the culture supernatant of the single expression cassette plasmid system E. coli BL21/pELipABa. This finding was somehow controversial to other reports. Peng et al. reported that no lipase activity was detected in the culture supernatant of the recombinant E. coli BL21(DE3) co-expressing the lipase and its chaperone from Pseudomonas aeruginosa CS-2 [23]. The lipase activity was also not detected in the culture supernatant by the recombinant E. coli JM109 harboring a plasmid containing both lipase and chaperone gene from P. aeruginosa LST-03 [27]. The secretion of subfamily I.1 and I.2 lipases did not proceed properly in heterologous hosts [28].
Functional lipase in soluble form
One question was raised where was the functional (in vivo folded) lipase present in the soluble form or in form of inclusion bodies? We have expressed the lipase LipA in the dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a and BL21/pELipAB-LipB3a and the single expression cassette plasmid system BL21/pELipAB1a. The cell pellets were sonified and the cell lysates were centrifuged to separate the cell lysate's supernatant (soluble fraction) and the cell lysate's precipitate (insoluble fraction). The lipase molecules LipA were retained most in the cell lysate's precipitate as inclusion bodies (Figure 2D, lane 3) and not observed in the cell lysate's supernatant as soluble form (Figure 2D, lane 2). However, the surprising finding was that the cell lysate's supernatant contained 88, 83, and 79% of the total functional lipase production in the dual expression cassette plasmid systems E. coli BL21/pELipAB-LipB1a and BL21/pELipAB-LipB3a and the single expression cassette plasmid system BL21/pELipAB, respectively, whereas the cell lysate's precipitate showed only 12, 17 and 21% of the total functional lipase production, respectively. This demonstrated that only a small fraction of the expressed lipase was folded in vivo and become active in soluble form whereas the major fraction of the expressed lipase was still not folded in vivo and thus maintained inactive as inclusion bodies. This study suggested that it is still a big potential to enhance functional production of the chaperone-dependent lipase.
Non-functional expression of lipase using dual expression cassette plasmid system
The expression level of the lipase expressed in the dual expression cassette plasmid systems E. coli BL21/pELipA-LipB1a (Figure 2B, lane 4) and BL21/pELipA-LipB3a (Figure 2B, lane 6) was relatively equal (~40% of total cellular protein) to that in BL21/pELipAB-LipB1a (Figure 2A, lane 10), BL21/pELipAB-LipB3a (Figure 2A, lane 7), as well as in the single expression cassette plasmid system BL21/pELipAa (Figure 2B, lane 2), and BL21/pELipABa (Figure 2A, lane 4). However, no lipase activity was detected in the cell culture supernatant as well as in the cell lysate of the dual expression cassette plasmid systems BL21/pELipA-LipB1a and BL21/pELipA-LipB3a. The expression level of the chaperone B1 and B3 in the dual expression cassette plasmid systems BL21/pELipA-LipB1a (Figure 2B, lane 4) and BL21/pELipA-LipB3a (Figure 2B, lane 6) was relatively equal to that in BL21/pELipAB-LipB1a (Figure 2A, lane 10) and BL21/pELipAB-LipB3a (Figure 2A, lane 7). But why did BL21/pELipA-LipB1a and BL21/pELipA-LipB3a form no active lipase? The reason might be that the lipase molecule expressed in these systems was LipA fused with the 6x histidine tag, so that the structure of the lipase molecule might be changed and led to be not properly folded in vivo and no active lipase was found. Although the level of the lipase and the chaperone expressed in these systems (Figure 2B) was quite similar to those in BL21/pELipAB-LipB1a and BL21/pELipAB-LipB3a (Figure 2A).
Effect of culture components on production of active lipase by dual expression cassette plasmid systems
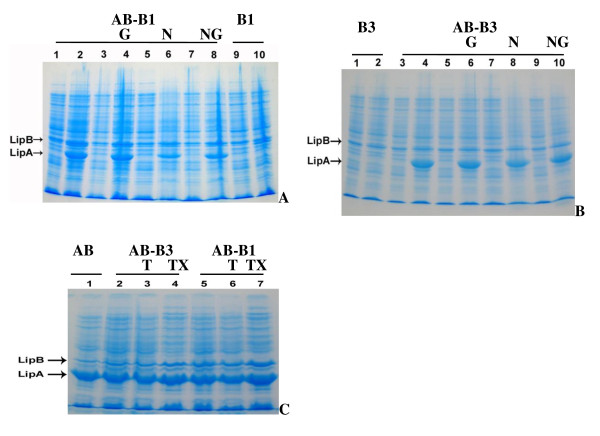
To figure out the effect of some additives including Neptune oil, gum Arabic, Tween 80, and Triton X-100 on the formation of the active lipase in vivo in the dual expression cassette plasmid systems BL21/pELipAB-LipB1a and pELipAB-LipB3a, we carried out the expression of the recombinant E. coli cells in LB medium by adding additives as showed in Table 3. The addition of the substrate Neptune oil and gum Arabic into the culture media did not enhance the expression level of the lipase LipA in the dual expression cassette plasmid system BL21/pELipAB-LipB1a (~30% of total cellular protein) (Figure 3A). The addition of Neptune oil decreased significantly the expression level of the lipase LipA (20-25% of total cellular protein) (Figure 3A, lane 6, 8). Gum Arabic (Figure 3A, lane 4), Tween 80 (Figure 3C, lane 6), and Triton X-100 (Figure 3C, lane 7) showed no different effect on the expression level of the lipase LipA (~30% of total cellular protein). The addition of 0.5% of Neptune oil plus 0.5% of gum Arabic showed a slight decrease of 2-10% in production of the active lipase by BL21/pELipAB-LipB1a, but the addition of 0.5% of Neptune oil plus 0.5% of gum Arabic, Tween 80 and Triton X-100 increased the active lipase production by 5-27% (Table 3).
Table 3.
Effect of 0.5% additives on the production level of the functional lipase LipA by the dual expression cassette plasmid systems BL21/pELipAB-LipB3a and BL21/pELipAB-LipB1a
| Additive (0.5%) supplemented to LB medium | BL21/pELipAB-LipB1a- | BL21/pELipAB-LipB3a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Culture supernatant | Whole cell lysate | Whole culture | Culture supernatant | Whole cell lysate | Whole culture | |||||||
| U/g | % | U/g | % | U/g | % | U/g | % | U/g | % | U/g | % | |
| LB medium | 5064.7 ± 11.2 | 100.0 ± 0.2 | 1408.3 ± 6.9 | 100.0 ± 0.5 | 6473.0 ± 18.0 | 100.0 ± 0.3 | 2930.8 ± 4.5 | 100 ± 0.2 | 1352.1 ± 4.2 | 100.0 ± 0.3 | 4282.9 ± 8.5 | 100.0 ± 0.2 |
| Neptune oil | 5520.9 ± 104.4 | 109.0 ± 2.1 | 831.3 ± 39.6 | 59.0 ± 2.8 | 6352.2 ± 85.9 | 98.1 ± 1.3 | 5683.9 ± 51.6 | 193.9 ± 1.8 | 2636.3 ± 39.3 | 195.0 ± 2.9 | 8320.2 ± 61.2 | 194.3 ± 1.4 |
| Gum Arabic | 5268.0 ± 121.0 | 104.0 ± 2.4 | 549.3 ± 62.0 | 39.0 ± 4.4 | 5817.3 ± 170.5 | 89.9 ± 2.6 | 3926.4 ± 20.9 | 134.0 ± 0.7 | 1149.3 ± 18.5 | 85.0 ± 1.4 | 5075.6 ± 39.3 | 118.5 ± 0.9 |
| Gum Arabic + Neptune oil | 5420.0 ± 85.9 | 107.0 ± 1.7 | 1366.1 ± 51.7 | 97.0 ± 3.7 | 6786.1 ± 95.4 | 104.8 ± 1.5 | 4249.5 ± 46.8 | 145.0 ± 1.6 | 1743.7 ± 43.2 | 129.0 ± 3.2 | 5993.1 ± 81.9 | 139.9 ± 1.9 |
| Tween 80 | 6179.2 ± 119.1 | 122.0 ± 2.4 | 1534.9 ± 55.2 | 109.0 ± 3.9 | 7714.0 ± 171.4 | 119.2 ± 2.6 | 3780.0 ± 70.1 | 129.0 ± 2.4 | 1014.5 ± 29.9 | 75.0 ± 2.2 | 4794.5 ± 57.6 | 111.9 ± 1.3 |
| Triton X-100 | 8003.5 ± 72.0 | 158.0 ± 1.4 | 225.5 ± 34.5 | 16.0 ± 2.5 | 8229.0 ± 74.3 | 127.1 ± 1.1 | 4864.5 ± 67.2 | 166.0 ± 2.3 | 122.8 ± 12.4 | 9.1 ± 0.9 | 4987.3 ± 72.5 | 116.4 ± 1.7 |
Figure 3.
SDS-PAGE of BL21 cell lysates harboring the recombinant plasmids. (A). pELipAB-LipB1a (1-8) and pELipB1k (9-10). LB medium without addititive (1-2), with 0.5% (v/v) of Neptune oil (5-6), 0.5% (w/v) of gum Arabic (3-4), 0.5% (w/v) of gum Arabic + 0.5% (v/v) of Neptune oil (7-8). Odd and even lanes indicated induction by IPTG for 0 and 4 hours, respectively. (B). pELipB3k (1-2) and pELipAB-LipB3a (3-10). LB medium without additive (3-4), with 0.5% (w/v) of gum Arabic (5-6), 0.5% (v/v) of Neptune oil (7-8), 0.5% (w/v) of gum Arabic + 0.5% (v/v) of Neptune oil (9-10). Odd and even lanes indicated induction by IPTG for 0 and 4 hours, respectively. (C). pELipABa (1); pELipAB-LipB3a (2-4), pELipAB-LipB1a (5-7). LB medium without additive (1, 2, 5), with 0.5% of Tween 80 (3, 6), and 0.5% of Triton X-100 (4, 7) after induction by IPTG for 4 hours.
In the dual expression cassette plasmid system BL21/pELipAB-LipB3a, the addition of Neptune oil (Figure 3B, lane 8), Neptune oil plus gum Arabic (Figure 3B, lane 10), gum Arabic (Figure 3B, lane 6), Tween 80 (Figure 3C, lane 3), Triton X-100 (Figure 3C, lane 4) showed no effect on the expression level of the lipase LipA (~30% of total cellular protein). However, the addition of 0.5% of Neptune oil and 0.5% of Neptune oil plus 0.5% of gum Arabic increased the production of the active lipase by the dual expression cassette plasmid system BL21/pELipAB-LipB3a by 94 and 40%, respectively. Whereas the addition of 0.5% of gum Arabic, Tween 80, and Triton X-100 increased only by 12-16% (Table 3). The addition of Triton X-100, Tween 80, and gum Arabic could solubilize and activate the membrane-bound lipase, so that it could help the cell to secrete more lipase.
Conclusions
The production of the active chaperone-dependent lipase from Ralstonia sp. M1 (lipase subfamily I.1) in heterologous host E. coli was enhanced up to 4 times by using two-plasmid co-expression systems and up to 19-29 times by using dual expression cassette plasmid systems in comparison to the single expression cassette plasmid systems. In dual expression cassette plasmid systems, the active lipase was secreted into the culture medium at the level of up to 29-51 times as high as the single expression cassette plasmid systems. Only a small fraction of the overexpressed lipase was in vivo folded into the functional lipase in soluble and secreted form and the main part was still inactive in form of inclusion bodies. The addition of Neptune oil, gum Arabic, Tween 80 and Triton X-100 led to double production and secretion of the active lipase in the dual expression cassette plasmid system E. coli. Our findings demonstrated that the dual expression cassette plasmid system E. coli could overproduce and secreted the active chaperone-dependent lipase (subfamilies I.1 and I.2) in vivo and an improved dual expression cassette plasmid system E. coli could be potentially applied for industrial-scale production of subfamily I.1 and I.2 lipases.
Materials and methods
Chemicals and reagents
Fast red TR, α-naphthyl acetate, tributyrin, olive oil, Triton X-100, Tween 80, and gum Arabic were purchased from Sigma-Aldrich Co. (St. Louis, USA). Bacto-tryptone and yeast extract were from Difco (Lawrence, USA). Restriction enzymes, CIAP, T4 ligase, and Taq polymerase were supplied by Fermentas (Thermo Fisher Scientific Inc., Waltham, USA). PCR mastermix was from Invitrogen Corp. (Carlsbad, USA). DNA Gel-Extraction Kit was from Qiagen (Venlo, Netherlands). All other reagents were of analytical grade unless otherwise stated.
Plasmids, bacterial strains and culture conditions
Escherichia coli DH5α (F-, ø80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk-, mk+), phoA, supE44, λ-, thi-1, gyrA96, relA1) and pCR2.1 TOPO (TOPO® TA cloning Kit, Invitrogen Corp., Carlsbad, USA) were used to subclone the lipase gene lipA and chaperone gene lipB. The vectors pET22b(+) and pET28a(+) (Novagen, Merck KGaA, Darmstadt, Germany) and Escherichia coli BL21 (DE3) cells (F- ompT gal dcm lon hsdSB(rB- mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) were used to express the lipase gene lipA and its chaperone gene lipB under the control of the T7-promoter induced by IPTG. LB medium (Luria-Bertani) containing 1% (w/v) bacto tryptone; 0.5% (w/v) yeast extract; 1% (w/v) NaCl; pH 7-7.5 was used for cultivation of E. coli DH5α and BL21 (DE3). LB agar contained additionally 2% (w/v) agar and 100 μg ampicillin/ml. Plasmids pELipABa, pELipAa, pELipB1a, and pELipB3a [25] were used to provide the lipase gene lipA and chaperone gene lipB. The plasmids used in this study are summarized in Table 4.
Table 4.
Plasmids used in this work
| Plasmid | Gene of interest | Reference |
|---|---|---|
| pET22b(+) | T7-promoter, Ampr, pelB, T7-terminator | Novagen, Merck KGaA, Darmstadt, Germany |
| pET28a(+) | T7-promoter, Kanr, T7-terminator | Novagen, Merck KGaA, Darmstadt, Germany |
| pCR2.1-TOPO | TOPO binding site, Ampr | Invitrogen Corp., Carlsbad, USA |
| pELipABa | lipA, lipB, T7-promoter, Ampr, pelB, T7-terminator | [25] |
| pELipAa | lipA, T7-promoter, Ampr, pelB, T7-terminator | [25] |
| pELipB1a | lipB1, T7-promoter, Ampr, pelB, T7-terminator | [25] |
| pELipB3a | lipB3, T7-promoter, Ampr, pelB, T7-terminator | [25] |
| pELipABa-LipB1a | lipA, lipB, lipB1, T7-promoter, Ampr, pelB, T7-terminator | In this study |
| pELipABa-LipB3a | lipA, lipB, lipB3, T7-promoter, Ampr, pelB, T7-terminator | In this study |
| pELipAa-LipB1a | lipA, lipB1, T7-promoter, Ampr, pelB, T7-terminator | In this study |
| pELipAa-LipB3a | lipA, lipB3, T7-promoter, Ampr, pelB, T7-terminator | In this study |
| pELipB1k | lipB1, T7-promoter, Kanr, T7-terminator | In this study |
| pELipB3k | lipB3, T7-promoter, Kanr, T7-terminator | In this study |
Ampr, ampicillin resistant; Kanr, kanamycin resistant; pelB, signal peptide; lipAB, gene cluster encoding LipA and LipB; lipA, gene encoding LipA; lipB1, gene encoding LipB1; lipB3, gene encoding LipB3.
DNA manipulations
Plasmid DNA isolation was carried out by the method as previously described [29]. DNA fragments and PCR products were excised from a 0.8% agarose gel and purified by a gel extraction kit (Qiagen, Venlo, Netherlands) according to the manufacturer's instructions. DNA sequencing was performed on an ABI PRISM 3100 Avant Genetic Analyzer (Applied Biosystems Inc., Foster City, USA). E. coli DH5α and BL21 were transformed using heat shock methods as previously described [29].
Construction of plasmids pELipB1k and pELipB3k
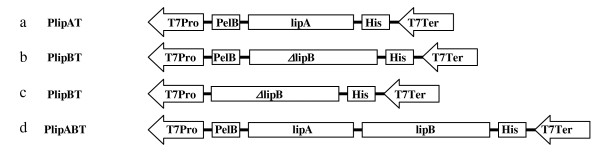
To co-express the lipase and chaperone using two different expression plasmids, two forms of the gene encoding chaperone LipB1 (N-terminal 56-aa truncated) and LipB3 (N-terminal 26-aa truncated) were isolated from the plasmid pELipB1a and pELipB3a, respectively, by using the restriction enzymes NcoI and HindIII. The gene fragments lipB1 and lipB3 were inserted into the expression vector pET28a(+) also digested with NcoI and HindIII resulting in expression plasmids pELipB1k and pELipB3k, respectively (Figure 4).
Figure 4.
Schematic representation of the expression cassettes in pELipAa (a), pELipB1a and pELipB3a (b), pELipB1k and pELipB3k (c), pELipABa (d), pELipA-B1a and pELipA-B3a (a,b), pELipAB-B1a and pELipAB-LipB3a (d,b), pLipABa + pELipB1k and pLipABa + pELipB1k (d,c), derived from vector pET22b(+) and pET28a(+) for E. coli BL21. T7Pro, T7 promoter; PelB, PelB signal peptide; lipA, mature lipase gene; His, 6xHis tag; T7Ter, T7 terminator; ΔlipB, truncated lipB ORF encoding a LipB derivative lacking the first 56 or 26 amino acids, corresponding to LipB1 and LipB3; lipB, the complete chaperone gene.
Construction of plasmids pELipAB-LipB1a and pELipAB-LipB3a
To construct plasmids harboring two expression cassettes promoter-gene-terminator, the expression plasmid pELipABa containing the expression cassette PlipABT (P: T7 promoter, lipase-chaperone gene cluster lipAB, T: T7 terminator, Figure 4) was used. The second expression cassette PlipB1T and PlipB3T (Figure 4) were amplified from the templates pELipB1a and pELipB3a, respectively. The designed specific primers PEF (5'-GGA GAT CTC GAT CCC GCG AAA TTA AT-3') and PER (5'-GGA GAT CTC AAA CCC CTC AAG AC-3') were complementary to upstream 5'-end of T7 promoter sequence and downstream 3'-end of T7 terminator sequence, respectively. Both primers contained the restriction enzyme BglII used to introduce the expression cassette PlipB1T and PlipB3T into the expression plasmid pELipABa. The PCR products were first subcloned into the cloning vector pCR2.1 and then digested with BglII and introduced into pELipABa which was also digested with BglII resulting in pELipAB-LipB1a and pELipAB-LipB3a harboring two independent expression cassettes. Each expression cassette contained the complete set of the T7 promoter and T7 terminator, so that the direction of each cassette does not influence on the expression level of the other protein. The PCR mixture contained 0.5 μl DNA template (50 ng); 1.2 μl each primer (10 pmol), 10 μl PCR master mix, supplemented with 7 μl of distilled water to a final volume of 20 μl. The thermocycler conditions were performed as follows: 94°C/5'; 35 cycles of 94°C/1', 61°C/1', 72°C/30″; 72°C/10'.
Construction of plasmids pELipA-LipB1a and pELipA-LipB3a
The cassettes PlipB1T and PlipB3T (Figure 4), amplified from the templates pELipB1a and pELipB3a, respectively, were inserted into the plasmid pELipAa resulting in the dual expression cassette plasmids pELipA-LipB1a and pELipA-LipB3a harboring two expression cassettes PlipAT + PlipB1T and PlipAT + PlipB3T, respectively. The construction of these plasmids was similar to the construction of the dual expression cassette plasmid pELipAB-LipB1a and pELipAB-LipB3a described in details as above mentioned.
Co-expression of lipase and chaperone
The transformants E. coli BL21 (pELipABa, pELipAB-LipB1a, pELipAB-LipB3a, pELipA-LipB1a, pELipA-LipB3a) and the transformants E. coli BL21 (pELipABa + pELipB1k, pELipABa + pELipB1k, pELipAa + pELipB1k, pELipAa + pELipB1k) were cultivated in 5 ml of LB medium containing only 5 μl of ampicillin (100 mg/ml) and 5 μl of ampicillin (100 mg/ml) with 5 μl kanamycin (10 mg/ml), respectively, overnight, at 37°C with agitation at 220 rpm. Fifty μl of the overnight culture were transferred into 50 ml of LB medium containing only 50 μl of ampicillin (100 mg/ml) and 50 μl of ampicillin (100 mg/ml) with 5 μl kanamycin (10 mg/ml), respectively. The cultures were grown at 37°C on orbital shaking incubator at 220 rpm for more 3 hours until OD600 nm reached 0.6, then supplemented with 500 μl of IPTG (100 mM). The cultures were incubated at 37°C on orbital shaking incubator at 220 rpm for 1-4 hours of induction. Cells were harvested by centrifugation at 6000 rpm for 10 min at 4°C.
Enzyme preparation
The cell pellet (of 250 mg cell wet weight) of 50 ml LB medium culture was harvested by centrifugation at 6000 rpm for 10 min and washed 3 times with distilled water. The cell pellet was resuspended in 1 ml of 50 mM potassium phosphate pH 8 and disintegrated by ultrasonic waves (3 × 1 min with 1 min pause). The cell lysate was centrifuged at 13000 rpm and 4°C for 15 min to separate the soluble fraction and precipitate. The pellet was resuspended in 1 ml of 50 mM potassium phosphate pH 8. The culture supernatant, cell lysate, cell lysate's supernatant (soluble fraction) and cell lysate's precipitate (inclusion bodies) were used for lipase activity estimation as well as for SDS-PAGE.
Lipase activity estimation
Lipase activity was estimated using olive oil as substrate in a pH-stat 718 (Metrohm) as described previously [17] with some modifications. One percent (v/v) of olive oil was emulsified in distilled water containing 0.5% (w/v) of gum Arabic as stabilizer using a homogenizer (Blender, USA) for 3 × 1 min at maximum speed. A 20 ml volume of the triglyceride solution was heated to 55°C and adjusted to pH 8. Autohydrolysis was measured in 10 min without addition of enzyme. After addition of 10 μl of the lipase solution, the activity was measured with a pH-stat 718 (Metrohm AG, Herisau, Switzerland) for 10 min. One unit of lipase activity was defined as the amount of enzyme, which released 1 μmol of fatty acid per min under standard assay conditions. All measurements were carried out three times and from values the average value was taken.
Gel electrophoresis
The expression level and molecular mass of the lipase and chaperone was determined by 12.5% SDS polyacrylamide gel electrophoresis with Biometra equipment (Göttingen, Germany) [30]. SDS-PAGE was usually performed with gels of 12.5% (w/v) of acrylamide according to the manufacturer's recommendations. An amount of 0.5 mg cell wet weight was loaded on each well. The expression level was determined by densitometry analysis Dolphin 1-D software operation (Wealtec Biosciences Co., Ltd, Taipei, Taiwan). For lipase activity staining [31], gels were incubated in 50 ml 0.1 M Tris-HCl buffer pH 7.5 containing 0.5% (w/v) of Triton X-100 for 30 min to renature the lipase. Then the gels were stained for 30 min in a mixture of 50 ml 0.1 M Tris-HCl buffer pH 7.5 containing 20 mg α-naphthyl acetate which was dissolved before in 5 ml acetone and 50 ml 0.1 M Tris-HCl buffer pH 7.5 containing 50 mg Fast Red TR.
DNA and amino acid sequence alignment
Homologies of the DNA and amino acid sequences were determined with the program MegAlign DNAStar.
Effect of culture factors on active lipase production
To figure out the effect of the substrate and detergents on the production of the active lipase, 0.5% (w/v) Neptune oil (substrate), gum Arabic, Tween 80 and Triton X-100 were supplemented to the LB medium. After induction by IPTG for 4 hours in different culture media supplemented without Neptune oil, with Neptune oil, gum Arabic, gum Arabic and Neptune oil, Tween 80, and Triton X-100, total proteins were applied to SDS-PAGE to estimate the expression level of the lipase in the dual expression cassette plasmid systems BL21/pELipAB-LipB1a and BL21/pELipAB-LipB3a.
Abbreviations
Protein/enzyme
LipA: Lipase from Ralstonia sp. M1; LipB: Chaperone from Ralstonia sp. M1; LipB1: 56-aa truncated chaperone LipB; LipB3: 26-aa truncated chaperone LipB; LipA6xHis: muture Lipase fused with 6x histidine tag
Gene
lipA: Lipase gene from Ralstonia sp. M1; lipB: Chaperone gene from Ralstonia sp. M1; lipB1: 168-nt truncated chaperone gene lipB; lipB3: 78-nt truncated chaperone lipB; lipAB: Lipase and chaperone gene cluster
Plasmid
pELipAa: pET22b(+) + lipA, a: ampicillin resistant marker; pELipABa: pET22b(+) + lipAB, a: ampicillin resistant marker; pELipB1a: pET22b(+) + lipB1, a: ampicillin resistant marker; pELipB3a: pET22b(+) + lipB3, a: ampicillin resistant marker; pELipB1k: pET28a(+) + lipB1, k: kanamycin resistant marker; pELipB3k: pET28a(+) + lipB3, k: kanamycin resistant marker; pELipA-LipB1a: pET22b(+) + lipA + T7 promoter + lipB1+ T7 terminator, a: ampicillin resistant marker; pELipA-LipB3a: pET22b(+) + lipA + T7 promoter + lipB3 + T7 terminator, a: ampicillin resistant marker; pELipAB-LipB1a: pET22b(+) + lipAB + T7 promoter + lipB1 + T7 terminator, a: ampicillin resistant marker; pELipAB-LipB3a: pET22b(+) + lipAB + T7 promoter + lipB3 + T7 terminator, a: ampicillin resistant marker
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DTQ designed the experimental setup, initiated the project, assisted with data analysis and manuscript preparation, read and approved the final manuscript. HCV performed experiments of plasmid construction and expression and TTGL performed experiments of influence of additives on expression. All authors read and approved the final manuscript.
Contributor Information
Thi Dinh Quyen, Email: quyen@ibt.ac.vn.
Chi Hai Vu, Email: haichivu@gmail.com.
Giang Thi Thu Le, Email: ltgiang1979@yahoo.com.
Acknowledgements
The study was supported by the Foundation of Fundamental Sciences (Vietnam Ministry of Science and Technology, Project No. 821104: Functional production of the lipase from Ralstonia M1 in E. coli). Dinh Thi Quyen gratefully acknowledged a grant for equipment from the International Foundation of Science (Sweden, 2008).
References
- Martinelle M, Holmquist M, Hult K. On the interfacial activation of Candida antarctica lipase A and B as compared with Humicola lanuginosa lipase. Biochim Biophys Acta. 1995;1258:272–276. doi: 10.1016/0005-2760(95)00131-u. [DOI] [PubMed] [Google Scholar]
- Zaks A, Klibanov AM. Enzymatic catalysis in nonaqueous solvents. J Biol Chem. 1988;263:3194–3201. [PubMed] [Google Scholar]
- Jaeger K-E, Eggert T. Lipases for biotechnology. Curr Opin Biotechnol. 2002;13(4):390–397. doi: 10.1016/S0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- Chihara-Siomi M, Yoshikawa K, Oshima-Hirayama N, Yamamoto K, Sogabe Y, Nakatani T, Nishioka T, Oda J. Purification, molecular cloning, and expression of lipase from Pseudomonas aeruginosa. Arch Biochem Biophys. 1992;296(2):505–513. doi: 10.1016/0003-9861(92)90604-U. [DOI] [PubMed] [Google Scholar]
- Wohlfarth S, Hoesche C, Strunk C, Winkler UK. Molecular genetics of the extracellular lipase of Pseudomonas aeruginosa PAO1. J Gen Microbiol. 1992;138(7):1325–1335. doi: 10.1099/00221287-138-7-1325. [DOI] [PubMed] [Google Scholar]
- Aoyama S, Yoshida N, Inouye S. Cloning, sequencing and expression of the lipase gene from Pseudomonas fragi IFO-12049 in E. coli. FEBS Lett. 1988;242(1):36–40. doi: 10.1016/0014-5793(88)80980-3. [DOI] [PubMed] [Google Scholar]
- Kok RG, van Thor JJ, Nugteren-Roodzant IM, Brouwer MB, Egmond MR, Nudel CB, Vosman B, Hellingwerf KJ. Characterization of the extracellular lipase, LipA, of Acinetobacter calcoaceticus BD413 and sequence analysis of the cloned structural gene. Mol Microbiol. 1995;15(5):803–818. doi: 10.1111/j.1365-2958.1995.tb02351.x. [DOI] [PubMed] [Google Scholar]
- Sullivan ER, Leahy JG, Colwell RR. Cloning and sequence analysis of the lipase and lipase chaperone-encoding genes from Acinetobacter calcoaceticus RAG-1, and redefinition of a proteobacterial lipase family and an analogous lipase chaperone family. Gene. 1999;230(2):277–286. doi: 10.1016/S0378-1119(99)00026-8. [DOI] [PubMed] [Google Scholar]
- Frenken LG, Egmond MR, Batenburg AM, Bos JW, Visser C, Verrips CT. Cloning of the Pseudomonas glumae lipase gene and determination of the active site residues. Appl Environ Microbiol. 1992;58(12):3787–3791. doi: 10.1128/aem.58.12.3787-3791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litthauer D, Ginster A, Skein E. Pseudomonas luteola lipase: a new member of the 320-residue Pseudomonas lipase family. J Enzyme Microb Technol. 2002;30(2):209–215. doi: 10.1016/S0141-0229(01)00469-0. [DOI] [Google Scholar]
- Jorgensen S, Skov KW, Diderichsen B. Cloning, sequence, and expression of a lipase gene from Pseudomonas cepacia: lipase production in heterologous hosts requires two Pseudomonas genes. J Bacteriol. 1991;173(2):559–567. doi: 10.1128/jb.173.2.559-567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipa MA, Liebeton K, Costa JV, Cabral JM, Jaeger KE. Lipase from Chromobacterium viscosum: biochemical characterization indicating homology to the lipase from Pseudomonas glumae. Biochim Biophys Acta. 1995;1256(3):396–402. doi: 10.1016/0005-2760(95)00052-e. [DOI] [PubMed] [Google Scholar]
- Quyen DT, Nguyen TT, Le TTG, Kim HK, Oh TK, Lee JK. A novel lipase/chaperone pair from Ralstonia sp. M1: analysis of the folding interaction and evidence for gene loss in R. solanacearum. Mol Gen Genomics. 2004;272:538–549. doi: 10.1007/s00438-004-1084-7. [DOI] [PubMed] [Google Scholar]
- Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343(1):177–183. doi: 10.1042/0264-6021:3430177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenken LG, Bos JW, Visser C, Muller W, Tommassen J, Verrips CT. An accessory gene, lipB, required for the production of active Pseudomonas glumae lipase. Mol Microbiol. 1993;9(3):579–589. doi: 10.1111/j.1365-2958.1993.tb01718.x. [DOI] [PubMed] [Google Scholar]
- Ihara F, Okamoto I, Akao K, Nihara T, Yamada Y. Lipase modulator protein (LimL) of Pseudomonas sp. strain 109. J Biacteriol. 1995;177:1245–1258. doi: 10.1128/jb.177.5.1254-1258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quyen DT, Schmidt-Dannert C, Schmid RD. High-level formation of active Pseudomonas cepacia lipase after heterologous expression of the encoding gene and its modified chaperone in Escherichia coli and rapid in vitro refolding. Appl Environ Microbiol. 1999;65(2):787–794. doi: 10.1128/aem.65.2.787-794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima-Hirayama N, Yoshikawa K, Takaaki N, Oda J. Lipase from Pseudomonas aeruginosa. Production in Escherichia coli and activation in vitro with a protein from the downstream gene. Eur J Biochem. 1993;215:239–246. doi: 10.1111/j.1432-1033.1993.tb18028.x. [DOI] [PubMed] [Google Scholar]
- Hobson AH, Buckley CM, Aamand JL, Jorgensen ST, Diderichsen B, McConnell DJ. Activation of a bacterial lipase by its chaperone. Proc Natl Acad Sci USA. 1993;90(12):5682–5686. doi: 10.1073/pnas.90.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kobayashi K, Iwasaki Y, Nakano H, Yamane T. In vitro analysis of roles of a disulfide bridge and a calcium binding site in activation of Pseudomonas sp. strain KWI 56 lipase. J Bacteriol. 2000;182:295–302. doi: 10.1128/JB.182.2.295-302.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S-s, Lin X-z, Shen J-h. Effects of co-expression of molecular chaperones on heterologous soluble expression of the cold-active lipase Lip-948. Protein Expr Purif. 2011;77:166–172. doi: 10.1016/j.pep.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Madan B, Mishra P. Co-expression of the lipase and foldase of Pseudomonas aeruginosa to a functional lipase in Escherichia coli. Appl Microbiol Biotechnol. 2010;85:597–604. doi: 10.1007/s00253-009-2131-4. [DOI] [PubMed] [Google Scholar]
- Peng R, Lin J, Wei D. Co-expression of an organic solvent-tolerant lipase and its cognate foldase of Pseudomonas aeruginosa CS-2 and the application of the immobilized recombinant lipase. Appl Biochem Biotechnol. 2011;165(3-4):926–937. doi: 10.1007/s12010-011-9309-9. [DOI] [PubMed] [Google Scholar]
- Narayanan N, Khan M, Chou CP. Enhancing functional expression of heterologous Burkholderia lipase in Escherichia coli. Mol Biotechnol. 2011;47(2):130–143. doi: 10.1007/s12033-010-9320-3. [DOI] [PubMed] [Google Scholar]
- Quyen DT, Le TTG, Nguyen TT, Oh TK, Lee JK. High-level heterologous expression and properties of a novel lipase from Ralstonia sp. M1. Protein Exp Purif. 2005;39:97–106. doi: 10.1016/j.pep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Quyen DT. PhD thesis. Stuttgart, Germany: University of Stuttgart; 1998. Recombinant lipase from Burkholderia cepacia: High-level expression in Escherichia coli and protein engineering. [Google Scholar]
- Ogino H, Katou Y, Akagi R, Mimitsuka T, Hiroshima S, Gemba Y, Doukyu N, Yasuda M, Ishimi K, Ishikawa H. Cloning and expression of gene, and activation of an organic solvent-stable lipase from Pseudomonas aeruginosa LST-03. Extremophiles. 2007;11(6):809–817. doi: 10.1007/s00792-007-0101-2. [DOI] [PubMed] [Google Scholar]
- Rosenau F, Jaeger KE. Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie. 2000;82(11):1023–1032. doi: 10.1016/S0300-9084(00)01182-2. [DOI] [PubMed] [Google Scholar]
- Quyen DT, Nguyen SLT, Dao TT. A novel esterase from Ralstonia sp. M1: gene cloning, sequencing, high-level expression and characterization. Protein Expr Purif. 2007;51(2):133–140. doi: 10.1016/j.pep.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structure proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Quyen DT, Schmidt-Dannert C, Schmid RD. High-level expression of a lipase from Bacillus thermocatenulatus BTL2 in Pichia pastoris and some properties of the recombinant lipase. Protein Expr Purif. 2003;28(1):102–110. doi: 10.1016/S1046-5928(02)00679-4. [DOI] [PubMed] [Google Scholar]