Abstract
Background
MicroRNAs (miRNAs) are endogenously encoded small RNAs that post-transcriptionally regulate gene expression. MiRNAs play essential roles in almost all plant biological processes. Currently, few miRNAs have been identified in the model food legume Phaseolus vulgaris (common bean). Recent advances in next generation sequencing technologies have allowed the identification of conserved and novel miRNAs in many plant species. Here, we used Illumina's sequencing by synthesis (SBS) technology to identify and characterize the miRNA population of Phaseolus vulgaris.
Results
Small RNA libraries were generated from roots, flowers, leaves, and seedlings of P. vulgaris. Based on similarity to previously reported plant miRNAs,114 miRNAs belonging to 33 conserved miRNA families were identified. Stem-loop precursors and target gene sequences for several conserved common bean miRNAs were determined from publicly available databases. Less conserved miRNA families and species-specific common bean miRNA isoforms were also characterized. Moreover, novel miRNAs based on the small RNAs were found and their potential precursors were predicted. In addition, new target candidates for novel and conserved miRNAs were proposed. Finally, we studied organ-specific miRNA family expression levels through miRNA read frequencies.
Conclusions
This work represents the first massive-scale RNA sequencing study performed in Phaseolus vulgaris to identify and characterize its miRNA population. It significantly increases the number of miRNAs, precursors, and targets identified in this agronomically important species. The miRNA expression analysis provides a foundation for understanding common bean miRNA organ-specific expression patterns. The present study offers an expanded picture of P. vulgaris miRNAs in relation to those of other legumes.
Background
Small non-coding RNAs (sncRNAs) have been recognized as an important class of gene expression regulators. MicroRNAs (miRNAs) are ~21 nucleotide (nt) sncRNAs that regulate a multitude of biological processes in plants, including development, metabolism, stress responses, defense against pathogens, and maintenance of genome integrity [1]. MiRNAs direct cleavage or translational inhibition of a specific messenger RNA (mRNA) based on base-pairing complementarity between the miRNA and the target mRNA. MiRNAs are derived from imperfectly matched stem-loop structures that are formed from single-stranded primary miRNA transcripts (pri-miRNAs). MiRNA genes are RNA polymerase II transcription units that give rise to pri-miRNAs [2]. Pri-miRNAs are preferentially processed in the nucleus by an RNaseIII-type enzyme DICER-LIKE1 (DCL1) to release the precursor miRNAs (pre-miRNAs) [3]. Most plant pre-miRNAs produce a single miRNA/miRNA* duplex; the exceptions are some miR159 and miR319 loci [4]. These small RNA duplexes are subsequently 2'-O-methylated by the nuclear HUA ENHANCER 1 (HEN1) protein, preventing miRNA turnover, and are exported to the cytoplasm by the plant homolog of exportin-5 HASTY (HST) [5-7]. Finally, one of the strands of each duplex is incorporated into the RNA-Induced Silencing Complex (RISC) to guide gene silencing [8]. RISCs contain a member of the ARGONAUTE (AGO) protein family to direct the endonucleolytic cleavage of target RNAs. AGO1 is the predominant carrier of plant miRNAs [9]. Thus, miRNAs are used by RISCs as templates for recognizing complementary mRNA to regulate gene expression.
Since the first plant miRNAs were reported in Arabidopsis thaliana in 2002, there has been an exponential growth of identified miRNAs in a diverse number of plant species [10]. The miRNA database miRBase (release 16, Sept 2010) contains 15,172 microRNA loci from 142 species. Plant miRNA loci belonging to 43 species correspond to 22.4% of the total mature miRNA entries. The plant species with the most miRNA loci identified are Oryza sativa, Medicago truncatula, Arabidopsis thaliana, Physcomitrella patens, Glycine max, Sorghum bicolor, Populus trichocarpa, and Zea mays. These 8 widely studied plant model species contribute 65.8% of the plant mature miRNA entries. On the other hand, 15 plant species, including Phaseolus vulgaris, have fewer than 10 reported miRNA loci in miRBase.
Identification of P. vulgaris microRNAs was first performed using an in silico approach [11]. The eight miRNA loci reported for P. vulgaris in miRBase were identified by Arenas-Huertero et al. (2009) in different organs and growth conditions, although the first mature miRNA sequence ever cloned and characterized for P. vulgaris was pvu-miR399a [12]. This group of eight miRNAs in miRBase comprises miR2118, miR159a, miR1514a, miR482, miR2119, miR166a, miR319c, and miR399a. Besides these miRNAs and their stem-loop precursors, another 19 mature miRNA sequences have been identified [13]. In addition, Valdés-López et al. (2010) analyzed the expression of 68 miRNAs under several abiotic stress conditions in leaves, roots and nodules of P. vulgaris based on a hybridization approach using miRNA macroarrays. The macroarrays contained probes for 9 miRNAs previously reported for common bean, 24 conserved miRNAs also identified in other legumes and 35 miRNAs found in soybean [11,13,14].
The first studies to identify plant miRNAs employed the traditional Sanger sequencing method. This was also the case for the study performed by Arenas-Huertero et al. (2009) to identify miRNA sequences in Phaseolus vulgaris. Despite the Sanger sequencing method's usefulness and importance in scientific research, it has several limitations with regard to miRNA identification [15,16]. For example, low-abundance miRNAs, such as many non-conserved miRNAs, are inefficiently detected by this small-scale sequencing method. Introduction of high-throughput next-generation sequencing (NGS) technologies has increased the number of miRNAs identified by allowing small RNA population analysis on a massive scale (RNA-seq). Deep-sequencing technologies have identified a larger number of novel miRNAs due to their ability to generate millions of reads with a determined length [17]. Moreover, the population of short sequence reads produced allows the identification of differential processing of stem-loop precursors by DICER-LIKE1, and mature miRNA isoforms within and between species. Likewise, relative abundance associated with small RNAs estimated by these technologies has permitted a higher level of confidence for miRNA annotation.
Legumes such as common bean are valuable crops for worldwide consumption because they are rich in protein and constitute a high calorie food source. Involvement of miRNAs in biological processes like nutrient balance, development, reproduction and plant-microbe interaction, makes its research crucial for improvement of staple crops. Up to now, identification and characterization of Phaseolus vulgaris miRNAs have been very limited. In this study, Illumina's sequencing by synthesis (SBS) technology was used to examine conserved, novel and species-specific common bean miRNAs comprehensively based on small RNA libraries generated from leaves, roots, seedlings and flowers. A detailed analysis of mature, mature-star (the complementary strand of the mature miRNA), isoforms and novel common bean miRNAs found in different organs is reported. Stem-loop precursors and target sequences for several common bean miRNAs were also determined from public databases. Overall, this work serves to extend our knowledge of the common bean miRNA population and to spotlight new miRNA variants found in different organs. It facilitates comparisons between common bean miRNAs and those found in other legumes and plants.
Results
Small RNA sequencing analysis
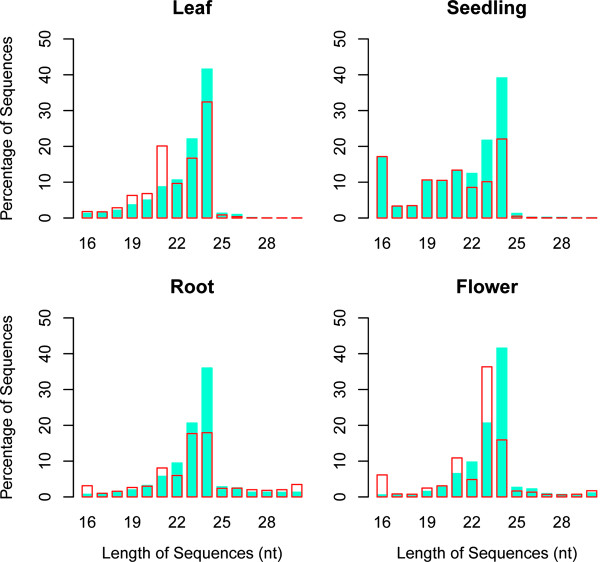
In order to identify novel and conserved miRNAs in common bean, we generated four small RNA libraries from leaves (LL), roots (RL), seedlings (SL) and flowers (FL) using the Genome Analyzer II and the Illumina Cluster Station (Illumina Inc, USA). Small RNA sequencing yielded more than eighty million raw sequence reads (Table 1). After removing low-quality sequences, adapters, and small sequences (< 16 nt long), 79.5% of the raw reads were left. These remaining sequences represent 3,372,753 (LL), 4,187,414 (RL), 4,015,702 (SL), and 3,453,543 (FL) unique sequence tags. The small RNA length distribution (16-30 nt) of each library showed that the most abundant and diverse species were those 21-24 nt in length, a typical size range for Dicer-derived products (Figure 1). The 24 nt class was the most diverse in all four libraries, followed by the 21 nt class, which was the second most abundant class in the LL library (20%), and the third most abundant class in the SL (13.4%), RL (8%), and FL (10.8%) libraries. Filtering sequences for removal of non-coding RNAs such as tRNA, rRNA, snoRNA or snRNA was performed using the RNA families database Rfam because of the limited availability of P. vulgaris genome sequences. After removing miRNA sequences from Rfam, the remainder of the database (Rfam 10.0) was used to eliminate fragments corresponding to non-coding RNAs. Alignment against the four small RNA libraries was carried out using BLASTN and only perfect matches were removed from the libraries (Table 1).
Table 1.
Summary of small RNA sequencing data analysis
| Leaves | Roots | Seedlings | Flowers | |
|---|---|---|---|---|
| Total raw reads | 16869046 | 20464127 | 17188077 | 27937376 |
| High-quality readsa | 11797480 | 16722005 | 16846110 | 20238107 |
| Unique sequence tags | 3372753 | 4187414 | 4015702 | 3453543 |
| Total Rfam matching sequences | 757788 | 5653553 | 5307793 | 9726833 |
| Unique Rfam matching sequences | 51227 | 60121 | 224240 | 96726 |
| Perfect miRNA matching sequencesb | 265161 | 307003 | 582159 | 1415225 |
| Total unfiltered isoform sequences | 798835 | 287027 | 467691 | 228556 |
| Total miRNA isoform sequences | 744348 | 200630 | 335274 | 60167 |
aSequences after quality, adapter and size filters. bTotal sequences from conserved miRNAs.
Figure 1.
Sequence length distribution of P. vulgaris sRNAs. Size distribution in small RNA libraries. Average percentage (Y-axis) of unique (filled blue bars) and redundant (non-filled red bars) sequences of 16-30 nt length (X-axis) for each of the four sequenced libraries.
Identification of conserved miRNAs
To identify conserved miRNAs in the common bean small RNA libraries, unique mature plant miRNA sequences were extracted from miRBase (Release 16). BLASTN and SSAHA2 (Sequence Search and Alignment by Hashing Algorithm) tools were used separately for alignment of small RNA datasets against mature miRNA sequences in search of previously reported miRNAs with exactly the same size and nucleotide composition [18,19]. Both alignment tools found the same number of hits. MiRNAs that were detected in just one library or that totalled fewer than 15 absolute appearances in all libraries combined were removed (Additional file 1).
In total, 109 conserved miRNAs (Table 2) belonging to 29 families were identified in common bean. Fourteen of the conserved miRNAs were mature-star miRNAs (Table 3). Ten were detected in just two libraries, twenty five in three libraries, and seventy four in all four libraries. Twenty five of the families detected are highly conserved among plant species (Figure 2). Nine conserved families identified in common bean, including miR156, miR159, miR319, miR160, miR166, miR171, miR408, miR390 and miR395, were present in the common ancestor of all embryophytes [20]. Additionally, the miR396 family identified in all tracheophytes, and a pair of miRNA families detected in seed plants, miR397 and miR398, were present in P. vulgaris. Another nine identified are present in all angiosperm lineages: miR162, miR164, miR167, miR168, miR169, miR172, miR393, miR394, and miR399. The miR2111 family, which has been identified only in eudicot species, and the miR2118 family which has been found in monocots and in Fabaceae, were also identified in common bean. Finally, among the deeply conserved miRNA families detected, miR157, closely related to miR156, is present in Brassicaceae, Solanaceae, Malvaceae and Fabaceae.
Table 2.
Conserved miRNAs from common bean
| miRNA family |
Reference miRNA |
Sequence (5'- 3') | Length (nt) | Reads | ||||
|---|---|---|---|---|---|---|---|---|
| LL | FL | RL | SL | Total | ||||
| 156 | ahy-miR156a | UGACAGAAGAGAGAGAGCAC | 20 | 56 | 15 | 8 | 41 | 120 |
| ahy-miR156c | UUGACAGAAGAGAGAGAGCAC | 21 | 94 | 3512 | 3 | 317 | 3926 | |
| ath-miR156a | UGACAGAAGAGAGUGAGCAC | 20 | 82529 | 63 | 3705 | 14478 | 100775 | |
| ath-miR156g | CGACAGAAGAGAGUGAGCAC | 20 | 74 | 0 | 3 | 10 | 87 | |
| bna-miR156a | UGACAGAAGAGAGUGAGCACA | 21 | 112 | 21 | 2 | 26 | 161 | |
| ptc-miR156k | UGACAGAAGAGAGGGAGCAC | 20 | 47 | 0 | 6 | 2 | 55 | |
| vvi-miR156e | UGACAGAGGAGAGUGAGCAC | 20 | 30 | 0 | 1 | 1 | 32 | |
| zma-miR156k | UGACAGAAGAGAGCGAGCAC | 20 | 183 | 0 | 14 | 10 | 207 | |
| 157 | ath-miR157a | UUGACAGAAGAUAGAGAGCAC | 21 | 3114 | 2787 | 4370 | 4839 | 15110 |
| ath-miR157d | UGACAGAAGAUAGAGAGCAC | 20 | 361 | 30 | 26 | 89 | 506 | |
| 159 | aqc-miR159 | UUUGGACUGAAGGGAGCUCUA | 21 | 33 | 312 | 43 | 89 | 477 |
| pvu-miR159a.1 | UUUGGAUUGAAGGGAGCUCUA | 21 | 67007 | 1082688 | 180739 | 453692 | 1784126 | |
| ath-miR159b | UUUGGAUUGAAGGGAGCUCUU | 21 | 3 | 438 | 135 | 178 | 754 | |
| gma-miR159c | AUUGGAGUGAAGGGAGCUCCG | 21 | 0 | 0 | 1044 | 25 | 1069 | |
| gma-miR159d | AGCUGCUUAGCUAUGGAUCCC | 21 | 78 | 148 | 202 | 240 | 668 | |
| osa-miR159a.1 | UUUGGAUUGAAGGGAGCUCUG | 21 | 37 | 224 | 31 | 148 | 440 | |
| osa-miR159f | CUUGGAUUGAAGGGAGCUCUA | 21 | 57 | 666 | 298 | 182 | 1203 | |
| pta-miR159a | UUGGAUUGAAGGGAGCUCCA | 20 | 0 | 3 | 0 | 25 | 28 | |
| 160 | ath-miR160a | UGCCUGGCUCCCUGUAUGCCA | 21 | 136 | 5160 | 5474 | 104 | 10874 |
| bdi-miR160 | UGCCUGGCUCCCUGUAUGCC | 20 | 9 | 212 | 225 | 5 | 451 | |
| osa-miR160f | UGCCUGGCUCCCUGAAUGCCA | 21 | 9 | 3 | 240 | 2 | 254 | |
| 162 | ath-miR162a | UCGAUAAACCUCUGCAUCCAG | 21 | 597 | 583 | 517 | 1192 | 2889 |
| zma-miR162 | UCGAUAAACCUCUGCAUCCA | 20 | 14 | 22 | 15 | 30 | 81 | |
| 164 | ath-miR164a | UGGAGAAGCAGGGCACGUGCA | 21 | 7245 | 5621 | 3628 | 1229 | 17723 |
| ath-miR164c | UGGAGAAGCAGGGCACGUGCG | 21 | 49 | 71 | 91 | 122 | 333 | |
| 166 | gma-miR166a | UCGGACCAGGCUUCAUUCCCC | 21 | 5333 | 53276 | 20481 | 15163 | 94253 |
| ctr-miR166 | UCGGACCAGGCUUCAUUCCCCC | 22 | 2 | 18 | 10 | 10 | 40 | |
| osa-miR166e | UCGAACCAGGCUUCAUUCCCC | 21 | 0 | 17 | 8 | 4 | 29 | |
| vvi-miR166a | UCGGACCAGGCUUCAUUCC | 19 | 93 | 205 | 166 | 382 | 846 | |
| zma-miR166h | UCGGACCAGGCUUCAUUCCC | 20 | 140 | 769 | 352 | 630 | 1891 | |
| 167 | ath-miR167a | UGAAGCUGCCAGCAUGAUCUA | 21 | 661 | 2883 | 3251 | 90 | 6885 |
| ath-miR167d | UGAAGCUGCCAGCAUGAUCUGG | 22 | 22003 | 1443 | 5196 | 626 | 29268 | |
| bna-miR167a | UGAAGCUGCCAGCAUGAUCUAA | 22 | 2 | 0 | 17 | 0 | 19 | |
| ccl-miR167a | UGAAGCUGCCAGCAUGAUCUGA | 22 | 2267 | 8164 | 2727 | 276 | 13434 | |
| osa-miR167d | UGAAGCUGCCAGCAUGAUCUG | 21 | 15021 | 20039 | 6209 | 378 | 41647 | |
| ptc-miR167f | UGAAGCUGCCAGCAUGAUCUU | 21 | 36 | 252 | 146 | 5 | 439 | |
| 168 | ath-miR168a | UCGCUUGGUGCAGGUCGGGAA | 21 | 382 | 991 | 605 | 43 | 2021 |
| 169 | ath-miR169a | CAGCCAAGGAUGACUUGCCGA | 21 | 0 | 11 | 0 | 44 | 55 |
| ath-miR169b | CAGCCAAGGAUGACUUGCCGG | 21 | 1339 | 8784 | 154 | 6351 | 16628 | |
| gma-miR169d | UGAGCCAAGGAUGACUUGCCGGU | 23 | 0 | 4 | 11 | 0 | 15 | |
| gma-miR169e | AGCCAAGGAUGACUUGCCGG | 20 | 133 | 47 | 62 | 316 | 558 | |
| mtr-miR169c | CAGCCAAGGGUGAUUUGCCGG | 21 | 3916 | 166 | 78 | 2 | 4162 | |
| mtr-miR169d | AAGCCAAGGAUGACUUGCCGG | 21 | 126 | 863 | 389 | 4 | 1382 | |
| osa-miR169e | UAGCCAAGGAUGACUUGCCGG | 21 | 1 | 29 | 1 | 16 | 47 | |
| 171 | ath-miR171b | UUGAGCCGUGCCAAUAUCACG | 21 | 119 | 790 | 0 | 37 | 946 |
| sly-miR171d | UUGAGCCGCGCCAAUAUCAC | 20 | 2 | 1 | 67 | 7 | 77 | |
| zma-miR171b | UUGAGCCGUGCCAAUAUCAC | 20 | 28 | 40 | 13 | 2 | 83 | |
| zma-miR171f | UUGAGCCGUGCCAAUAUCACA | 21 | 62 | 245 | 139 | 68 | 514 | |
| 172 | ath-miR172a | AGAAUCUUGAUGAUGCUGCAU | 21 | 43 | 38 | 24 | 1 | 106 |
| ptc-miR172g | GGAAUCUUGAUGAUGCUGCAG | 21 | 353 | 18985 | 200 | 51 | 19589 | |
| sbi-miR172b | GGAAUCUUGAUGAUGCUGCA | 20 | 1 | 193 | 7 | 0 | 201 | |
| zma-miR172a | AGAAUCUUGAUGAUGCUGCA | 20 | 344 | 565 | 622 | 36 | 1567 | |
| 319 | ath-miR319a | UUGGACUGAAGGGAGCUCCCU | 21 | 0 | 1672 | 192 | 320 | 2184 |
| mtr-miR319 | UUGGACUGAAGGGAGCUCCC | 20 | 21 | 66392 | 4936 | 40730 | 112079 | |
| osa-miR319a | UUGGACUGAAGGGUGCUCCC | 20 | 0 | 19 | 5 | 3 | 27 | |
| ppt-miR319a | CUUGGACUGAAGGGAGCUCC | 20 | 1 | 1622 | 14 | 106 | 1743 | |
| ppt-miR319c | CUUGGACUGAAGGGAGCUCCC | 21 | 16 | 11934 | 643 | 3082 | 15675 | |
| pta-miR319 | UUGGACUGAAGGGAGCUCC | 19 | 0 | 1917 | 201 | 1139 | 3257 | |
| ptc-miR319e | UUGGACUGAAGGGAGCUCCU | 20 | 0 | 22 | 58 | 45 | 125 | |
| vvi-miR319e | UUUGGACUGAAGGGAGCUCCU | 21 | 0 | 55 | 2 | 1 | 58 | |
| vvi-miR319g | UUGGACUGAAGGGAGCUCCCA | 21 | 0 | 285 | 23 | 53 | 361 | |
| 390 | ath-miR390a | AAGCUCAGGAGGGAUAGCGCC | 21 | 299 | 1264 | 153 | 63 | 1779 |
| gma-miR390b | AAGCUCAGGAGGGAUAGCACC | 21 | 46 | 25 | 193 | 124 | 388 | |
| 393 | ath-miR393a | UCCAAAGGGAUCGCAUUGAUCC | 22 | 51 | 295 | 441 | 9 | 796 |
| osa-miR393 | UCCAAAGGGAUCGCAUUGAUC | 21 | 13 | 65 | 101 | 0 | 179 | |
| 394 | ath-miR394a | UUGGCAUUCUGUCCACCUCC | 20 | 71 | 7473 | 339 | 482 | 8365 |
| vvi-miR394a | UUGGCAUUCUGUCCACCUCCAU | 22 | 0 | 112 | 7 | 0 | 119 | |
| 395 | tae-miR395b | UGAAGUGUUUGGGGGAACUC | 20 | 5 | 25 | 4 | 8 | 42 |
| 396 | ath-miR396a | UUCCACAGCUUUCUUGAACUG | 21 | 760 | 44655 | 10489 | 601 | 56505 |
| ath-miR396b | UUCCACAGCUUUCUUGAACUU | 21 | 1 | 185 | 33 | 16 | 235 | |
| gma-miR396d | AAGAAAGCUGUGGGAGAAUAUGGC | 24 | 0 | 61 | 22 | 77 | 160 | |
| gma-miR396e | UUCCACAGCUUUCUUGAACUGU | 22 | 0 | 29 | 6 | 0 | 35 | |
| ptc-miR396f | UUCCACGGCUUUCUUGAACUG | 21 | 0 | 11 | 4 | 1 | 16 | |
| vvi-miR396a | UUCCACAGCUUUCUUGAACUA | 21 | 0 | 23 | 2 | 1 | 26 | |
| vvi-miR396b | UUCCACAGCUUUCUUGAACU | 20 | 742 | 19924 | 2108 | 823 | 23597 | |
| 397 | ath-miR397a | UCAUUGAGUGCAGCGUUGAUG | 21 | 2057 | 13 | 5276 | 70 | 7416 |
| zma-miR397a | UCAUUGAGCGCAGCGUUGAUG | 21 | 5 | 0 | 23 | 0 | 28 | |
| 398 | ahy-miR398 | UGUGUUCUCAGGUCACCCCU | 20 | 0 | 25 | 350 | 0 | 375 |
| osa-miR398b | UGUGUUCUCAGGUCGCCCCUG | 21 | 3760 | 57 | 1717 | 7 | 5541 | |
| 399 | ath-miR399a | UGCCAAAGGAGAUUUGCCCUG | 21 | 15 | 12 | 4 | 0 | 31 |
| ath-miR399b | UGCCAAAGGAGAGUUGCCCUG | 21 | 35 | 121 | 22 | 2 | 180 | |
| osa-miR399e | UGCCAAAGGAGAUUUGCCCAG | 21 | 49 | 45 | 3 | 1 | 98 | |
| 408 | ath-miR408 | AUGCACUGCCUCUUCCCUGGC | 21 | 2083 | 3718 | 9050 | 167 | 15018 |
| bdi-miR408 | AUGCACUGCCUCUUCCCUGG | 20 | 5 | 25 | 67 | 0 | 97 | |
| osa-miR408 | CUGCACUGCCUCUUCCCUGGC | 21 | 3 | 5 | 17 | 0 | 25 | |
| ppt-miR408b | UGCACUGCCUCUUCCCUGGCU | 21 | 0 | 5 | 55 | 0 | 60 | |
| 482 | gso-miR482a | UCUUCCCUACACCUCCCAUAC | 21 | 7 | 318 | 85 | 2 | 412 |
| pvu-miR482 | UCUUCCCAAUUCCGCCCAUUCC | 22 | 5 | 291 | 54 | 0 | 350 | |
| 1511 | gma-miR1511 | AACCAGGCUCUGAUACCAUG | 20 | 978 | 625 | 449 | 1529 | 3581 |
| 1514 | pvu-miR1514a | UUCAUUUUGAAAAUAGGCAUUG | 22 | 188 | 5481 | 1282 | 311 | 7262 |
| 1515 | gma-miR1515 | UCAUUUUGCGUGCAAUGAUCUG | 22 | 27 | 853 | 341 | 86 | 1307 |
| 2111 | ath-miR2111a | UAAUCUGCAUCCUGAGGUUUA | 21 | 48 | 211 | 255 | 0 | 514 |
| 2118 | mtr-miR2118 | UUACCGAUUCCACCCAUUCCUA | 22 | 8 | 7 | 1 | 1 | 17 |
| pvu-miR2118 | UUGCCGAUUCCACCCAUUCCUA | 22 | 238 | 17300 | 13707 | 1771 | 33016 | |
| 2119 | gma-miR2119 | UCAAAGGGAGUUGUAGGGGAA | 21 | 85 | 42 | 1448 | 6 | 1581 |
Conserved miRNAs in common bean identified in leaves (LL), roots (RL), seedlings (SL) and flowers (FL).
Table 3.
Conserved mature-star miRNAs from common bean
| miRNA family |
Reference miRNA |
Sequence (5'- 3') | Length (nt) | Reads | ||||
|---|---|---|---|---|---|---|---|---|
| LL | FL | RL | SL | Total | ||||
| 166 | aly-miR166b* | GGACUGUUGUCUGGCUCGAGG | 21 | 233 | 0 | 5 | 3 | 241 |
| aly-miR166g* | GGAAUGUUGUUUGGCUCGAGG | 21 | 38 | 143 | 186 | 257 | 624 | |
| zma-miR166a* | GGAAUGUUGUCUGGCUCGGGG | 21 | 26 | 3 | 11 | 7 | 47 | |
| gma-miR166a-5p | GGAAUGUUGUCUGGCUCGAGG | 21 | 36866 | 3839 | 6253 | 23902 | 70860 | |
| zma-miR166m* | GGAAUGUUGGCUGGCUCGAGG | 21 | 55 | 655 | 50 | 3363 | 4123 | |
| 167 | ahy-miR167-3p | AGAUCAUGUGGCAGUUUCACC | 21 | 155 | 16 | 84 | 11 | 266 |
| 168 | aly-miR168a* | CCCGCCUUGCAUCAACUGAAU | 21 | 80 | 22 | 8 | 3 | 113 |
| 171 | aly-MIR171a* | UAUUGGCCUGGUUCACUCAGA | 21 | 179 | 710 | 1 | 543 | 1433 |
| 172 | aly-miR172c* | GGAGCAUCAUCAAGAUUCACA | 21 | 10 | 352 | 19 | 1 | 382 |
| 390 | aly-miR390a* | CGCUAUCCAUCCUGAGUUUCA | 21 | 149 | 524 | 69 | 10 | 752 |
| gma-miR390a-3p | CGCUAUCCAUCCUGAGUUUC | 20 | 14 | 3 | 0 | 2 | 19 | |
| 396 | aly-miR396a* | GUUCAAUAAAGCUGUGGGAAG | 21 | 267 | 306 | 3931 | 791 | 5295 |
| 482 | gma-miR482a-5p | AGAAUUUGUGGGAAUGGGCUGA | 22 | 0 | 17 | 1 | 1 | 19 |
| pvu-miR482* | GGAAUGGGCUGAUUGGGAAGCA | 22 | 1185 | 6 | 440 | 10 | 1641 | |
Conserved mature-star miRNAs in common bean identified in leaves (LL), roots (RL), seedlings (SL) and flowers (FL).
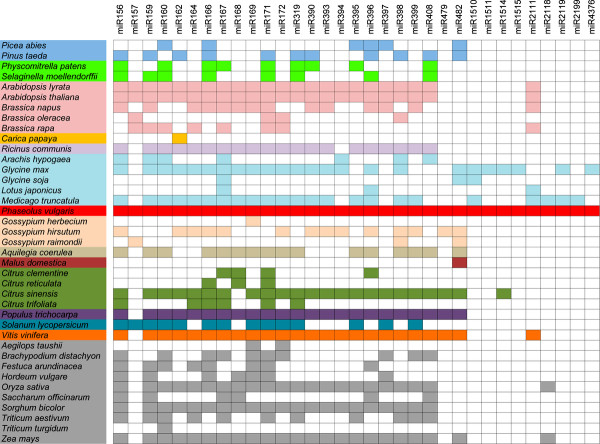
Figure 2.
Conserved miRNA families in P. vulgaris and across-species. Evolutionary conservation of thirty-three miRNA families identified in P. vulgaris within plant species reported in miRBase (v.16). Closely related species are shown in the same color. Colored boxes denote the presence of the indicated miRNA family.
Four other, less conserved miRNA families that were identified have been reported mainly in legumes, specifically in Glycine max. This is the case for miR1511, which had been previously detected only in soybean, although recent P. vulgaris miRNA studies pointed out its presence in common bean [13,14,21,22]. Here, it was confirmed that soybean and common bean share exactly the same sequence for this miRNA and that it is one of the most abundant miRNAs in these legumes (Table 2). The miR1514 and miR2119 families have been reported exclusively in legumes such as Glycine max, Medicago truncatula and Phaseolus vulgaris. Surprisingly, miR1515 was reported only in two species: G. max and Citrus Sinensis.
The P. vulgaris miRNAs with the most reads in all four libraries (ath-miR159a, mtr-miR319, ath-miR156a, ath-miR166a, zma-miR166c*, ath-miR396a, osa-miR167d, pvu-miR2118 and ath-miR167d) generally corresponded to the most abundant miRNAs detected in legumes like M. truncatula, G. max, and in other plants like Vitis vinifera [23-25]. It is worth noting that among these ten most abundant miRNAs there is a mature-star miRNA gma-miR166a-5p that has also been highly detected by both deep sequencing and microarray expression analysis in the shoot apical meristem (SAM) of soybean [26]. Also, it has been demonstrated that gma-miR166a and its mature-star miRNA have different expression patterns in the SAM, suggesting that they may play different roles in regulating leaf development [26]. The high number of reads detected for other mature-star sequences found in this study may also imply particular regulatory roles of gene expression conditioned by these small RNAs since inhibitory activity of mature-star species has been reported in animals and plants (Table 3) [27,28].
The miRNA families comprising the most miRNAs in common bean are miR156, miR159, miR166, miR167, miR169, miR319 and miR396. These highly conserved families, which generally have the most abundant miRNAs, have variable numbers of loci among the model legumes G. max and M. truncatula (Table 4) (miRBase, release 16). According to mature miRNA sequence differences, the predicted numbers of loci for P. vulgaris miRNA families have a higher Pearson's correlation coefficient with the numbers of loci reported for soybean (0.78) than with those identified in M. truncatula (0.21).
Table 4.
Predicted number of loci in common bean miRNA families
| miRNA family | Number of loci | miRNA family | Number of loci | ||||
|---|---|---|---|---|---|---|---|
| Pvu | Gma | Mtr | Pvu | Gma | Mtr | ||
| 156 | 6 | 7 | 9 | 396 | 5 | 5 | 2 |
| 157 | 1 | 0 | 0 | 397 | 2 | 0 | 0 |
| 159 | 6 | 4 | 1 | 398 | 1 | 2 | 3 |
| 160 | 2 | 1 | 5 | 399 | 3 | 0 | 17 |
| 162 | 1 | 1 | 1 | 408 | 2 | 0 | 0 |
| 164 | 2 | 1 | 4 | 479 | 1 | 0 | 0 |
| 166 | 2 | 2 | 8 | 482 | 3 | 2 | 0 |
| 167 | 5 | 7 | 1 | 1510 | 1 | 2 | 2 |
| 168 | 1 | 1 | 1 | 1511 | 1 | 1 | 0 |
| 169 | 6 | 5 | 17 | 1514 | 1 | 2 | 0 |
| 171 | 3 | 3 | 7 | 1515 | 1 | 1 | 0 |
| 172 | 2 | 3 | 1 | 2111 | 1 | 0 | 19 |
| 319 | 4 | 3 | 2 | 2118 | 2 | 0 | 1 |
| 390 | 3 | 3 | 1 | 2119 | 1 | 1 | 1 |
| 393 | 1 | 3 | 2 | 2199 | 2 | 0 | 1 |
| 394 | 1 | 2 | 0 | 4376 | 1 | 1 | 0 |
| 395 | 1 | 0 | 18 | ||||
Abbreviations stand for: Pvu, Phaseolus vulgaris; Gma, Glycine max; Mtr, Medicago truncatula.
Identification of miRNA isoforms
Small non-coding RNAs such as microRNAs were initially thought to have a specific sequence of a defined length. Identification of more miRNAs from different species has revealed that there is variation in pre-miRNA processing. The miRBase (release16) dataset for plant miRNAs consists of small sequences of 17 (.08%), 18 (.13%), 19 (.88%), 20 (9.96%), 21 (67.06%), 22 (13.85%), 23 (1.10%), and 24 (6.91%) nucleotides in length. Ebhardt et al. (2010) demonstrated that a fifth of the annotated Arabidopsis thaliana miRNAs (miRBase, release 14) have a stable miRNA isoform of one or two nucleotides longer [29]. Also, several studies have revealed that the biological function of miRNA isoforms may differ from the function of their previously reported miRNAs due to differential associations with AGO proteins [30,31]. The legumes M. truncatula and G. max, which account for the majority of entries for the Fabaceae family in miRBase, present mature miRNA sequences ranging from 20-22 and 19-24 nucleotides in length, respectively.
With the aim of identifying P. vulgaris miRNA length isoforms and species-specific miRNA variants, all small RNA sequences remaining from previous analyses were aligned against miRBase, allowing at most two mismatches and/or two nucleotides in length difference. The total number of variants found for each library (Table 1) was subjected to a filter that consisted of choosing variants that had a total number of reads 50% greater than the number of total reads of their reference miRNA previously reported, so that low-abundance and probable non-functional variants were discarded. MiRNA isoforms were classified as length variants, non-conserved miRNA variants, or conserved miRNA variants (Table 5). Seven variants based only on length were detected. In all four P. vulgaris libraries, the 21-nt variant of gma-miR1511 is clearly predominant. In the case of the variant for gso-miR482a, it was highly expressed in the flower (FL) and root (RL) libraries. Unexpectedly, a variant for miRNA mtr-miR171c, a miRNA that was absent in all libraries, was detected mainly in flower. The high abundance of pvu-miR482* in common bean compared with other plant species was previously corroborated by northern blot [13]. As Table 5 shows, the high number of reads detected for this mature-star miRNA in all organs was produced by the 21-nt variant and not by the one reported in miRBase. As mentioned before, some miR159 and miR319 loci produce two miRNA/miRNA* duplexes. This was the case for the P. vulgaris miR159a stem-loop. The variant for pvu-miR159a.2 that is found closer to the loop is mainly a 19 nt miRNA, and its number of reads does not equate to the abundance of pvu-miR159a.1. The length variant gma-miR4376 increases the number of P. vulgaris miRNA families identified in this study because of its high number of reads. The miR4376 family had been identified only in soybean (Figure 2), and was registered in miRBase as a 22 nt miRNA [21]. The group of non-conserved miRNA variants contains nine variants from eight non-conserved miRNA families (Table 5). Considering the number of reads and that mature-miRNA homologs may present one or two mismatches, another three common bean miRNA families were identified: miR1510, miR479, and miR2199. For the miR1510 family, two variants were identified that can be produced from the same locus. This family has been reported only in soybean and M. truncatula, and was proposed to be conserved in common bean due to a 20 nt isoform [13,14,23]. The variant of csi-miR479, a miRNA detected in Citrus sinensis, Vitis vinifera, Populus trichocarpa and Gossypium hirsutum was particularly abundant in roots [25,32-34]. Interestingly, in this study two variants with the same length were detected for mtr-miR2199, one that was detected in all four libraries (A17 > G) and the other (C12 > T) expressed mostly in roots. A previous study in M. truncatula where this family was detected, reported the mature miRNA as the variant A17 > G and the genome derived-hairpin as A17, which accounts for its annotation in miRBase with an adenine in position 17 [35]. The miRNA sequence conserved in Lotus japonicus was the variant A17 > G also found in P. vulgaris [35]. The ten miRNA variants classified in the conserved miRNA variants group constitute candidates for new miRNAs of previously identified miRNA families (Table 5). The miRNA candidates with the most reads were the variants of osa-miR156l, csi-miR393, sly-miR171c, and mtr-miR169p.
Table 5.
MiRNA isoforms from P.vulgaris
| Variant group |
Reference miRNA |
Sequence (5'- 3') | Length (nt) | Reads | ||||
|---|---|---|---|---|---|---|---|---|
| LL | FL | RL | SL | Total | ||||
| Length variants | gma-miR1511 | AACCAGGCTCTGATACCATG | 20 | 978 | 625 | 449 | 1529 | 3581 |
| pvu-isomiR 1511 | AACCAGGCTCTGATACCATGA | 21 | 14902 | 13725 | 7707 | 17239 | 53573 | |
| gso-miR482a | TCTTCCCTACACCTCCCATAC | 21 | 7 | 318 | 85 | 2 | 412 | |
| pvu-isomiR 482a | TCTTCCCTACACCTCCCATACC | 22 | 369 | 17616 | 5564 | 305 | 23854 | |
| mtr-miR171c | TGATTGAGCCGTGCCAATATT | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR 171a | TGATTGAGCCGTGCCAATA | 19 | 115 | 1390 | 60 | 57 | 1622 | |
| pvu-miR482* | GGAATGGGCTGATTGGGAAGCA | 22 | 1185 | 6 | 440 | 10 | 1641 | |
| pvu-isomiR 482* | GGAATGGGCTGATTGGGAAGC | 21 | 714044 | 3806 | 173153 | 1 | 1207244 | |
| pvu-miR159a.2 | CTTCCATATCTGGGGAGCTTC | 21 | 1 | 0 | 0 | 0 | 1 | |
| pvu-isomiR 159a | CTTCCATATCTGGGGAGCT | 19 | 13 | 263 | 157 | 23 | 456 | |
| gma-miR4376 | TACGCAGGAGAGATGACGCTGT | 22 | 1 | 1 | 0 | 0 | 2 | |
| pvu-isomiR 4376 | TACGCAGGAGAGATGACGCTG | 21 | 3841 | 1090 | 0 | 398 | 5329 | |
| mtr-miR171b | TGATTGAGCCGCGTCAATATC | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR171b | TCTGATTGAGCCGCGTCAATA | 21 | 101 | 1 | 79 | 10 | 191 | |
| Non conserved variants | ath-miR858 | TTTCGTTGTCTGTTCGACCTT | 21 | 0 | 0 | 0 | 0 | 0 |
| pvu-isomiR 858 | CTCGTTGTCTGTTCGACCTTG | 21 | 37 | 351 | 0 | 26 | 414 | |
| csi-miR479 | TGTGATATTGGTTCGGCTCATC | 22 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR479 | TGTGATATTGGTTTGGCTCA | 20 | 58 | 1 | 1588 | 11 | 1658 | |
| gma-miR1510a-3p | TTGTTGTTTTACCTATTCCACC | 22 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR1510a | TGTTGTTTTTCCTATTCCACC | 21 | 463 | 877 | 902 | 28 | 2270 | |
| gma-miR1510b | TGTTGTTTTACCTATTCCACC | 21 | 0 | 0 | 4 | 0 | 4 | |
| pvu-isomiR1510b | TTGTTTTTCCTATTCCACCAA | 21 | 3313 | 17893 | 8413 | 222 | 29841 | |
| mtr-miR2199 | TGATACACTAGCACGGATCAC | 21 | 8 | 0 | 0 | 0 | 8 | |
| pvu-isomiR 2199a | TGATACACTAGCACGGGTCAC | 21 | 16 | 1323 | 799 | 59 | 2197 | |
| pvu-isomiR 2199b | TGATACACTAGTACGGATCAC | 21 | 2586 | 0 | 0 | 5 | 2591 | |
| mtr-miR2597 | TTTGGTACTTCGTCGATTTGA | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR2597 | TTTGGTACTTCCTTGATTTGA | 21 | 0 | 22 | 253 | 0 | 275 | |
| ppt-miR894 | CGTTTCACGTCGGGTTCACC | 20 | 1 | 3 | 1 | 0 | 5 | |
| pvu-isomiR894 | CGTTTCACGTCAGGTTCACCA | 21 | 15 | 6 | 0 | 0 | 21 | |
| pta-miR1310 | GGCATCGGGGGCGTAACGCCCCT | 23 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR1310 | GGCATCGGGGGCGCAACGCCC | 21 | 33 | 5 | 0 | 0 | 38 | |
| Conserved variants | ctr-miR166a | TCGGACCAGGCTTCATTCCCCC | 22 | 2 | 18 | 10 | 10 | 40 |
| pvu-isomiR166a | TCGGACCAGGCTTCCTTCCCC | 21 | 113 | 371 | 241 | 63 | 788 | |
| osa-miR156l | CGACAGAAGAGAGTGAGCATA | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR 156a | TGACAGAAGAGAGTGAGCA | 19 | 2682 | 7 | 334 | 222 | 3245 | |
| mtr-miR164d | TGGAGAAGCAGGGCACATGCT | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR164a | TGGAGAAGCAGGACACATGC | 20 | 58 | 64 | 535 | 12 | 669 | |
| vvi-miR156e | TGACAGAGGAGAGTGAGCAC | 20 | 30 | 0 | 1 | 1 | 32 | |
| pvu-isomiR 156b | TGACAGACGAGAGTGAGCAC | 20 | 261 | 0 | 2 | 2 | 265 | |
| csi-miR393 | ATCCAAAGGGATCGCATTGATC | 22 | 1 | 0 | 0 | 0 | 1 | |
| pvu-isomiR393 | TTCCAAAGGGATCGCATTGA | 20 | 911 | 581 | 667 | 4 | 2163 | |
| ctr-miR171 | TTGAGCCGCGTCAATATCTCC | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR171c | TTGAGCCGCGTCAATATCTCA | 21 | 30 | 151 | 129 | 58 | 368 | |
| mtr-miR169p | TGAGCCAGGATGGCTTGCCGG | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR169a | TGAGCCGGGATGGCTTGCCGG | 21 | 2 | 228 | 26 | 97 | 353 | |
| ptc-miR396f | TTCCACGGCTTTCTTGAACTG | 21 | 0 | 11 | 4 | 1 | 16 | |
| pvu-isomiR396a | TTCCACCGCTTTCTTGAACTG | 21 | 11 | 10 | 3 | 0 | 24 | |
| zma-miR398a | TGTGTTCTCAGGTCGCCCCCG | 21 | 2 | 0 | 2 | 0 | 4 | |
| pvu-isomiR398a | TGTGTTCTCAGGCCGCCCCTG | 21 | 21 | 0 | 18 | 0 | 39 | |
| sly-miR171c | TATTGGTGCGGTTCAATGAGA | 21 | 0 | 0 | 0 | 0 | 0 | |
| pvu-isomiR171d | TATTGGTCCGGTTCAATGAGA | 21 | 353 | 386 | 0 | 192 | 931 | |
Phaseolus vulgaris miRNA isoforms were classified as length variants, non-conserved miRNA variants, or conserved miRNA variants. Variants were named with the prefix pvu-isomiR followed by the miRNA family number of the reference miRNA. Underlined bases denote differences in length and/or composition of bases between variants and reference miRNAs.
Identification of stem-loop precursors
Plants have a more diverse population of sncRNAs than do animals mainly because of particular RNA polymerases. A criterion that supports miRNA annotation is the identification of a stem-loop precursor from which the duplex of miRNA/miRNA* is excised. A search for potential miRNA precursors involves secondary structure prediction analysis of genomic DNA or EST (Expressed Sequence Tag) sequences that match perfectly with determined miRNAs. Although plant miRNA stem-loops are more variable in length and structure than are animal pre-miRNAs, several characteristics are conserved among plant precursors. To identify common bean stem-loop precursors, miRNAs from Table 2, Table 3, Additional file 1, and Table 5 were aligned against all P. vulgaris ESTs and GSSs (Genomic Survey Sequences) from NCBI in search of perfect alignments. EST and GSS candidates were subjected to secondary structure prediction analysis using the mfold program with its default parameters [36]. Only the lowest energy structures were selected as described by Reinhart et al. (2002).
Secondary structure prediction analysis of P. vulgaris sequences resulted in the identification of eleven new stem-loop precursors belonging to eight conserved miRNA families (Figure 3 for an example and Additional file 2 for complete set). The precursor for the miR171 family was the precursor for the variant of ctr-miR171 (pvu-isomiR171c). The other miRNA families with a precursor identified were: miR166, miR167, miR156, miR157, miR398, miR408, and miR168. Taking into account previously reported P. vulgaris precursors, a total of 39 miRNAs can be predicted by the processing of common bean precursors, including pvu-isomiR171c (Additional file 2) [13]. The miRNAs ath-miR319c, osa-miR156k, gma-miR156f, ath-miR398a, and gma-miR482a-3p, discarded because they appeared in just one library or gave rise to fewer than 15 reads in all libraries, aligned perfectly with their respective precursors. Pvu-miR482*, aly-miR168a* and gma-miR166a-5p were also found in the precursor sequences. Therefore, common bean isoforms pvu-isomiR159a and pvu-isomiR482* were most likely derived also from these precursors.
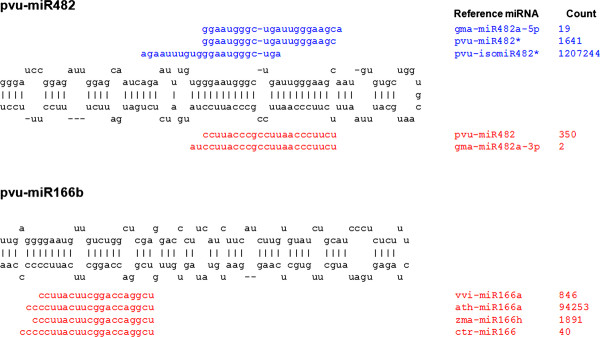
Figure 3.
Differential processing of P. vulgaris pre-miRNAs. Stem-loop precursors of pvu-miR482 (acc.: MI0010702) and novel pvu-miR166b (gi: 171606821) pre-miRNAs were aligned against possible derived mature (red) and mature-star (blue) miRNA sequences. Count data number represents the total number of reads found in all four libraries (right column).
Identification of novel microRNAs
From previous analyses, P. vulgaris miRNAs identical to those already present in miRBase or close homologs were identified. Numerous studies have described novel miRNAs in several plant species based on small RNA high-throughput sequencing results. However, this analysis is usually performed based on a reference genome that allows for mapping of a given small RNA to a genomic location and retrieval of adjoining sequence to help with secondary structure prediction of a miRNA precursor. In the present study, the sequencing reads remaining after removing known miRNAs were employed to scan the collection of ESTs and genomic sequences available from NCBI and PlantGDB in search of potential miRNAs using the miRDeep software [37]. This program searches for those small RNAs present in the sequencing reads that can be mapped to a given reference sequence. Selected sequences are searched for their potential RNA secondary structure to identify those that can be folded as hairpin structures typical of plant miRNA precursors. Additionally, the mapped small RNAs should be located in the stem region of the folded RNA, and a higher probability score is awarded to a given candidate if there are sequencing reads corresponding to the predicted miRNA* region.
The analysis for identification of novel miRNAs was carried out with 95030, 74759, 87956 and 65741 unique sequences from the leaf, root, seedling and flower small RNA libraries, respectively, and a total of 124894 ESTs and genomic sequences. A total of 29 candidates for new miRNA precursors were identified (Figure 4 for examples and Additional file 3 for complete set of candidate miRNAs). Four of the candidate precursors included reads for a mature miRNA and a putative miRNA* and the other 25 had only reads consistent with the predicted mature miRNA. The small number of ESTs and amount of genomic data available for P. vulgaris limits the number of candidates recovered by this analysis. The forthcoming availability of a complete genome should reveal a more comprehensive picture of the miRNA genes present in the common bean genome.
Figure 4.
Predicted stem-loop precursors of two novel miRNAs. Secondary structure prediction for candidate novel miRNAs pvu_6 and pvu_29 were carried out with mfold. Mature (red) and mature-star (blue) miRNAs correspond to predicted mature and mature star miRNA sequences from miRDeep.
Identification of miRNA targets in the genus Phaseolus
Plant miRNAs typically have higher sequence complementarity with their target mRNAs compared with miRNA-target interactions in animals [38]. MiRNA target prediction in plants is based on this high degree of complementarity between miRNAs and targets [39]. Many plant miRNA:mRNA duplexes exhibit paired nucleotides in the microRNA 5' region. However, some conserved plant miRNA-target interactions with unpaired nucleotides in the 'seed' region have been identified, for instance miR398a-CSD2 and miR396a-GRL7/GRL8/GRL9 in A. thaliana [39]. In this study, the RNAhybrid program as described by Alves et al. (2009) for prediction of Phaseolus miRNA targets was used (See methods) [40]. RNAhybrid predicts the energetically most favorable miRNA:mRNA hybrids according to user preferences [41]. All Phaseolus ESTs (NCBI) were evaluated as targets of miRNAs from Table 2, Table 3, and Table 5. Identification analysis for P. vulgaris miR390 non-coding transcript target TAS3 was done separately (See methods) [42]. Once miRNA:mRNA hybrids were obtained, ESTs were aligned against plant sequences of the UniProt Knowledgebase (UniProtKB) (release 2011_01) using BLASTX for annotation.
Target prediction analysis identified 194 ESTs annotated as established target gene families in plants (Additional file 4). Thirty seven ESTs were previously reported by Arenas-Huertero et al. (2009) and most are ESTs with small minimum free energies (MFEs) (data not shown). The conserved miRNA families for which conserved targets were found are: miR156/miR157, miR160, miR164, miR167, miR168, miR169, miR171, miR172, miR319, miR393, miR395, miR397, miR398 and miR408. The number of ESTs found for each gene family are: SBP(7), ARF(18), NAC(1), AGO1(1), NFY(14), SCL(2), AP2(8), TCP(6), TIR/F-box-AFB(5), ATP sulfurylases(14), Laccases(1), COX/SOD(66) and Plastocyanins(11). The miR156 and miR157 families share the same target EST sequences for Squamosa Binding Proteins (SBP). The MFEs for the conserved miRNA:mRNA hybrids range from -35.0 to -51.2 kcal/mol. Finally, among conserved miRNA targets, three ESTs homologous to AtTAS3, a target of miR390 family were identified (Additional file 4). In addition, 325 ESTs as putative targets for P. vulgaris conserved miRNAs, mature-star sequences and miRNA isoforms were identified (Additional file 5). Target prediction analysis for novel candidate miRNAs generated candidate targets for ten novel miRNAs represented by 177 ESTs (Additional file 6).
Organ-specific expression of P. vulgaris miRNA families
Specific miRNA expression patterns are proposed to be a consequence of tissue-specific, cell-specific and/or stress-specific regulatory elements in promoters of plant microRNA (MIRNA) genes [43-45]. The expression of some conserved MIRNA genes and the relative abundance patterns of mature miRNAs in different organs or in different developmental stages are essential for proper cell differentiation and organ developmental regulation. Modification of MIRNA gene expression leads to severe developmental defects. Understanding expression patterns of microRNAs in plant organs is necessary to discern miRNA-mediated regulatory pathways. The frequency of miRNAs detected by high throughout sequencing methods serves for relative miRNA expression estimation [46]. Moldovan et al. (2009), comparing Arabidopsis root miRNA frequencies against leaf and whole plant reads represented in the Arabidopsis Small RNA Project database (ASRP), found that most miRNA families have organ-specific expression [47,48].
To explore organ-specific miRNA expression in Phaseolus vulgaris, the open-source R/Bioconductor software package DESeq was employed [49,50]. The DESeq package, supported by a model based on negative binomial distribution, estimates variance-mean dependence in count data from high-throughput sequencing assays and tests for differential expression. The miRanalyzer tool bases its differential expression module on the DESeq package and has been used to test for miRNA differential expression [51,52]. To explore organ-specific expression for miRNA families as a whole, frequencies for miRNAs of the same family were pooled together for each library and employed as input data. The five miRNAs that aligned perfectly with precursor sequences and were discarded because they appeared in just one library or totalled fewer than 15 reads in all libraries, as well as those newly identified from the isoform analysis, were also considered. Mature-star miRNAs were discarded. Count data was transformed with the DESeq package and hierarchical cluster analysis (HCA) was performed (Figure 5, see methods).
Figure 5.
Hierarchical cluster analysis (HCA) of P. vulgaris miRNA families. Frequencies of conserved miRNA families were used to analyze relative miRNA family expression in leaf, seedling, root and flower. Hierarchical cluster analysis was performed on thirty-three conserved miRNA families (right). The color scale denotes abundant (green) and non-abundant (red) miRNA families according to transformed count data. Density plots are included in blue lines.
MiRNA families were clustered in two major groups: the group with fewer miRNAs consists of miRNA families that were abundantly expressed in all four libraries, and the other group contains miRNA families with differential expression patterns among organs. The accumulation patterns of miRNA families in leaf and root organs were more similar to each other than to the other two samples. The most abundant families of the analysis, especially in seedlings, were miR159, miR319, miR396, miR166, miR408, and miR482. The miR159 family has previously been reported as an abundant and widespread family in all plant organs. It is important to mention that the expression of miR319 was abundant in all organs except for leaf (Figure 5). Similar to miR319, in this study miR396 was less abundant in leaf than in the other three organs. Other quite abundant miRNA families were: miR1510, miR164, miR167, miR169, miR157 and miR156. Interestingly miR1510, a miRNA family detected only in legumes, was found in all four libraries.
In the group of families with a particular expression pattern, miR2119, miR2111, and miR479 were highly accumulated specifically in one organ. MiR2119 was detected especially in roots. This expression pattern correlates with previous northern blot analysis of roots, leaves, immature embryos and dry embryos from P. vulgaris [13]. Expression of miR2111 was observed mainly in roots and miR399 was only slightly expressed in leaves. Both miRNAs are induced under phosphorus (P) limiting conditions [12,53]. The miR479 family was preferentially expressed in seedlings. On the other hand, there were some interesting families that were poorly expressed in one particular organ. For example, miR168 was poorly expressed in seedlings, miR1515 in leaves, miR2199 in roots, miR394 in leaves, miR4375 in roots, and miR397 in flowers. Finally, HCA showed that miR395 was poorly expressed in all four small RNA libraries, consistent with its induction upon low sulphate conditions [54].
Discussion
Identification of microRNAs in species with or without fully sequenced genomes has been revolutionized by high-throughput sequencing methods. High-throughput sequencing miRNA analysis in Phaseolus vulgaris will facilitate more particular and specific bean miRNA studies as well as legume sncRNAs research in this rapidly growing field. Although deep-sequencing experiments have become the major source supporting microRNA annotations, technical variations, transcript length bias and mapping bias have been reported with this approach for transcriptome analysis of coding RNAs (mRNAs) [17,55,56]. Studies to elucidate the number of miRNA molecules sequenced from a small RNA library are still needed for more accurate small RNA profiling studies. In terms of reads, the small RNA libraries sequenced here yielded a larger number of total raw reads to work with than did many other studies in which plant miRNAs have been identified; however, the total number of reads of identified miRNAs, including non-abundant and miRNA isoforms, constitutes only 0.047% of total raw reads. Further studies are needed to understand to what extent the 24-nt class representing siRNA populations, usually the most abundant and diverse class of sncRNAs sequenced in small RNA libraries, masks miRNA populations. Also, coverage analyses with fully sequenced genomes are needed to elucidate sequenced sample proportions of small non-coding RNAs such as tRNA, rRNA, snoRNA or snRNA.
Most miRNA families identified in this study in P. vulgaris are evolutionarily conserved in Fabaceae, particularly in the best-studied plants M. truncatula and G. max. MiRNA family miR157 has not been reported in these two legumes, probably because of the similarity of its sequence to that of miR156. Other miRNA families not reported in miRBase in G. max and M. truncatula are miR397 and miR408. The only Fabaceae where miR408 has been previously found was Arachis hypogaea. One miRNA of the bean miR397 family with the same sequence as ath-miR397 was detected herein with 7416 reads in total, so it is likely to be present in other legumes as well. MicroRNA miR2199 has been reported in miRBase only for M. truncatula and the variant found here for this miRNA in all four libraries of P. vulgaris was previously reported as miRS1 by Arenas-Huertero et al. (2009) because mtr-miR2199 was not yet reported. As well as in Lotus japonicus, the A17- > G (miRS1) variant was found in peanuts (Arachis hypogaea L.) [35,57].
It is interesting that miRNA families such as miR1511, miR1514 and miR1515 first detected in soybean, have not been found in other legumes besides P. vulgaris. In this regard, other miRNA families reported specifically in soybean (gma-miR1524, gma-miR1532, gma-miR1526, gma-miR1516 and gma-miR1513 and gma-miR1508) were detected by Valdés-López et al. (2010) using a hybridization approach (macroarrays) under several abiotic stress conditions in leaves, roots and nodules of P. vulgaris. These miRNAs from soybean were not detected by high-throughput sequencing, with the exception of two reads for gma-miR1508. In the Valdés-López et al. (2010) analysis, expression was detected for miRNAs miR1524, miR1526, miR1532 and miR1508 in common bean plants grown under sufficient nutrient conditions or stressed conditions. Family miRNAs gma-miR1513 and gma-miR1516 were detected only under stressed conditions. In addition, expression was detected in the macroarrays for an oligonucleotide probe designated as pvu-miR1509. This probe was based on a variant detected by Arenas-Huertero et al. (2009) that was not detected here because it has more than two mismatches with the reference miRNA gma-miR1509. The miR170 family, for which Valdés-López et al. (2010) detected expression in P. vulgaris, was neither found in this study. MiR170 is very similar to miR171, so it is possible that the macroarray probes hybridized with the same miRNA family.
MiRNA variants are considered to be a consequence of inaccuracies in Dicer pre-miRNA processing. Smaller variants with missing bases and low frequencies are viewed as degradation products or pyrophosphate sequencing errors. Herein, small RNA sequences from libraries were classified as miRNA isoforms only if they were similar to a reference miRNA reported in miRBase and had a significantly greater number of reads compared to those found for the reference miRNA in all four organs. From this isoform identification analysis, four more miRNA families (miR1510, miR2199, miR4376 and miR479) were added to the total number of conserved miRNA families identified in common bean. All of the variants found here for these four families were highly abundant in all four libraries. It is probable that miR1510 and miR2199 miRNA families are part of the P. vulgaris miRNA population, based on the two variants identified for each of these two families, along with their conservation in other species (see results) and previously experimental expression analyses [13,22].
Another family highly likely to be present in common bean is the miR4376 family. The variant found here for miR4376 was very abundant in common bean and has only one nucleotide missing relative to its reference miRNA found in soybean. On the other hand, further experimental and genomic sequences analyses are still needed to validate the variant identified in common bean for the miR479 family, which has two nucleotides missing and one mismatch compared with its reference miRNA csi-miRNA479. That is also the case for other less conserved miRNA families identified based on the isoform analysis, like miR858, miR2597, miR894 and miR1310. Most of the reference miRNAs for these less conserved miRNA family variants were poorly or not detected in the libraries.
In plant and animal microRNAs, 3' heterogeneity is the most frequent length variation. Most of the variants identified from the length variant group exhibit 3' heterogeneity. Sequence length heterogeneity for plant microRNAs has been proven to be essential for correct plant development and environmental responses. It is known that miR168 in Arabidopsis thaliana is produced in length variants of 21 and 22 nucleotides [31]. A decrease in abundance of the 21 nt variant reduces miR168 homeostasis and leads to developmental defects. The large number of reads detected for some variants in P. vulgaris suggests significant regulatory roles like detected for miR168 in A. thaliana. Using the pre-miRNA of pvu-miR482 (Figure 4) as an example, three variants generated for "mature-star" miRNAs (5p) and two variants generated for the mature miRNAs (3p) were observed. Considering the number of reads found for these variants, mature-star sequence variants were by far greater in abundance than their corresponding mature sequence variants. In particularl two mature-star sequence length variants, one previously reported in miRBase as pvu-miR482* of 22 nucleotides and the other here denoted as pvu-isomiR482* of 21 nucleotides, had 1641 and 1207244 total reads respectively. It is important to take into account that sequences for reference miRNA gso-miR482a and its highly abundant variant pvu-isomiR482a were detected, and that these variants necessarily have to be excised from another stem-loop precursor not yet identified. For this reason, it is possible that the variants pvu-miR482* and pvu-isomiR482* were generated from different loci, as actually happens for the two miR168 variants in A. thaliana. In addition to miRNAs previously reported in other plants and those present in closely related species such as soybean or Medicago, common bean may encode species-specific miRNAs. To address this question, sequencing reads were explored using the miRDeep algorithm [37]. Those candidates were favoured where in addition to a mature miRNA present in the stem region of the hairpin precursor, there was also evidence of a miRNA* sequence recovered. Next, candidate miRNAs having a plausible stem-loop precursor but without any miRNA* sequences in the libraries were obtained. It will be interesting to see to what extent these candidates can be confirmed as genuine miRNAs and whether they are involved in biological processes specific to P. vulgaris. As in other examples shown here, the availability of a fully-sequenced genome will provide a more complete picture of novel miRNAs.
Currently, microarray hybridization approaches, real time quantitative PCR (RT-qPCR) analyses and high-throughput sequencing technologies are widely used microRNA profiling methods. MiRNA regulatory functions in different organs, different stress conditions, and different developmental stages are still unknown. MiRNA profiling studies are important first approximations to analyze miRNA functions according to their different expression patterns. In this work miRNA expression was analyzed in order to find important differences in miRNA family expression levels within common bean organs. Biological replicates are essential to determine if differences observed are caused by conditions and not just by experimental variations. Because of this, cluster analysis was employed. The DESeq package allows users to work without replicates with the caveat that the test will lose strength. It assumes that most transcripts will have approximately the same levels within replicates under the different conditions, and that the estimated variance should not change too much.
Experiments designed to explore miRNA and mRNA expression are subjected to many different technical and biological biases. Nevertheless, differential processing of stem-loop precursors, small RNA duplexes, and, in general, miRNA biochemical properties challenge expression analysis methods usually used to analyze mRNAs. The majority of conserved miRNA families identified were expressed in all organs at different levels. Conserved miRNA families seem to be transcribed in almost all plant organs. Which of those conserved miRNAs have an essential role in legumes for certain developmental stages or stress responses is not completely understood. Perhaps less conserved miRNAs families that are highly abundant in legumes such as miR1510, miR1511, miR1514, miR1515, miR2118 and miR2199, play essential roles in characteristic processes of legumes such as nodulation, as altered expression of miR482 and miR1515 has been proven to increase soybean nodulation [58].
The present study contributes, together with previous common bean miRNA studies, to characterizing the P. vulgaris miRNA population. It represents a unique analysis of P. vulgaris miRNAs performed with high-throughput next-generation DNA sequencing (NGS) technologies. Shortly, full genome sequence and transcriptome datasets from different P. vulgaris cultivars will be available. The analysis of novel common bean genome sequence information will benefit from the tools presented here to expand important small RNA research needed in this critical worldwide crop.
Conclusions
109 miRNAs belonging to 29 conserved families in Phaseolus vulgaris were identified using high-throughput sequencing. In addition, twenty six miRNA isoforms were characterized. MiRNA variant analysis identified four highly abundant miRNA families. Eleven new stem-loop precursors belonging to eight conserved miRNA families were determined. Thirty nine miRNAs identified can be explained by processing of characterized common bean precursors. In addition, twenty nine novel miRNA candidates were predicted based on small RNA reads and precursor prediction. Evidence for miRNA* sequences for four of these precursors was found. Target prediction analysis identified 157 new Phaseolus ESTs as established miRNA target genes in plants. Candidate targets for miRNA families derived from Phaseolus ESTs were proposed. The common bean miRNA families identified were differentially expressed in leaves, roots, seedlings and flowers. This work provides an important global view of conserved and novel Phaseolus vulgaris miRNAs, their precursors and their targets.
Methods
Plant material and growth conditions
Phaseolus vulgaris L. cv. Negro Jamapa and cv. Pinto Villa were used in this study. Common bean (cv. Negro Jamapa) seeds were surface sterilized by an initial treatment with 100% ethanol for 1 min, rinsed with sterile water, and treated with 20% sodium hypochlorite for 5 min. Sterilized seeds were transferred to sterile trays containing wet paper towels. Trays were covered with foil and held at 28°C for 72 hours until seed germination. Sprouts then were transferred to small plastic pots containing vermiculite watered with B&D solution supplemented with 8 mM KNO3 [59]. Incubation was performed in a chamber with 16 h of light and 8 h of dark at 28°C. Plants were watered every third day until organ collection. Roots (15 d old) and flower buds (35-40 d old) were collected in liquid N2 and stored at - 80°C.
P. vulgaris cv. Pinto Villa seeds were surface-sterilized in 50% (v/v) sodium hypochlorite and 0.5% (v/v) Tween-20 for 3 min, and rinsed with distilled water for 10 min. Seeds were transferred to trays containing wet paper towels. Whole seedlings 1-4 days old were collected in liquid N2 and stored at -80°C. For leaf collection, 4-day-old seedlings were transferred from trays to plastic pots containing 40% vermiculite, 30% Metro-Mix soil and 30% Agrolite. Once the first trifolium appeared, plants were kept well-watered. A pool of leaves from 10 and 20 days old was harvested for RNA purification.
RNA isolation and small RNA library sequencing
Total RNA was isolated from frozen roots, seedlings, flower buds and leaves using the Trizol reagent according to manufacturer's instructions (Invitrogen, Carlsbad, CA). The RNA Integrity Number (RIN) was larger than seven and the 28S/18S ratio was larger than 1.6 for all sample organs. Ten micrograms of each sample (roots, flower buds, seedlings, and leaves) were prepared for Deep Sequencing following Illumina's Small RNA alternative sample preparation protocol v1.5. Small RNA fragments ranging from 18-30 nt were selected for the construction of the small RNA libraries. Complementary DNA libraries were separately Single Read-sequenced using the Genome Analyzer IIx (GAIIx)(36 bp) and the Illumina Cluster Station (Illumina Inc, USA) at the Instituto de Biotecnología (Universidad Nacional Autónoma de México).
Small RNA sequencing analysis
Raw reads from the Illumina Pipeline 1.4 for the four small RNA libraries were cleaned of sequence adapters, low quality tags and small sequences (< 16 nt long). Quality analysis per cycle was performed for each library. Later, sequences were converted to FASTA format grouped in unique sequence tags with their respective frequencies. The R/Bioconductor software package ShortRead (version 1.10.4) was used for RNA sequence length distribution analysis [60]. Mature miRNA sequences from miRBase (release 16) were removed from the RNA families database (Rfam 10.0). Libraries were aligned against miRNA cleaned Rfam database using BLASTN. Exact matches identified with a Perl script for parsing blast results were then removed from libraries.
Identification of conserved, isoform, and novel miRNAs
Mature and mature-star miRNA sequences from plants were extracted from miRBase (release 16). Sequences were grouped into unique sequences with a reference miRNA identifier. Small RNA libraries cleaned with the Rfam database were aligned against the unique mature and mature-star miRNA sequences using BLASTN and SSAHA2 (version 2.5.3). Alignment results were processed to obtain small RNA sequences that corresponded exactly in size and nucleotide composition to reported plant miRNA sequences. MiRNAs that were detected in just one library or that totalled fewer than 15 absolute appearances in all libraries were separated. Correlation coefficient analysis was done with the number of loci reported in miRBase (v.16) for Medicago truncatula and Glycine max, and the number of possible loci for P. vulgaris based on mature miRNA differences. With the purpose of identifying miRNA isoforms, sequences left from Rfam and miRBase filters that presented a total frequency higher than 10 in all libraries were BLASTN aligned against miRBase (mature and mature-star plant miRNA sequences) allowing at most two mismatches and/or two different nucleotides in length. The total number of variants found for each library was subjected to an abundance filter which consisted of choosing variants that had a total number of reads 50% greater than the total reads of their reference miRNA previously reported. If no reference miRNA for a variant was previously detected in all libraries, the variant with the highest frequency was considered. Isoforms detected for conserved miRNAs that belong to a miRNA family were analyzed to discard the possibility of being simply another miRNA of the same family. MiRNA isoforms were classified as length variants, non-conserved miRNA variants or conserved miRNA variants. Identification of novel miRNAs was performed with sequencing reads that remained after known miRNAs and miRNA isoforms were removed. EST and genomic sequences available from NCBI and PlantGDB were used to search for potential miRNAs using the miRDeep software [37]. Novel miRNA candidates were further aligned against nucleotide and protein NCBI databases to discard possible RNA degradation products.
Identification of stem-loop precursors and targets
To identify stem-loop precursors, P. vulgaris ESTs and GSSs http://www.ncbi.nlm.nih.gov/projects/dbEST/; http://www.ncbi.nlm.nih.gov/projects/dbGSS/from NCBI were aligned against mature miRNAs, mature-star sequences and miRNA isoforms (Table 2, Table 3, Table 5 and Additional file 1) [61]. Sequence candidates with 100% coverage and identity were tested with the secondary structure program mfold for pre-miRNA structures [36]. Only the lowest energy structures generated for each sequence candidate were analyzed. Target prediction analysis was performed for all ESTs of the genus Phaseolus from NCBI. EST sequences of the genus Phaseolus were confirmed to be targets of mature miRNAs, mature-star sequences and miRNA isoforms (Table 2, Table 3, and Table 5) using the RNAhybrid program (version 2.1) as described by Alves et al. (2009). Potential EST targets were then aligned against plant sequences of the UniProt Knowledgebase (UniProtKB) (release 2011_01) using BLASTX for annotation. Identification analysis for common bean miR390 non-coding transcript target TAS3 was done separately. The AtTAS3 (locus: AT3G17185.1) sequence from the Arabidopsis Information Resource (TAIR) was used for BLASTN alignment against Phaseolus ESTs. Later, miR390 conserved binding sites were analyzed on EST candidates. Only novel mature and mature-star miRNA sequences detected in the libraries were used for target prediction analysis.
Organ-specific expression analysis
As first step for organ-specific expression analysis, frequencies of miRNAs of the same miRNA family were added. MiRNAs considered for this analysis were the conserved miRNAs (Table 2), the five miRNAs from Additional file 1 that aligned with common bean stem-loop precursors, and the new four miRNA families proposed from the miRNA isoform study. Mature-star sequences were not considered. To test for miRNA differential expression the open-source R/Bioconductor software package DESeq was used [49]. The effective library size for all small RNA library data was estimated with the estimateSizefactors function. This function is used to normalize frequencies of transcripts in relation to library sizes. Then, estimateVarianceFunctions was used to predict the variance from the mean for each organ sample. Within these functions, the method = "blind" parameter for comparisons without replicates was employed. Eventually the count data was transformed with the getVarianceStabilizedData function such that its variance becomes independent of the mean producing a homoscedastic version of the data. Heatmap was performed using the tool heatmap.2 from the gplopts package of open source R software. Hierarchical cluster analysis was performed with the hclust function from the stats package.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JL and PP conceived of and coordinated the study. PP wrote the manuscript. PP, MS and LP carried out the bioinformatic analyses. GE and JL carried out plant growth, RNA extraction and preparation, and helped drafting the manuscript. JL, AA and FE participated in its design, helped to draft the manuscript and provided intellectual suggestions. All authors read and approved the final manuscript.
Supplementary Material
Conserved miRNAs detected only in one library and/or with fewer than 15 reads. MiRNAs in common bean identified in leaves (LL), roots (RL), seedlings (SL) and flowers (FL) detected in just one library and/or with fewer than 15 total reads.
Stem-loop miRNA precursors in common bean. EST fragments (new conserved pre-miRNAs) and fragments of stem-loop sequences reported in miRBase (v. 16) for P. vulgaris where aligned against miRNA sequences (blue sequences). Count data number represents the total number of reads in all four libraries. Count data number represents the total number of reads in all four libraries.
Novel miRNAs detected in Phaseolus vulgaris. Novel miRNAs in common bean identified in leaves (LL), roots (RL), seedlings (SL) and flowers (FL) using miRDeep.
Predicted conserved targets in Phaseolus. Predicted conserved miRNA targets based on ESTs of the genus Phaseolus. GenBank accession numbers are used for EST identification. Calculated MFEs (kcal/mol) using RNAhybrid are shown. MiRNA families 156 and 157 share the same predicted conserved targets. MicroRNA sequences in 3'-5' sense were used to represent miRNA:target pairing. Crosses (x) and asterisks (*) denote one-nucleotide wobbles and mismatches, respectively. G:U base pairing is not considered a mismatch (|)
Predicted Putative targets in Phaseolus. Predicted putative miRNA targets based on ESTs of the genus Phaseolus. GenBank accession numbers are used for EST identification. Calculated MFEs (kcal/mol) using RNAhybrid are shown. MicroRNA sequences in 3'-5' sense were used to represent miRNA:target pairing. Crosses (x) and asterisks (*) denote one-nucleotide wobbles and mismatches, respectively. G:U base pairing is not considered a mismatch (|)
Putative targets for novel miRNAs identified in Phaseolus. Predicted putative targets for novel miRNAs in common bean based on EST of the genus Phaseolus. MicroRNA sequences in 3'-5' sense were used to represent miRNA:target pairing. Crosses (x) and asterisks (*) denote one-nucleotide wobbles and mismatches, respectively. G:U base pairing is not considered a mismatch (|)
Contributor Information
Pablo Peláez, Email: ppelaez@ibt.unam.mx.
Minerva S Trejo, Email: susami@ibt.unam.mx.
Luis P Iñiguez, Email: liniguez@lcg.unam.mx.
Georgina Estrada-Navarrete, Email: geo@ibt.unam.mx.
Alejandra A Covarrubias, Email: crobles@ibt.unam.mx.
José L Reyes, Email: jlreyes@ibt.unam.mx.
Federico Sanchez, Email: federico@ibt.unam.mx.
Acknowledgements
We thank the Unidad Universitaria de Secuenciación Masiva de DNA (UUSMD) of the Universidad Nacional Autónoma de México (UNAM) for sequencing services. We also are grateful to the Instituto de Biotecnología-UNAM for giving us access to its computer cluster. Financial support for this research was provided by the International Atomic Energy Agency (research contract No. 16601/R0), Consejo Nacional de Ciencia y Tecnología 83324 and Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica IN214909-3 grants.
References
- Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet. 2006;38(Suppl):S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Bologna NG, Mateos JL, Bresso EG, Palatnik JF. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009;28(23):3646–3656. doi: 10.1038/emboj.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol. 2005;15(16):1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci. 2005;102(10):3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci. 2005;102(33):11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008;31(12):1834–1843. doi: 10.1111/j.1365-3040.2008.01883.x. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero C, Pérez B, Rabanal F, Blanco-Melo D, De la Rosa C, Estrada-Navarrete G, Sanchez F, Covarrubias AA, Reyes JL. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol. 2009;70(4):385–401. doi: 10.1007/s11103-009-9480-3. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics. 2008;9:160. doi: 10.1186/1471-2164-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18(10):1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011. pp. D152–7. [DOI] [PMC free article] [PubMed]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11(10):1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011;23(2):431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi T, Yan Z, Libault M, Jeong DH, Park S, Green PJ, Sherrier DJ, Farmer A, May G, Meyers BC, Xu D, Stacey G. Prediction of novel miRNAs and associated target genes in Glycine max. BMC Bioinformatics. 2010;11(Suppl 1):S14. doi: 10.1186/1471-2105-11-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Yang SS, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernández G. MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 2010;187(3):805–818. doi: 10.1111/j.1469-8137.2010.03320.x. [DOI] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. 2008;9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011;11:5. doi: 10.1186/1471-2229-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mica E, Piccolo V, Delledonne M, Ferrarini A, Pezzotti M, Casati C, Del Fabbro C, Valle G, Policriti A, Morgante M, Pesole G, Pè ME, Horner DS. Correction: High throughput approaches reveal splicing of primary microRNA transcripts and tissue specific expression of mature microRNAs in Vitis vinifer. BMC Genomics. 2010;11:109. doi: 10.1186/1471-2164-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Zhao YT, Wang XJ, Croft L, Wang ZH, Haerizadeh F, Mattick JS, Singh MB, Carroll BJ, Bhalla PL. MicroRNAs in the shoot apical meristem of soybean. J Exp Bot. 2011;62(8):2495–2506. doi: 10.1093/jxb/erq437. [DOI] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3' UTR evolution. Nat Struct Mol Biol. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers EA, Branscheid A, May P, Krajinski F. Stars and Symbiosis: MicroRNA- and MicroRNA*-Mediated Transcript Cleavage Involved in Arbuscular Mycorrhizal Symbiosis. Plant Physiol. 2011;156(4):1990–2010. doi: 10.1104/pp.111.172627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Fedynak A, Fahlman RP. Naturally occurring variations in sequence length creates microRNA isoforms that differ in argonaute effector complex specificity. Silence. 2010;1(1):12. doi: 10.1186/1758-907X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell. 2008;133(1):116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS One. 2009;4(7):e6442. doi: 10.1371/journal.pone.0006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics. 2010;11:246. doi: 10.1186/1471-2164-11-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M, Woodward AW, Agarwal V, Guan X, Ha M, Ramachandran V, Chen X, Triplett BA, Stelly DM, Chen ZJ. Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.) Genome Biol. 2009;10:R122. doi: 10.1186/gb-2009-10-11-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G, Zheng Y, Li YF, Shukla LI, Matts J, Hoyt P, Macmil SL, Wiley GB, Roe BA, Zhang W, Sunkar R. Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 2009;184(1):85–98. doi: 10.1111/j.1469-8137.2009.02915.x. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5(9):R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Alves L Jr, Niemeier S, Hauenschild A, Rehmsmeier M, Merkle T. Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res. 2009;37(12):4010–4021. doi: 10.1093/nar/gkp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006. pp. W451–W454. [DOI] [PMC free article] [PubMed]
- Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG. MicroRNA promoter element discovery in Arabidopsis. RNA. 2006;12(9):1612–1619. doi: 10.1261/rna.130506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18(18):2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válóczi A, Várallyay E, Kauppinen S, Burgyán J, Havelda Z. Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 2006;47(1):140–151. doi: 10.1111/j.1365-313X.2006.02766.x. [DOI] [PubMed] [Google Scholar]
- 't Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36(21):e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: the Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005. pp. D637–D640. [DOI] [PMC free article] [PubMed]
- Moldovan D, Spriggs A, Yang J, Pogson BJ, Dennis ES, Wilson IW. Hypoxia-responsive microRNAs and trans-acting small interfering RNAs in Arabidopsis. J Exp Bot. 2010;61(1):165–177. doi: 10.1093/jxb/erp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Rodríguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucleic Acids Res. 2011. pp. W132–W138. [DOI] [PMC free article] [PubMed]
- Dhahbi JM, Atamna H, Boffelli D, Magis W, Spindler SR, Martin DI. Deep sequencing reveals novel microRNAs and regulation of microRNA expression during cell senescence. PLoS One. 2011;6(5):e20509. doi: 10.1371/journal.pone.0020509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009;150(3):1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;52(2):313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- Frith MC, Wan R, Horton P. Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res. 2010;38(7):e100. doi: 10.1093/nar/gkq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CZ, Xia H, Frazier TP, Yao YY, Bi YP, Li AQ, Li MJ, Li CS, Zhang BH, Wang XJ. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.) BMC Plant Biol. 2010;10:3. doi: 10.1186/1471-2229-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng Y, Wu T, Subramanian S, Yu O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 2010;153(4):1759–1770. doi: 10.1104/pp.110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. Control of leghaemoglobin synthesis in snake beans. Biochem J. 1971;125(4):1075–1080. doi: 10.1042/bj1251075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25(19):2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The National Center for Biotecnological information. [ http://www.ncbi.nlm.nih.gov/projects/dbEST][ http://www.ncbi.nlm.nih.gov/projects/dbGSS]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conserved miRNAs detected only in one library and/or with fewer than 15 reads. MiRNAs in common bean identified in leaves (LL), roots (RL), seedlings (SL) and flowers (FL) detected in just one library and/or with fewer than 15 total reads.
Stem-loop miRNA precursors in common bean. EST fragments (new conserved pre-miRNAs) and fragments of stem-loop sequences reported in miRBase (v. 16) for P. vulgaris where aligned against miRNA sequences (blue sequences). Count data number represents the total number of reads in all four libraries. Count data number represents the total number of reads in all four libraries.
Novel miRNAs detected in Phaseolus vulgaris. Novel miRNAs in common bean identified in leaves (LL), roots (RL), seedlings (SL) and flowers (FL) using miRDeep.
Predicted conserved targets in Phaseolus. Predicted conserved miRNA targets based on ESTs of the genus Phaseolus. GenBank accession numbers are used for EST identification. Calculated MFEs (kcal/mol) using RNAhybrid are shown. MiRNA families 156 and 157 share the same predicted conserved targets. MicroRNA sequences in 3'-5' sense were used to represent miRNA:target pairing. Crosses (x) and asterisks (*) denote one-nucleotide wobbles and mismatches, respectively. G:U base pairing is not considered a mismatch (|)
Predicted Putative targets in Phaseolus. Predicted putative miRNA targets based on ESTs of the genus Phaseolus. GenBank accession numbers are used for EST identification. Calculated MFEs (kcal/mol) using RNAhybrid are shown. MicroRNA sequences in 3'-5' sense were used to represent miRNA:target pairing. Crosses (x) and asterisks (*) denote one-nucleotide wobbles and mismatches, respectively. G:U base pairing is not considered a mismatch (|)
Putative targets for novel miRNAs identified in Phaseolus. Predicted putative targets for novel miRNAs in common bean based on EST of the genus Phaseolus. MicroRNA sequences in 3'-5' sense were used to represent miRNA:target pairing. Crosses (x) and asterisks (*) denote one-nucleotide wobbles and mismatches, respectively. G:U base pairing is not considered a mismatch (|)