Abstract
Background
Flower bilateral symmetry (zygomorphy) has evolved multiple times independently across angiosperms and is correlated with increased pollinator specialization and speciation rates. Functional and expression analyses in distantly related core eudicots and monocots implicate independent recruitment of class II TCP genes in the evolution of flower bilateral symmetry. Furthermore, available evidence suggests that monocot flower bilateral symmetry might also have evolved through changes in B-class homeotic MADS-box gene function.
Methods
In order to test the non-exclusive hypotheses that changes in TCP and B-class gene developmental function underlie flower symmetry evolution in the monocot family Commelinaceae, we compared expression patterns of teosinte branched1 (TB1)-like, DEFICIENS (DEF)-like, and GLOBOSA (GLO)-like genes in morphologically distinct bilaterally symmetrical flowers of Commelina communis and Commelina dianthifolia, and radially symmetrical flowers of Tradescantia pallida.
Results
Expression data demonstrate that TB1-like genes are asymmetrically expressed in tepals of bilaterally symmetrical Commelina, but not radially symmetrical Tradescantia, flowers. Furthermore, DEF-like genes are expressed in showy inner tepals, staminodes and stamens of all three species, but not in the distinct outer tepal-like ventral inner tepals of C. communis.
Conclusions
Together with other studies, these data suggest parallel recruitment of TB1-like genes in the independent evolution of flower bilateral symmetry at early stages of Commelina flower development, and the later stage homeotic transformation of C. communis inner tepals into outer tepals through the loss of DEF-like gene expression.
Keywords: B class genes, Commelinaceae, CYCLOIDEA, homeotic change, monocots, tepals, teosinte branched1
Background
Evolutionary transitions between flower radial symmetry (polysymmetry and actinomorphy) and bilateral symmetry (monosymmetry and zygomorphy) have occurred multiple times independently across angiosperms, and are associated with increased pollinator specialization and speciation rates [1-6]. Indeed, some of the largest angiosperm families have species with predominantly bilaterally symmetrical flowers, including the legumes (Leguminosae, rosids, core eudicots), daisies (Asteraceae, asterids, core eudicots) and orchids (Orchidaceae, monocots) [7]. Recent functional studies in distantly related core eudicots - including Antirrhinum majus (Plantaginaceae), Iberis amara (Brassicaceae), Pisum sativum and Lotus japonicus (Leguminosae) and Gerbera hybrida (Asteraceae) - have demonstrated a role for class II TEOSINTE BRANCHED1 (TB1)/CYCLOIDEA (CYC)/PROLIFERATING CELL NUCLEAR ANTIGEN GENE-CONTROLLING ELEMENT BINDING FACTOR (PCF) (TCP) transcription factors in establishing flower symmetry by specifying identity to the dorsal (adaxial) region of the flower [8-13]. Since bilateral symmetry has evolved independently in these lineages, genetic data suggest parallel recruitment of class II TCP genes in the evolution of a convergent trait [11,14-16]. Whether homologous TCP genes have been similarly utilized in monocot flower bilateral symmetry remains largely untested [but see [17,18]].
The genetic basis of flower bilateral symmetry is best understood in the model species A. majus, and involves the action of four asymmetrically expressed transcription factors [8,9,19-21]. Dorsal identity is specified by the class II TCP genes CYCLOIDEA (CYC) and DICHOTOMA (DICH), and the MYB gene RADIALIS (RAD), whereas ventral (abaxial) identity is conferred by the MYB gene DICHOTOMA (DICH). CYC and DICH are derived from a recent duplication event at the base of the Antirrhineae tribe [22], and have distinct but overlapping functions, as inferred from their mutant phenotypes [8,9]. Whereas wild type A. majus plants have five petals, four stamens and a dorsal staminode, cyc single mutants often have extra dorsal petals that are reduced in size, and a fully developed dorsal stamen. By contrast, dich single mutants only lack the internal asymmetry of wild type dorsal petals. Together with the fully ventralized, and, hence, radially symmetrical flowers of cyc:dich double mutants, these data demonstrate a role for CYC in dorsal stamen abortion, petal growth and organ number determination, and a role for DICH in shaping dorsal petal growth [8,9].
A similar range of dorsal identity functions has been assigned to CYC-like genes of other core eudicots, including Linaria vulgaris, P. sativum, I. amara, Lotus japonicus, and G. hybrida [10-13,20,23]. Furthermore, although monocots do not have a strict CYC gene ortholog due to two separate duplication events at the base of core eudicots [24], the observation that the class II TCP gene RETARDED PALEA1 (REP1) in Oryza sativa (rice, Poaceae) is expressed only in the dorsally positioned palea, suggests further recruitment of TCP genes in the evolution of monocot flower bilateral symmetry [17]. Indeed, it was recently found that bilaterally symmetrical flowers of Costus spicatus (Costaceae; Zingiberales) and Heliconia stricta (Heliconiaceae) have asymmetric TCP gene (CsTB1a and HsTBL2b, respectively) expression at early to late stages of flower development [18]. However, in contrast to the core eudicots, CsTB1a and HsTBL2b expression is restricted to the ventral, rather than the dorsolateral, side of the flower [18].
In addition to a hypothesized role for TCP genes, the floral homeotic B-class genes DEFICIENS (DEF) and GLOBOSA (GLO) have been implicated in the multiple independent derivations of flower bilateral symmetry in monocots [25-29]. Evidence supporting this comes largely from the observation that DEF-like gene paralogs are differentially expressed in the morphologically distinct outer tepals, dorsolateral inner tepals and ventral inner tepal (lip or labellum) of Phalaenopsis equestris (Orchidaceae) [25,26]. In both core eudicots and monocots, DEF- and GLO-like proteins function as obligate heterodimers to confer identity to the second and third whorls, although there are exceptions [30,31]. In the second whorl of Arabidopsis thaliana (Brassicaceae) and A. majus APETALA3 (AP3)/DEF and PISTILLATA (PI)/GLO form tetramers with SEPALLATA (SEP) and APETALA1 (AP1)/SQUAMOSA (SQUA) MADS-box proteins, resulting in the activation of downstream genes that confer petal identity [32-36]. Thus, since DEF/GLO-, and to a lesser extent SEP- and SQUA-like, gene function is largely conserved across angiosperms [37-45], shifts between radial and bilateral perianth symmetry could be explained by changes in expression of these floral homeotic genes, resulting in the loss, gain or modification of second whorl identity.
The monocot order Commelinales contains species with both radially and bilaterally symmetrical flowers, is sister to the Zingiberales and comprises five families: Commelinaceae (for example, Commelina and Tradescantia), Hanguanaceae (Hanguana only), Philydraceae (for example, Philydrella and Helmholtzia), Haemodoraceae, and Pontederiaceae [46,47] (Figure 1). Phylogenetic analyses suggest that showy, insect-pollinated and bilaterally symmetrical flowers are plesiomorphic to the Commelinales/Zingiberales clade [47]. However, since Hanguana and Cartonema (Commelinaceae) are both progressively sister to the two major Commelinaceae clades, and have radially symmetrical flowers, it has been suggested that radial flower symmetry is ancestral to Commelinaceae [29,48]. Based on the prevalence of radial flower symmetry in its sister families - including Joinvilleaceae and Flagellariaceae - an ancestral state of bilateral flower symmetry has similarly been invoked for Poaceae [48-50]. Thus, flower bilateral symmetry probably evolved independently in the Commelinaceae (Figure 1), Philydraceae, Zingiberales and Poaceae, as well as in other monocots outside the commelinid clade (for example, Orchidaceae) [29]. A critical question in evolutionary developmental biology is whether similar changes at the level of gene networks, genes and/or gene regions underlie these convergent shifts in floral form.
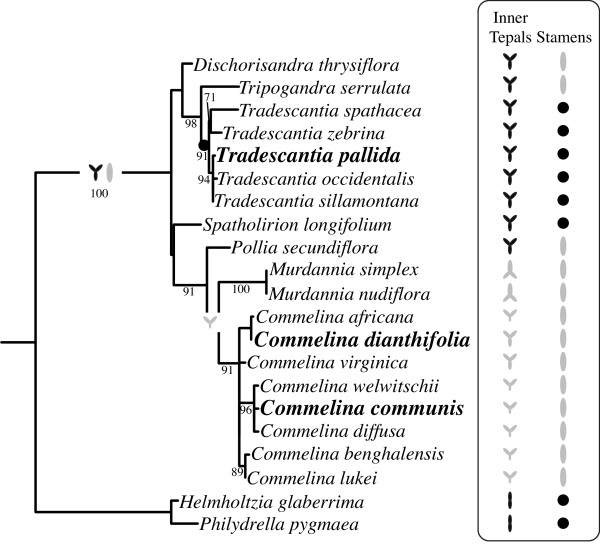
Figure 1.
Evolution of inner tepal and stamen symmetry in Commelinaceae. Maximum likelihood (ML) trnL phylogeny reconstruction with support values for 500 ML bootstrap replicates based on Burns et al. (2011), using Philydraceae species as outgroups [55]. Focal taxa are in bold. Inner tepal and stamen morphology is depicted for each species as bilaterally symmetrical (gray) or radially symmetrical (black). The ancestral state of Commelinaceae inner tepal and stamen symmetry is reconstructed as radial and bilateral, respectively. A shift to radial stamen symmetry is inferred at the base of Tradescantia, whereas a late-developmental shift to weak bilateral symmetry is inferred for C. dianthifolia and C. virginica.
Flower bilateral symmetry in the genus Commelina involves differential organ development in all of the whorls (Figures 1 and 2) [51-55]. In C. communis and C. dianthifolia, each hermaphroditic flower comprises two whorls of three tepals, two whorls of three stamens/staminodes and a whorl of three fused carpels (Figure 2A, B) [54]. In the first whorl of both species, all three outer tepals are membranous, but the lateroventral two are distinct from the dorsal one in both size and shape (Figure 2G). In the second whorl of C. dianthifolia all inner tepals are large and showy, but vary slightly in overall size and shape along the dorsoventral axis (Figure 2E, H). By contrast, in the second whorl of C. communis, only the dorsolateral inner tepals are large and showy; the ventral inner tepal is small and membranous, similar to outer tepals (Figure 2D, G) [54]. In the stamen whorls of both species, the dorsal (outer whorl) or dorsolateral (inner whorl) organs are underdeveloped staminodes that function to attract pollinators, but produce few sterile pollen grains (Figure 2D, G, H). By contrast, the long lateroventral stamens in the outer whorl, and the medium ventral stamen in the inner whorl are showy and produce viable pollen (Figure 2D, G, H). Finally, in the fifth whorl of both species, the dorsal carpel is underdeveloped and sterile, while the two ventral carpels are fertile [54].
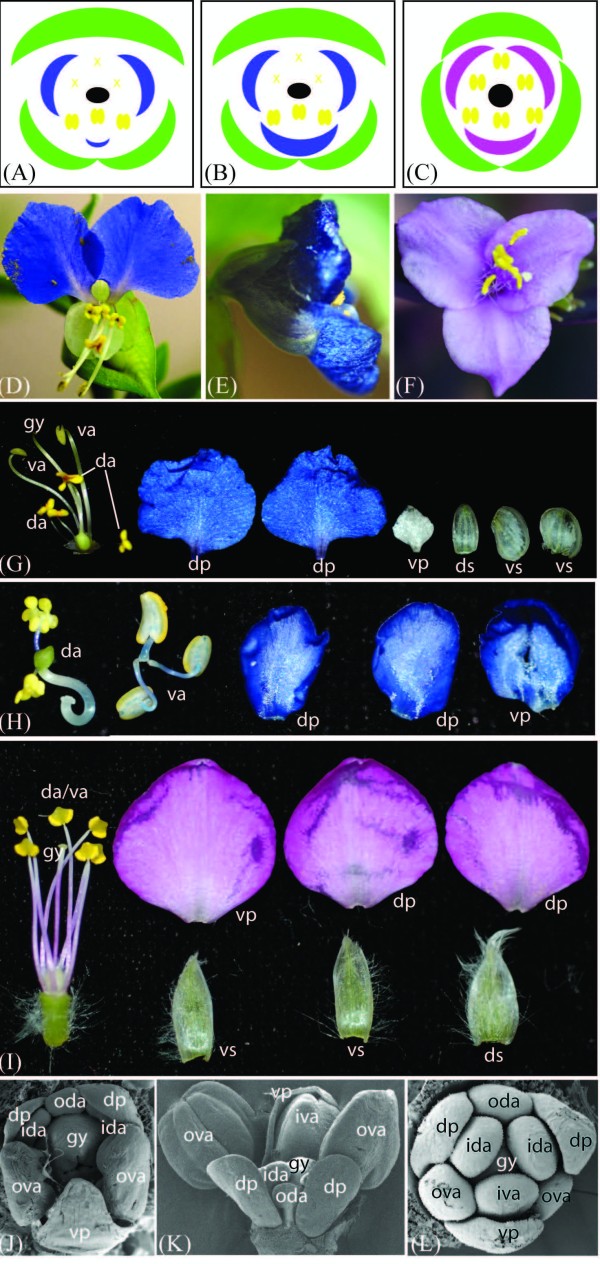
Figure 2.
Flower morphological diversity in representative Commelinaceae species. (A-C) Floral diagrams of mature C. communis (A), C. dianthifolia (B) and T. pallida (C) flowers. Stamens (yellow) and gynoecia (black) of both Commelina species and inner tepals (blue or pink) of C. communis are strongly bilaterally symmetrical, outer tepals (green) of both Commelina species and inner tepals of C. dianthifolia are weakly bilaterally symmetry, and all organ whorls of T. pallida are nearly radially symmetry. (D) Front view of a bilaterally symmetrical Commelina communis flower showing the two large dorsal inner tepals (blue), reduced ventral inner tepal, reduced dorsal staminodes (upper yellow organs), and pollen-producing ventral stamens (lower yellow organs). (E) Side view of a C. dianthifolia flower showing the two large dorsal inner tepals (upper blue) and the even larger ventral inner tepal (lower blue). (F) Front view of a radially symmetrical Tradescantia pallida flower showing the three equal pink inner tepals, and six equal stamens. (G) Individual organs of post-anthesis C. communis flowers. (H) Individual organs of post-anthesis C. dianthifolia flowers; outer tepals are not shown. (I) Individual organs of post-anthesis T. pallida flowers. (J) Scanning electron micrograph (SEM) of a mid-stage C. communis flower; the ventral region develops faster than the dorsal region (this stage), and then the ventral inner tepal arrests development (later stage). (K) SEM of a late stage C. dianthifolia flower. The ventral region develops faster than the dorsal region from early to late stages. (L) SEM of an early stage T. pallida flower showing that radial symmetry is established early in development. ds, dorsal outer tepal; ventral outer tepal; dp, dorsal inner tepal; vp, ventral inner tepal; oda, outer dorsal androecium; ida, inner dorsal androecium; iva, inner ventral androecium; ova, outer ventral androecium; da; dorsal androecia; va, ventral androecia; gy, gynoecium.
Based on detailed analyses of flower development, and the known role of class II TCP genes in core eudicots, Hardy et al. (2009) recently hypothesized a role for TB1-like TCP genes in establishing bilateral symmetry in early developmental stages of Commelina flower development [54]. Furthermore, we propose a second, non-exclusive hypothesis, that dorsal-specific expression of DEF/GLO-like genes defines the dorsoventral axis of symmetry in mid- to late-stage C. communis corollas. In this study, we use gene expression and micromorphological data to test three predictions of these hypotheses that: (1) TB1-like genes are asymmetrically expressed in bilaterally symmetrical C. communis and C. dianthifolia, but not radially symmetrical Tradescantia (formerly Setcreasea) pallida, corollas consistent with a role in differential organ growth, (2) DEF and GLO orthologs are co-expressed in staminodes, stamens and the dorsolateral inner tepals, but not in outer tepals or the outer tepal-like ventral inner tepal of C. communis, and (3) cellular morphology of the C. communis ventral inner tepal is more similar to outer tepals than dorsolateral inner tepals, suggesting asymmetric loss of inner tepal identity in the second whorl.
Methods
Plant material
Inflorescence material was harvested from wild-collected, flowering individuals of C. communis and T. pallida in Missouri and Kansas (USA) during the summer months, and from C. dianthifolia plants grown under standard greenhouse conditions at the University of Kansas. For scanning electron microscopy (SEM) and in situ hybridization, whole inflorescences were fixed overnight in formalin acetic alcohol (FAA: 47.5% (v/v) ethanol, 5% (v/v) acetic acid, 3.7% (v/v) formaldehyde), and gradually taken through an alcohol series to 100% ethanol. For quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), flower organs from 10 to 16 flowers were individually dissected twice from 3 to 5 mm long buds, pooled according to identity/position, and snap-frozen in liquid nitrogen. Total RNA was extracted using TriReagent (Applied BioSystems, Carlsbad, CA, USA), DNA contamination was removed using TURBO DNase (Applied BioSystems), and 1 μg of RNA was reverse transcribed using the iScript Select cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Genomic DNA was extracted from leaf material using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA).
SEM
Ethanol-dehydrated inflorescences were dissected to reveal internal floral organs as necessary. Tissues were critical point dried in a Tousimis critical point drier, mounted on stubs, sputter-coated with gold and viewed with a D. Leo field emission scanning electron microscope (VTT, Espoo, Finland). For micromorphological analyses of inner and outer tepals, both adaxial and abaxial surfaces were analyzed at regions proximal, medial and distal to the floral axis.
Gene isolation and phylogenetic analysis
In order to isolate and sequence all TB1-like orthologs from genomic DNA of C. communis, C. dianthifolia and T. pallida, we used the degenerate forward primers, CYCF1, CYCF2, CYC73aaF, CYC73bF, which were previously designed to amplify CYC/TB1 genes from a wide range of angiosperm taxa, in combination with the reverse primers CYCR1 and CYCR2 [56,57]. Amplicons were cloned into the pGEM-T vector (Bio-Rad), and 5 to 10 colonies per successful PCR reaction were sequenced. To determine if the Commelinaceae sequences corresponded to CYC/REP/TB1-like genes, and to determine orthology and copy number, amino acid sequences spanning the TCP to R domain from C. communis, C. dianthifolia and T. pallida were aligned by MAAFT [58,59] and then by eye in MacClade [60] with previously designated CINCINATTA (CIN)-like, CYC-like, REP-like and TB1-like TCP genes [24,61]. Nucleotide alignments were subjected to maximum likelihood (ML) analyses in GARLI 0.951 and Bayesian analyses in MrBayes 3.1.2 following model optimization in MrModelTest [62-64]. ML analyses were run using 10 random addition sequences with 500 bootstrap replicates. Bayesian analyses were run twice for 10 million generations, sampling every 1,000 generations, with 25% of trees discarded as burn-in. Newly generated TB1-like sequences were submitted to Genbank with accession numbers [Genbank: JQ622131-JQ622135 and JQ622142].
DEF and GLO orthologs were isolated and sequenced from cDNA of C. communis, C. dianthifolia and T. pallida using previously described degenerate primers [65]. Amplicons were cloned and sequenced as previously described, and aligned nucleotide sequences were subjected to ML and Bayesian analyses as described for the TB1-like genes. Newly generated DEF- and GLO-like genes were submitted to Genbank with accession numbers [Genbank: JQ622136-JQ622141].
qRT-PCR
To compare patterns of TB1-like and B-class gene expression in mid-stage floral organs of T. pallida, C. dianthifolia and C. communis, qRT-PCR analyses using DyNAzyme II Hot Start DNA Polymerase (GE Healthcare, Pittsburgh, PA, USA) and SYBR Green I (Invitrogen, Carlsbad, CA, USA) were conducted on a DNA Engine Opticon 2 real-time PCR machine (MJ Research. Waltham, MA, USA) as previously described [66]. Two primers pairs per target gene were designed in Primer 3, and the most efficient primer pair was selected for expression analysis (Additional file 1) [67]. Where possible, primer pairs were designed to span introns to rule out DNA contamination. β-ACTIN (ACT) and EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 ALPHA 1 (EF1alpha) showed little transcriptional variation across different tissues of C. communis and C. dianthifolia, and T. pallida, respectively, and were, therefore, selected as reference genes. A negative cDNA control containing RNA from dorsolateral inner tepals and lacking the reverse transcriptase enzyme was initially used as a negative control template for all qRT-PCR analyses. Furthermore, when no transcription was detected in floral material, genomic DNA was used as a positive control. For each gene the mean and standard deviation was determined for three to four technical replicates. Results were analyzed separately for one (C. dianthifolia) or two (C. communis and T. pallida) biological replicates to account for differences in developmental staging of the pooled material, and compared for consistent results between organ type and organ position. Analyses were not carried out for C. dianthifolia outer tepals due to difficulty obtaining good quality RNA from these organs.
In situ hybridization
To better determine the spatio-temporal pattern of gene expression, in situ hybridization was carried out in wax-embedded inflorescence tissues of the most closely related species, C. communis (DEF, GLO and TB1a) and C. dianthifolia (DEF and TB1a). Antisense and sense gene-specific probes of CcDEF (for both species), CcGLO (C. communis only), CcTB1a (C. communis only) and CdTB1a (C. dianthifolia only) were generated using T7 and SP6 RNA Taq polymerase (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer's instructions. Each probe spanned ca. 400 bps, and included part of the C-terminal domain and the 3'-untranscribed region. As an experimental control, an antisense probe was generated for CcHistone4 as described previously [57]. In situ hybridization was performed as described previously [68,69].
Results
Flower development and perianth micromorphology
Comparative morphological analyses revealed a similar progression of flower development for C. communis and C. dianthifolia at early to mid stages. In both species, development proceeded asymmetrically, with the ventral/lateroventral organs developing more rapidly than the dorsal/dorsolateral organs (Figure 2J, K). In the outer tepal and stamen whorls, this asymmetry was maintained into late stage development for both species (Figure 2D, E, G, J, K). By contrast, at mid to late stages of development the ventral inner tepal of C. communis, but not C. dianthifolia, arrested prematurely. This resulted in strong asymmetry in the mature corolla of C. communis, with the ventral inner tepal resembling the small colorless outer tepals (Figure 2D, G versus 2E, H). Unlike C. communis and C. dianthifolia, flowers of T. pallida were radially symmetrical from early to late stages of development (Figure 2F, I, L).
To determine if the reduced outer tepal-like ventral inner tepal of C. communis also resembled outer tepals at the micromorphological level, detailed SEM analyses were conducted on mature floral organs at the cellular level. C. communis outer tepals, dorsolateral inner tepals and ventral inner tepals were all characterized by long wavy cells on both the adaxial and abaxial surfaces (Figure 3A). Additionally, the abaxial surface of both outer tepals and ventral inner tepals had stomata; no stomata were found on either surface of the dorsolateral inner tepals, or the adaxial surface of outer tepals and ventral inner tepals (Figure 3A). To distinguish between the alternative possibilities that stomata define outer tepal identity or determine ventral inner tepal identity, similar analyses were conducted on mature C. dianthifolia (both outer and inner tepals) and T. pallida (only inner tepals) flowers (Figure 3B, C). As in C. communis, C. dianthifolia outer tepals were marked by stomata on the abaxial surface. However, unlike C. communis ventral inner tepals, no stomata were evident on either surface of the dorsolateral and ventral inner tepals in C. dianthifolia or T. pallida (Figure 3B, C).
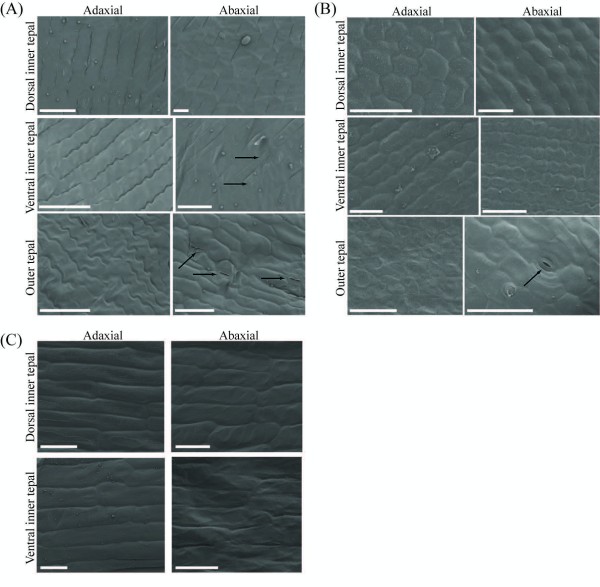
Figure 3.
Cellular micromorphology of Commelina tepals. (A) The adaxial surfaces of Commelina communis inner and outer tepals are characterized by long cells with wavy margins; whilst the abaxial surfaces have cells with straighter edges. The abaxial surfaces of the ventral inner tepal and the outer tepals are similar regarding the presence of stomata, which are absent from the abaxial surface of dorsal inner tepals. (B) Stomata are only found on the abaxial surface of outer tepals from Commelina dianthifolia. (C) Stomata are lacking from the abaxial and adaxial surfaces of both dorsal and ventral inner tepals of Tradescantia pallida. Scale bars denote 50 μm. Arrows indicate closed (C. communis ventral inner tepals), partially closed (C. communis outer tepals), and open stomata (C. dianthifolia outer tepals).
Gene duplication in the TB1-like gene lineage
Cloning and phylogenetic analyses revealed two CYC/REP/TB1-like homologs (hereafter TB1a and TB1b) in C. communis, C. dianthifolia and T. pallida that formed two distinct clades (Figure 4). Maximum likelihood and Bayesian analyses yielded single trees with similar topologies that largely tracked the known species phylogeny of monocots. The Commelinaceae TB1a genes formed a well-supported clade (73% ML bootstrap (MB); 99% posterior probability (PP)) sister to the TBL2 genes of Zingiberales species (100% PP), and together these clades were sister to the REP1 genes in Poaceae (98% PP) (Figure 4). Thus, orthology between Commelinacae TB1a, Zingiberales TBL2 and Poaceae REP1 genes was strongly supported by Bayesian PPs. Commelinaceae TB1b genes similarly formed a well-supported clade (99% PP) sister to the TB1-like genes of Zingiberales, Poaceae and Liliaceae species. However, the relationship between genes of each family was not well-supported (Figure 4). On a broader scale, both REP1 and TB1 clades grouped with CYC-like genes from eudicots (100% MB; 90% PP) (Figure 4), even when the more distantly related PCF-like TCP genes were used as outgroups [61] (data not shown).
Figure 4.
Best maximum likelihood phylogeny of monocot TB1-like genes rooted with core eudicot CIN-like genes. Maximum likelihood bootstrap values above 69% (left) and Bayesian posterior probabilities above 89% (right) are shown for each branch. Asterisks denote 100% support. Dashes indicate bootstrap support below 70% only when the corresponding posterior probability is shown. The TB1-like and CIN-like gene clades are shaded.
Differential expression of TB1-like genes
In order to test the prediction that TB1 orthologs are expressed asymmetrically in mid stage bilaterally symmetrical C. communis and C. dianthifolia tepals, but not radially symmetrical T. pallida tepals, expression analyses were carried out on dissected flower buds using qRT-PCR (Figure 5 and Additional file 2). In C. communis, TB1a expression was significantly higher in ventral versus dorsolateral outer tepals (average 10.4-fold difference (SD = 9.7) for two biological replicates), and ventral versus dorsolateral inner tepals (average 12.6-fold difference (SD = 9.7) for two biological replicates) (Figure 5 and Additional file 2). Similarly, in C. dianthifolia, TB1a expression was significantly higher in ventral versus dorsolateral inner tepals (9.1-fold difference for one biological replicate) (Figure 5). No TB1a expression was detected in either outer or inner tepals of T. pallida for either biological replicate (Figure 5 and Additional file 2).
Figure 5.
Relative quantitative gene expression in dissected flower tissues. TB1a expression is significantly stronger in ventral versus dorsal tepals of C. communis and C. dianthifolia but not T. pallida, and is expressed asymmetrically in different directions in stamens of all three species (top panel). TB1b expression is low or absent in floral tissues, but is expressed strongly in leaves (second top panel). GLO expression is significantly stronger in staminodes versus stamens of C. dianthifolia and C. communis, and is low to undetectable in outer tepals and gynoecia (second bottom panel). DEF expression is significantly stronger in dorsal versus ventral inner tepals of C. communis, being undetectable in the latter (bottom panel). β-ACT and EF1a were used as internal controls. Experiments were conducted with three to four technical replicates. Bars show mean and standard deviation. Second biological replicates were analyzed separately and are shown in Additional file 2. White bars, whole organs; blacks bars, dorsal organs; gray bars, ventral organs. Asterisks denote significant differences between dorsal and ventral organ comparisons. nd, not detected.
Unlike tepals, the correlation between TB1a expression and stamen differentiation was only weakly supported. In C. communis, although TB1a was asymmetrically expressed in dorsal staminodes versus ventral stamens, the direction of differential expression contrasted between replicates (Figure 5 and Additional file 2). This suggests dynamic expression of TB1a at mid stage stamen development. By contrast, TB1a expression was weak but consistently higher in dorsal staminodes versus ventral stamens of C. dianthifolia (2.3-fold difference for biological replicate one (Figure 5); detectable in dorsal staminodes but undetectable in ventral stamens for biological replicate two (data not shown)), and was not significantly different between dorsal and ventral stamens of T. pallida (1.2-fold difference for one biological replicate) (Figure 5). In contrast to qRT-PCR analyses, no TB1a expression was detectable in early to late stage flowers of C. communis, C. dianthifolia or T. pallida using in situ hybridization (data not shown). This suggests that TB1a transcripts are below the level of detection using this method.
QRT-PCR analyses of TB1b genes revealed different expression patterns relative to TB1a paralogs (Figure 5 and Additional file 2). In T. pallida, TB1b was undetectable in all floral organs and leaves, despite amplification of genomic DNA (Figure 5 and Additional file 2). In C. communis, TB1b expression was highest in leaves, with low (Figure 5) to no (Additional file 2) detectable expression in all other organs. For both replicates, no significant differences in transcript levels were detected between dorsal and ventral organs (Figure 5 and Additional file 2). Finally, for C. dianthifolia, no TB1b expression was detected in any floral organ, again despite amplification of genomic DNA (Figure 5).
Differential expression of B-class genes
To test the prediction that B-class gene expression is exclusively reduced or absent from the outer tepal-like ventral inner tepal of C. communis, DEF and GLO genes were isolated from all three focal species, and both qRT-PCR and in situ hybridization analyses were carried out on inflorescence tissues. ML and Bayesian analyses supported the isolation of both DEF and GLO orthologs from each species, and relationships among a larger set of DEF and GLO genes largely tracked the known species tree for monocots (Additional file 3). As predicted, GLO was expressed in inner tepals and stamens of C. communis, C. dianthifolia and T. pallida (Figure 5 and Additional file 2). Furthermore, analyses revealed low (T. pallida based on one biological replicate) to no (C. communis based on two biological replicates) expression in outer tepals of representative Tradescantia and Commelina species (Figure 5 and Additional file 2). In C. communis, GLO was expressed similarly in dorsolateral and ventral inner tepals, with expression being significantly higher in dorsal staminodes than ventral stamens (2.1-fold difference for two biological replicates (SD = 0.27)) (Figure 5 and Additional file 2). A similar asymmetric expression pattern was detected in the stamen whorls of C. dianthifolia (Figure 5). However, in T. pallida there was no significant difference in expression between ventral versus dorsal inner tepals and stamens for one biological replicate (Figure 5 and Additional file 2).
Consistent with qRT-PCR analyses, in situ hybridization analyses revealed GLO expression in the inner tepal and stamen whorls of young C. communis flower buds, with transcripts being absent from the center of the flower (Figure 6A). The second to fourth whorl expression was maintained during mid to late stages of C. communis flower development, with transcripts becoming detectable in the gynoecium from mid to late stages (Figure 6B, C). Since qRT-PCR for all three species, and in situ hybridization for C. communis, revealed no correlation between GLO expression and inner tepal morphology, in situ hybridization experiments were not carried out for C. dianthifolia or T. pallida GLO orthologs.
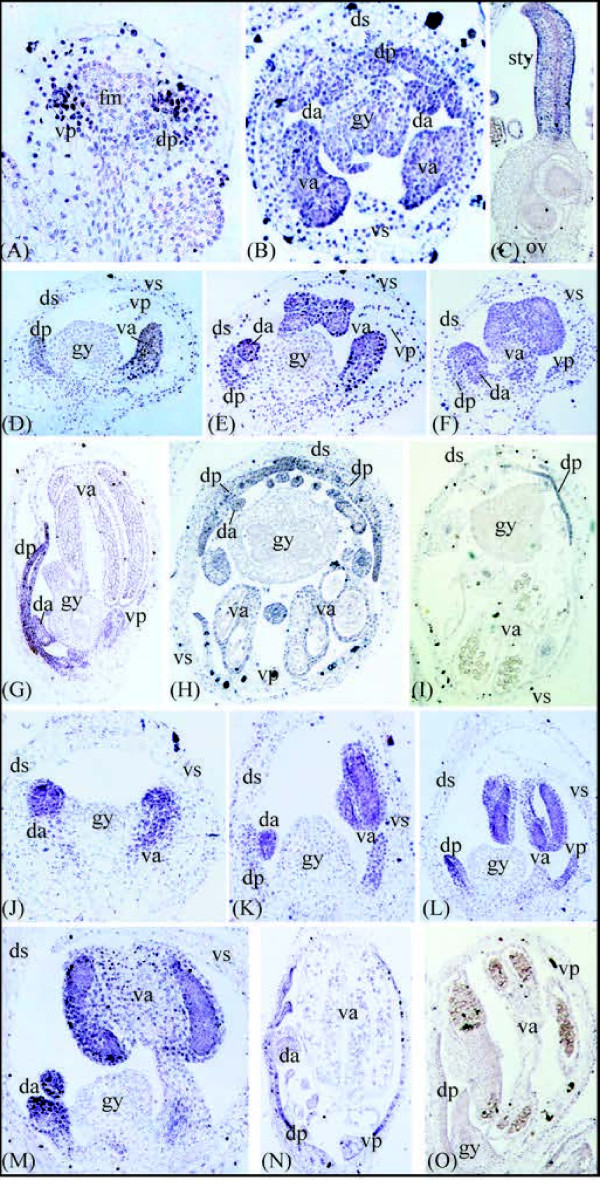
Figure 6.
In situ hybridization of B-class genes in Commelina communis and C. dianthifolia. (A-C) C. communis flower sections showing expression of GLOBOSA (GLO) in early to late stages of inner tepal and stamen/staminode development, and late stages of gynoecial development. (D-F) Mid-stage C. communis flowers sectioned longitudinally showing DEFICIENS (DEF) expression in the dorsal inner tepals, dorsal staminodes, and ventral stamens. No transcripts were detected in the outer tepals, ventral inner tepals, or gynoecium. (G) Late-stage longitudinal section of a C. communis flower showing DEF expression in the dorsal inner tepals and staminodes. (H-I) Transverse sections through late-stage C. communis flowers showing DEF expression in the dorsal inner tepals, dorsal staminodes, and ventral stamens. (J) Early-stage C. dianthifolia flower showing comparable DEF expression in the dorsal and ventral regions of the stamen primordia. (K-M) Mid-stage C. dianthifolia flowers showing DEF expression in the dorsal inner tepals, dorsal staminodes, ventral inner tepals, and ventral staminodes. (N) Late-stage C. dianthifolia flower showing DEF expression in all inner tepals and the dorsal staminodes. (O) Sense DEF control in C. communis shows little to no staining. fm, floral meristem; ds, dorsal outer tepal; vs, ventral stamen; dp, dorsal inner tepal; vp, ventral inner tepal; da, dorsal androecium; va, ventral androecium; gy, gynoecium; sty, style; ov, ovary.
QRT-PCR analyses revealed DEF gene transcripts in inner tepals and stamens of C. communis, C. dianthifolia and T. pallida (Figure 5 and Additional file 2), with low to no expression of DEF in outer tepals and gynoecia of C. communis and T. pallida (Figure 5 and Additional file 2). In C. communis, DEF expression was significantly higher in dorsal staminodes versus ventral stamens (3.5-fold difference for two biological replicates (SD = 0.32)), and in contrast to dorsolateral inner tepals, was completely absent from ventral inner tepals for two biological replicates (Figure 5 and Additional file 2). In C. dianthifolia, DEF expression was not significantly different between dorsal staminodes and ventral stamens. However, in striking contrast to C. communis, DEF was significantly higher in ventral versus dorsolateral inner tepals (2.3-fold difference for one biological replicate). Finally, in T. pallida, DEF expression was not significantly different between dorsal and ventral stamens, and was highly variable in dorsolateral versus ventral inner tepals between two biological replicates (Figure 5 and Additional file 2).
Consistent with qRT-PCR, in situ hybridization analyses of DEF showed much higher expression levels in dorsal compared to ventral inner tepals of early to late stage C. communis flower buds (Figure 6D-I). However, in the stamen whorls, this dorsoventral gradient of DEF expression only became evident following late-stage differentiation of the ventral stamens (Figure 6D-I). Comparable analyses in C. dianthifolia revealed DEF expression in both dorsolateral and ventral inner tepals, dorsal staminodes and ventral stamens during early to midstages of development (Figure 6J-M). Strong expression was maintained in both the dorsolateral and ventral inner tepals into late development (Figure 5N). However, DEF expression became gradually weaker in ventral stamens relative to dorsal staminodes during late stage anther differentiation (Figure 6M, N). No antisense transcripts of DEF were detectable in outer tepals or gynoecia of C. communis or C. dianthifolia. Furthermore, no staining was observed in control sections of either species using DEF or GLO sense probes (Figure 6O and data not shown).
Discussion
A major conclusion from this study is that TB1-like genes have been independently recruited in establishing Commelinaceae corolla bilateral symmetry in early development, similar to several core eudicot lineages and rice [8-13,17]. Furthermore, micromorphological evidence and expression of the B-class MADS-box gene CcDEF suggests that late stage asymmetry between the dorsolateral and ventral inner tepals of C. communis is due to a homeotic transformation of the ventral inner tepal into an outer tepal. In the sections below, we discuss the implications of these results as they relate to our understanding of convergent trait evolution.
Parallel evolution of TB1-like genes across angiosperms
Bilateral flower symmetry can be achieved through the asymmetric loss or reduction of organs (structural zygomorphy) or the modification of organs (presentation zygomorphy) within one or more whorls, and can be present in early and/or late stages of development [48]. Although both types of zygomorphy are found within the monocots, structural zygomorphy is particularly prevalent within the Asparagales (for example, Orchidaceae), Arecaceae, Dasypogonaceae, Zingiberales (for example, Zingiberaceae), Commelinales (for example, Commelinaceae) and Poales (for example, Poaceae) [18,27,29,48,70]. In most cases, bilateral symmetry affects the inner tepals and stamen whorls. However, outer tepal, inner tepal, stamen and gynoecial zygomorphy have all evolved in the monocots multiple times independently [48].
Maximum parsimony character state reconstructions support a gain of tepal bilateral symmetry within the Commelina-containing Commelineae clade of Commelinaceae (Figure 1). Available data suggest that members of the Commelineae have flowers that are strongly bilaterally symmetrical in early development, but vary in the strength of structural tepal zygomorphy in late development [54,71] (this study). For example, although all are strongly zygomorphic in early development, at anthesis C. communis inner tepals are highly differentiated along the dorsoventral axis, whereas C. dianthifolia and Murdannia nudiflora inner tepals are only slightly differentiated in size and shape. Based on evidence from core eudicots, it has been hypothesized that asymmetric expression of CYC/TB1-like genes underlies early stage structural zygomorphy in the Commelineae [54]. Consistent with this hypothesis, TB1a is expressed asymmetrically in tepals of C. communis and C. dianthifolia, but not T. pallida, at mid stages of flower development. However, in contrast to the asymmetric expression of CYC and DICH in the model core eudicot A. majus and REP1 in O. sativa, expression is significantly higher in the ventral, as opposed to dorsolateral, tepals [8,9,17] (this study). It is predicted that the asymmetrical expression of TB1a in C. communis and C. dianthifolia is initiated in early flower development and has no effect on late stage development when the ventral inner tepal of C. communis arrests growth (see next section).
In A. majus expression of CYC in the dorsal floral meristem represses growth; a similar repressive function has been assigned to IaCYC of the rosid core eudicot Iberis amara (Brassicaceae) in mid to late stage dorsal petal development [8,9,11]. By contrast, at mid to late stages of A. majus flower development, CYC expression in the dorsal petals actually increases cell proliferation and elongation relative to the ventral petal [8,9]. Together with results from Commelinaceae, these data support developmentally and taxonomically distinct roles for CYC in establishing early to late stage perianth organ growth, and suggest that parallel recruitment of CYC/TB1 genes in floral bilateral symmetry is not limited to the dorsal side of the flower. Indeed, a ventral pattern of expression was recently demonstrated for CsTBL1a in bilaterally symmetrical C. spicatus (Costaceae, monocot) flowers [18].
Further studies testing the involvement of CYC/TB1 genes in transitions from radial to bilateral flower symmetry will require functional tests, and should aim to elucidate whether changes in CYC/TB1 expression are due to cis-regulatory or upstream changes. In A. majus, the NAC family protein CUPULIFORMIS (CUP) has been implicated in the positive regulation of CYC [72]. Thus, expression analyses of CUP-like genes in perianth organs of Commelinaceae and other monocots varying in flower symmetry might be a good starting point to address this question.
Parallel evolution of B-class genes in monocots
It has been hypothesized that changes in the expression of organ identity genes can explain the evolution of flower bilateral symmetry in certain monocots by altering the identity of a subset of organs within a floral whorl. In the case of orchids, one of several DEF-like genes evolved an asymmetric expression pattern following gene duplication, and is implicated in modification of the ventrally positioned inner tepal into the characteristic lip [25,27,73-75]. Our data also support a role for DEF-like gene evolution in modification of the C. communis ventral inner tepal. However, unlike orchids, the modification of DEF-like gene expression in C. communis was not preceded by gene duplication and is presumably not associated with changes in protein function at the biochemical level. Furthermore, unlike C. dianthifolia and T. pallida, the ventral inner tepals of C. communis flowers have stomata on their abaxial surface similar to outer tepals. This micromorphological marker correlates both with the general outer tepal-like appearance of the ventral inner tepal and the complete absence of DEF-like gene expression during early to late stages of development.
Together, these studies suggest parallel evolution of DEF-like genes in the independent origins of monocot flower bilateral symmetry. Future studies in other monocot families will be required to determine the generality of these results, and to test whether evolution of DEF-like gene expression can explain petaloidy in the stamen whorl [29]. Furthermore, a key question remains as to whether evolution of DEF-like gene expression can be explained by cis-regulatory or upstream changes. In A. thaliana, SUPERMAN (SUP), FLORAL ORGAN NUMBER1 (FON1), CUP-SHAPED COTYLEDON1 (CUC1), and CUC2 proteins negatively regulate both B-class genes in the sepal whorl [76]. Thus, expression of these proteins in the ventral inner tepal of C. communis could explain the loss of DEF-like expression in this organ. However, this scenario would require that SUP-, FON1- and CUC1/2-like proteins do not regulate the C. communis GLO-like gene, which is expressed normally in the ventral inner tepal.
Conclusions
Together with other studies, our gene expression data on three morphologically diverse species of Commelinaceae suggest a parallel role for two major transcription factors in the independent evolution of angiosperm flower bilateral symmetry. In the case of class II TCP genes, changes in expression are correlated with early developmental shifts from radial to bilateral flower symmetry in several core eudicots and commelinid monocots (rice, Costus and Commelina). This supports a conserved role for class II TCP genes in organ growth across angiosperms, and suggests either evolutionary constraint on the flower symmetry gene network or the involvement of few genes in the establishment of floral meristem symmetry. Evolution of DEF-like genes is also implicated in shifts from radial to bilateral flower symmetry, at least within divergent monocots (Phaelaenopsis and C. communis). However, in contrast to class II TCP genes, changes in both function (Phaelaenopsis) and regulation (C. communis) of B-class genes are implicated in late developmental shifts in within-whorl organ identity. Further studies are required to test the generality of class II TCP and B-class gene evolution in diversification of monocot flowers, and to decipher whether/why the same genes have been the targets of repeated selection across millions of years of angiosperm diversification.
Abbreviations
ACT: β-ACTIN; DEF: DEFICIENS; CIN: CINCINATTA; CUC1: CUP-SHAPED COTYLEDON1; CUP: CUPULIFORMIS; CYC: CYCLOIDEA; DICH: DICHOTOMA; EF1alpha: EUKARYOTIC TRANSLATION ELONGATION FACTOR 1 ALPHA 1; FAA: formalin acetic alcohol; FON1: FLORAL ORGAN NUMBER1; GLO: GLOBOSA; MB: maximum likelihood bootstrap; ML: maximum likelihood; PCF: PROLIFERATING CELL NUCLEAR ANTIGEN GENE-CONTROLLING ELEMENT BINDING FACTOR; PP: Bayesian posterior probability; qRT-PCR: quantitative reverse transcriptase polymerase chain reaction; RAD: RADIALIS; REP1: RETARDED PALEA 1; SEM: SCANNING ELECTRON MICROSCOPY; SD: standard deviation; SUP: SUPERMAN; TB1: teosinte branched1.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JCP designed and performed the experiments. JCP and LCH wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Primers used for RT-PCR. Primer sequences used for amplification of target and housekeeping genes from Commelina communis, C. dianthifolia and Tradescantia pallida are listed in table format.
Second biological replicate for relative quantitative gene expression in dissected flower tissues. In agreement with the first biological replicate (Figure 5), GLO expression is significantly higher in dorsal staminodes versus ventral stamens of Commelina communis, but is similar across inner tepals of both Tradescantia pallida and C. communis (left). DEF is again undetectable in ventral, but not dorsal, inner tepals of C. communis. By contrast, DEF expression is higher in ventral versus dorsal inner tepals of Tradescantia pallida (second from left). Similar to the first biological replicate, TB1a is undetectable in T. pallida inflorescence tissues, and is expressed significantly more strongly in both outer and inner ventral versus dorsal tepals of C. communis (second from right). TB1b is undetectable in inflorescence organs of C. communis, although the primers amplify genomic DNA, and is undetectable in inner tepals and gynoecia of T. pallida (right). β-ACT and EF1a were used as internal controls. Experiments were conducted with three to four technical replicates. Bars show mean and standard deviation. White bars, whole organs; blacks bars, dorsal organs; gray bars, ventral organs. Asterisks denote significant differences between dorsal and ventral organ comparisons. gDNA, genomic DNA; Inf, whole inflorescence; Dot, dorsal outer tepal; Vot, ventral outer tepal; Dit, dorsal inner tepal; Vit, ventral inner tepal; Dst, dorsal stamens/staminodes; VSt, ventral stamens; Gyn, gynoecia; nd, not detected.
Best maximum likelihood phylogeny of monocot B-class genes. Maximum likelihood bootstrap values above 69% (left) and Bayesian posterior probabilities above 89% (right) are shown for each branch. Asterisks denote 100% support. Dashes indicate bootstrap support below 70% only when the corresponding posterior probability is shown. The focal genes of this study are highlighted in gray boxes.
Contributor Information
Jill C Preston, Email: jcpxt8@ku.edu.
Lena C Hileman, Email: lhileman@ku.edu.
Acknowledgements
We thank Iván Jiménez for help collecting Commelina communis material and two anonymous reviewers for comments on a previous version of the manuscript. This work was supported by a National Science Foundation grant (IOS-0616025) to LCH.
References
- Donoghue MJ, Ree RH, Baum DA. Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci. 1998;3:311–317. doi: 10.1016/S1360-1385(98)01278-3. [DOI] [Google Scholar]
- Ree RH, Donoghue MJ. Inferring rates of change in flower symmetry in asterid angiosperms. Syst Biol. 1999;48:633–641. doi: 10.1080/106351599260201. [DOI] [Google Scholar]
- Endress PK. Symmetry in flowers: diversity and evolution. Int J Plant Sci. 1999;160:S3–S23. doi: 10.1086/314211. [DOI] [PubMed] [Google Scholar]
- Sargent RD. Floral symmetry affects speciation rates in angiosperms. Proc R Soc Lond Biol Sci. 2004;271:603–608. doi: 10.1098/rspb.2003.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S. On 'various contrivances': pollination, phylogeny and flower form in the Solanaceae. Philos Transact R Soc Lond Biol Sci. 2010;365:449–460. doi: 10.1098/rstb.2009.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamosi JC, Vamosi SM. Key innovations within a geographical context in flowering plants: towards resolving Darwin's abominable mystery. Ecol Lett. 2010;13:1270–1279. doi: 10.1111/j.1461-0248.2010.01521.x. [DOI] [PubMed] [Google Scholar]
- Cubas P. Floral zygomorphy, the recurring evolution of a successful trait. Bioessays. 2004;26:1175–1184. doi: 10.1002/bies.20119. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/S0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, Chen J, Zheng L, Guo X, Luo J, Sato S, Tabata S, Ma W, Cao X, Hu X, Sun C, Luo D. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA. 2006;103:4970–4975. doi: 10.1073/pnas.0600681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Zachgo S. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc Natl Acad Sci USA. 2007;104:16714–16719. doi: 10.1073/pnas.0705338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA. 2008;105:9117–9122. doi: 10.1073/pnas.0801359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Luo Y, Li X, Wang L, Xu S, Yang J, Weng L, Sato S, Tabata S, Ambrose M, Rameau C, Feng X, Hu X, Luo D. Genetic control of floral zygomorphy in pea (Pisum sativum L.) Proc Natl Acad Sci USA. 2008;105:10414–10419. doi: 10.1073/pnas.0803291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk QC. Legume flowers bear fruit. Proc Natl Acad Sci USA. 2006;103:4801–4802. doi: 10.1073/pnas.0601298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Cubas P. An expanded evolutionary role for flower symmetry genes. J Biol. 2009;8:90. doi: 10.1186/jbiol193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Hileman LC. Developmental genetics of floral symmetry evolution. Trends Plant Sci. 2009;14:147–154. doi: 10.1016/j.tplants.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Gao S, Xue DW, Luo D, Li LT, Ding SY, Yao X, Wilson ZA, Qian Q, Zhang DB. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 2009;149:235–244. doi: 10.1104/pp.108.128231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett ME, Specht CD. Changes in expression pattern of the TEOSINTE BRANCHED1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. AmJ Bot. 2011;98:1–17. doi: 10.3732/ajb.1000247. [DOI] [PubMed] [Google Scholar]
- Galego L, Almeida J. Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev. 2002;16:880–891. doi: 10.1101/gad.221002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MMR, Fox S, Hanna AI, Baxter C, Coen E. Evolution of regulatory interactions controlling floral asymmetry. Development. 2005;132:5093–5101. doi: 10.1242/dev.02085. [DOI] [PubMed] [Google Scholar]
- Almeida J, Rocheta M, Galego L. Genetic control of flower shape in Antirrhinum majus. Development. 1997;124:1387–1392. doi: 10.1242/dev.124.7.1387. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Baum DA. Why do paralogs persist? Molecular evolution of CYCLOIDEA and related floral symmetry genes in Antirrhineae (Veronicaceae) Mol Biol Evol. 2003;20:591–600. doi: 10.1093/molbev/msg063. [DOI] [PubMed] [Google Scholar]
- Cubas P. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- Howarth DG, Donoghue MJ. Phylogenetic analysis of the "ECE" (CYC/TB1) clade reveals duplications predating the core eudicots. Proc Natl Acad Sci USA. 2006;103:9101–9106. doi: 10.1073/pnas.0602827103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W-C, Kuoh C-S, Chuang M-H, Chen W-H, Chen H-H. Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 2004;45:831–844. doi: 10.1093/pcp/pch095. [DOI] [PubMed] [Google Scholar]
- Tsai W-C, Pan Z-J, Hsiao Y-Y, Yeng M-F, Wu T-F, Chen W-H, Chen H-H. Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol. 2008;49:814–824. doi: 10.1093/pcp/pcn059. [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Theissen G. MADS about the evolution of orchid flowers. Trends Plant Sci. 2008;13:51–59. doi: 10.1016/j.tplants.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Hiese L, Harter A, Koch MA, Theissen G. Positive selection and ancient duplications in the evolution of class B floral homeotic genes of orchids and grasses. BMC Evol Biol. 2009;9:81–106. doi: 10.1186/1471-2148-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett ME, Specht CD. Evidence for the involvement of GLOBOSA-like gene duplications and expression divergence in the evolution of floral morphology in the Zingiberales. New Phytol. 2010;187:521–541. doi: 10.1111/j.1469-8137.2010.03279.x. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA. 1996;93:4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF. Nuclear localization of the Arabidopsis APETALA1 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 1996;10:1812–1821. doi: 10.1101/gad.10.14.1812. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. Plant biology. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- Melzer R, Verelst W, Theissen G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in 'floral quartet'-like complexes in vitro. Nucleic Acids Res. 2009;37:144–157. doi: 10.1093/nar/gkn900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muino JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, Parcy F, Xu L, Carles CC, Angenent GC, Kaufmann K. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Beltran J, Huijser P, Pape H, Lonnig W, Saedler H, Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell. 2000;5:569–579. doi: 10.1016/S1097-2765(00)80450-5. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development. 2004;131:6083–6091. doi: 10.1242/dev.01523. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Zanis MJ, Kellogg EA, Schmidt RJ. Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc Natl Acad Sci USA. 2006;104:1081–1086. doi: 10.1073/pnas.0606434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S, Hileman LC, de Martino G, Irish VF. Functional analyses of genetic pathways controlling petal specification in poppy. Development. 2007;134:4157–4166. doi: 10.1242/dev.013136. [DOI] [PubMed] [Google Scholar]
- Hileman LC, Irish VF. More is better: the uses of developmental genetic data to reconstruct perianth evolution. Am J Bot. 2009;96:83–95. doi: 10.3732/ajb.0800066. [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA. SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 2005;10:427–435. doi: 10.1016/j.tplants.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Litt A, Kramer EM. The ABC model and the diversification of floral organ identity. Semin Cell Dev Biol. 2010;21:129–137. doi: 10.1016/j.semcdb.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Davis JI, Petersen G, Seberg O, Stevenson DW, Hardy CR, Simmons MP, Michelangeli FA, Goldman DH, Campbell LM, Specht CD, Cohen JL. Are mitochondrial genes useful for the analysis of monocot relationships? Taxon. 2006;55:857–870. doi: 10.2307/25065681. [DOI] [Google Scholar]
- Givinish TJ, Pires JC, Graham SW, McPherson MA, Prince LM, Patterson TB, Rai HS, Roalson EH, Evans TM, Hahn WJ, Millam KC, Meerow AW, Molvray M, Kores PJ, O'Brien HE, Hall JC, Kress WJ, Sytsma KJ. Phylogenetic relationships of monocots based on the highly informative plastid gene ndhF: evidence for widespread concerted convergence. Aliso. 2006;22:28–51. [Google Scholar]
- Rudall PJ, Bateman RM. Evolution of zygomorphy in monocot flowers: iterative patterns and developmental constraints. New Phytol. 2004;162:25–44. doi: 10.1111/j.1469-8137.2004.01032.x. [DOI] [Google Scholar]
- GPWG (Grass Phylogeny Working Group) Phylogeny and subfamilial classification of the grasses (Poaceae) Ann Mo Bot Gard. 2001;88:373–457. doi: 10.2307/3298585. [DOI] [Google Scholar]
- Michelangeli F, Davis JI, Stevenson D. Phylogenetic relationships among Poaceae and related families as inferred from morphology, inversions in the plastid genome, and sequence data from the mitochondrial and plastid genomes. Am J Bot. 2003;90:93–106. doi: 10.3732/ajb.90.1.93. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Evans TM, Pires JC, Sytsma KJ. Polyphyly and convergent morphological evolution in Commelinales and Commelinidae: evidence from rbcL sequence data. Mol Phylogenet Evol. 1999;12:360–385. doi: 10.1006/mpev.1999.0601. [DOI] [PubMed] [Google Scholar]
- Evans TM, Sytsma KJ, Faden RB, Givnish TJ. Phylogenetic relationship in the Commelinaceae: II. A cladistics analysis of rbcL sequences and morphology. Syst Bot. 2003;28:270–292. [Google Scholar]
- Ushimaru A, Dohzono I, Takami Y, Hyodo F. Flower orientation enhances pollen transfer in bilaterally symmetrical flowers. Oecologia. 2009;160:667–674. doi: 10.1007/s00442-009-1334-9. [DOI] [PubMed] [Google Scholar]
- Hardy CR, Sloat LL, Faden RB. Floral organogenesis and the developmental basis for pollinator deception in the Asiatic dayflower, Commelina communis (Commelinaceae) Am J Bot. 2009;96:1236–1244. doi: 10.3732/ajb.0800344. [DOI] [PubMed] [Google Scholar]
- Burns JH, Faden RB, Steppan SJ. Phylogenetic studies in the Commelinaceae subfamily Commelinoideae inferred from nuclear ribosomal and chloroplast DNA sequences. Syst Bot. 2011;36:268–276. doi: 10.1600/036364411X569471. [DOI] [Google Scholar]
- Howarth DG, Donoghue MJ. Duplications in CYC-like genes from Dipsacales correlate with floral form. Int J Plant Sci. 2005;166:357–270. doi: 10.1086/428634. [DOI] [Google Scholar]
- Preston JC, Kost MA, Hileman LC. Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol. 2009;182:751–762. doi: 10.1111/j.1469-8137.2009.02794.x. [DOI] [PubMed] [Google Scholar]
- Katoh T, Kuma M. MAAFT: a novel method for rapid multiple sequence alignment based on fast Fourier Transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T. Recent developments in the MAAFT multiple sequence alignment program. Brief Bioinform. 2008;9:212. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4: analysis of phylogeny and character evolution. Sinauer, Sunderland, MA; 2003. version 4.0.6. [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M, Trontin C. High time for a roll call: gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann Bot. 2011;107:1533–1544. doi: 10.1093/aob/mcr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. Evolutionary Biology Centre. Uppsala University; 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Zwickl D. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis. University of Texas at Austin, Section of Integrative Biology, Texas, USA; 2006. [Google Scholar]
- Ochiai T, Nakamura T, Mashiko Y, Fukuda T, Yokoyama J, Kanno A, Kameya T. The differentiation of sepal and petal morphologies in Commelinaceae. Gene. 2004;343:253–262. doi: 10.1016/j.gene.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Preston JC, Hileman LC. SQUAMOSA-PROMOTER BINDING PROTEIN 1 initiates flowering in Antirrhinum majus through activation of meristem identity genes. Plant J. 2010;62:704–712. doi: 10.1111/j.1365-313X.2010.04184.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editor. New York, NY: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Jackson D. In: Molecular Plant Pathology: a Practical Approach. Bowles DJ, Gurr SJ, McPherson M, editor. Oxford UK: Oxford University Press; 1991. In situ hybridization in plants; pp. 163–174. [Google Scholar]
- Preston JC, Kellogg EA. Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analyses. Plant J. 2007;52:69–81. doi: 10.1111/j.1365-313X.2007.03209.x. [DOI] [PubMed] [Google Scholar]
- Endress PK. Evolution of flower symmetry. Curr Opin Plant Biol. 2001;4:86–91. doi: 10.1016/S1369-5266(00)00140-0. [DOI] [PubMed] [Google Scholar]
- Hardy CR, Stevenson DW. Floral organogenesis in some species of Tradescantia and Callisia (Commelinaceae) Int J Plant Sci. 2000;161:551–562. doi: 10.1086/314279. [DOI] [Google Scholar]
- Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development. 2004;131:915–922. doi: 10.1242/dev.00993. [DOI] [PubMed] [Google Scholar]
- Hsu H-F, Yang C-H. An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol. 2002;43:1198–1209. doi: 10.1093/pcp/pcf143. [DOI] [PubMed] [Google Scholar]
- Xu Y, Teo LL, Zhou J, Kumar PP, Yu H. Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 2006;46:54–68. doi: 10.1111/j.1365-313X.2006.02669.x. [DOI] [PubMed] [Google Scholar]
- Chang YY, Kao NH, Li JY, Hsu WH, Liang YL, Wu JW, Yang CH. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 2010;152:837–853. doi: 10.1104/pp.109.147116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC. Evolutionary genetics of core eudicot inflorescence and flower development. Int J Plant Dev Biol. 2010;4:17–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for RT-PCR. Primer sequences used for amplification of target and housekeeping genes from Commelina communis, C. dianthifolia and Tradescantia pallida are listed in table format.
Second biological replicate for relative quantitative gene expression in dissected flower tissues. In agreement with the first biological replicate (Figure 5), GLO expression is significantly higher in dorsal staminodes versus ventral stamens of Commelina communis, but is similar across inner tepals of both Tradescantia pallida and C. communis (left). DEF is again undetectable in ventral, but not dorsal, inner tepals of C. communis. By contrast, DEF expression is higher in ventral versus dorsal inner tepals of Tradescantia pallida (second from left). Similar to the first biological replicate, TB1a is undetectable in T. pallida inflorescence tissues, and is expressed significantly more strongly in both outer and inner ventral versus dorsal tepals of C. communis (second from right). TB1b is undetectable in inflorescence organs of C. communis, although the primers amplify genomic DNA, and is undetectable in inner tepals and gynoecia of T. pallida (right). β-ACT and EF1a were used as internal controls. Experiments were conducted with three to four technical replicates. Bars show mean and standard deviation. White bars, whole organs; blacks bars, dorsal organs; gray bars, ventral organs. Asterisks denote significant differences between dorsal and ventral organ comparisons. gDNA, genomic DNA; Inf, whole inflorescence; Dot, dorsal outer tepal; Vot, ventral outer tepal; Dit, dorsal inner tepal; Vit, ventral inner tepal; Dst, dorsal stamens/staminodes; VSt, ventral stamens; Gyn, gynoecia; nd, not detected.
Best maximum likelihood phylogeny of monocot B-class genes. Maximum likelihood bootstrap values above 69% (left) and Bayesian posterior probabilities above 89% (right) are shown for each branch. Asterisks denote 100% support. Dashes indicate bootstrap support below 70% only when the corresponding posterior probability is shown. The focal genes of this study are highlighted in gray boxes.