Abstract
We report an approach to the rapid release of DNA based on the application of electrochemical potentials to surfaces coated with polyelectrolyte-based thin films. We fabricated multilayered polyelectrolyte films (or ‘polyelectrolyte multilayers’, PEMs) using plasmid DNA and a model hydrolytically degradable cationic poly(β-amino ester) (polymer 1) on stainless steel substrates using a layer-by-layer approach. The application of continuous reduction potentials in the range of -1.1 to -0.7 V (vs. a Ag/AgCl electrode) to film-coated electrodes in PBS at 37 °C resulted in the complete release of DNA over a period of 1-2 minutes. Film-coated electrodes incubated under identical conditions in the absence of applied potentials required 1-2 days for complete release. Control over the magnitude of the applied potential provided control over the rate at which DNA was released. The results of these and additional physical characterization experiments are consistent with a mechanism of film disruption that is promoted by local increases in pH at the film/electrode interface (resulting from electrochemical reduction of water or dissolved oxygen) that disrupt ionic interactions in these materials. The results of cell-based experiments demonstrated that DNA was released in a form that remains intact and able to promote transgene expression in mammalian cells. Finally, we demonstrate that short-term (i.e., non-continuous) electrochemical treatments can also be used to promote faster film erosion (e.g., over 1-2 h) once the potential is removed. Past studies demonstrate that PEMs fabricated using polymer 1 can promote surface-mediated transfection of cells and tissues in vitro and in vivo. With further development, the electrochemical approaches reported here could thus provide new methods for the rapid, triggered, or spatially patterned transfer of DNA (or other agents) from surfaces of interest in a variety of fundamental and applied contexts.
Keywords: Layer-by-Layer, Thin Films, DNA, Rapid Release, Electrochemical Methods
Introduction
Materials that provide control over the release of DNA from surfaces are important in a variety of contexts, ranging from the development of new research tools to the development of gene-based therapies. Methods for the immobilization of DNA on surfaces provide straightforward ‘physical’ approaches to defining the locations at which DNA is made available (e.g., by local delivery to cells residing in the vicinity of a film-coated implant or to cells growing on patterned features in an assay plate, etc.).1-4 Many different approaches to the immobilization of DNA on surfaces5-8 and the encapsulation of DNA in polymer-based films, coatings, and matrices2,6,9-13 have been developed for this purpose. Much of the success with which these approaches can be translated for use in many applied contexts, however, will also depend upon the ability to exert simultaneous (and often variable) levels of control over the timing with which DNA is released (that is, to develop mechanisms that also allow for temporal control). For example, although sustained release may be desirable for some applications, others may require methods that can be used to promote rapid transfer or the triggered/on-demand release of DNA and other agents. Although many materials can promote sustained release of DNA from the surfaces of implants and interventional devices, the development of materials platforms that permit rapid or triggered release remains a general challenge. The work reported here takes a step toward addressing this broader goal by developing electrochemical approaches that can be used to promote the rapid release of plasmid DNA from surfaces coated with ultrathin polyelectrolyte-based films.
The approach reported here exploits methods for the ‘layer-by-layer’ fabrication of thin polyelectrolyte-based films (called ‘polyelectrolyte multilayers’, or PEMs) on surfaces.14,15 This aqueous-based approach can be used to design films using a broad range of different polyelectrolytes, including charged proteins, viruses, and DNA.16-22 In the context of drug delivery, these methods offer several practical advantages for the encapsulation and release of therapeutic agents. These advantages include: (i) precise, nanometer-scale control over film thickness and drug loading (by control over the number of layers incorporated), (ii) control over the relative locations of individual layers in a film (permitting the design of hierarchical films and films that can be used to release multiple agents),23-27 and (iii) the ability to fabricate thin films on surfaces with complex shapes.3,20-22 Provided that they can be constructed in ways that permit subsequent disassembly, PEMs also provide a unique platform for control over the release of macromolecular agents that are otherwise too large to be released by diffusion. Different approaches to promoting film disruption have been reviewed recently,3,4,18,20-22 and include (i) films that respond to changes in environmental conditions (e.g., pH or ionic strength),28-31 (ii) films fabricated using hydrolytically-,23,32,33 enzymatically-,34-36 and reductively-degradable37-39 polyelectrolytes, and (iii) films that respond to the application of external stimuli (e.g., light, electrochemical potentials, etc.).40-45
Several groups have demonstrated that the approaches outlined above can be used to design PEMs that promote the release of DNA.3 Work in our group has focused on the design of PEMs fabricated using plasmid DNA and hydrolytically degradable cationic poly(β-amino ester)s such as polymer 1.33,46,47 We and others have demonstrated that films fabricated using DNA and polymer 1 (referred to hereafter as ‘polymer 1/DNA films’) erode in aqueous environments and promote the release of DNA.33,46-52 These studies have also demonstrated that these materials can be used to promote the localized and surface-mediated delivery of DNA to cells and tissues in vitro46,47 and in vivo.50,52
Polymer 1.

In general, polymer 1/DNA films erode and release DNA gradually when incubated in physiologically relevant media [e.g., over a period of ∼2-4 days when incubated in phosphate-buffered saline (pH= 7.4) at 37 °C].33,47 We have also reported the design of films that exhibit more extended release profiles (e.g., over weeks or months),25,26,53,54 but we have, in general, found it difficult to design PEMs that erode and release DNA more rapidly (e.g., over seconds or minutes). As a first step toward the design of faster-releasing films, we recently reported that incorporation of layers of poly(acrylic acid) (a pH-dependent weak polyelectrolyte) into polymer 1/DNA films leads to films that release DNA over 3-6 hours in physiologically relevant media.55 Other groups have reported PEMs that can be induced to undergo rapid, triggered disassembly to release DNA or oligonucleotides upon exposure to chemical reducing agents (e.g., by contact of films containing disulfide functionality with thiol-based reducing agents).38,39,56 This approach is well-suited to affecting the intracellular release of DNA or in other situations where chemical reducing agents (e.g., glutathione) are present. This current investigation sought to develop approaches that could be used to promote the rapid release of DNA in response to an externally applied stimulus (e.g., the application of an electrochemical potential) that would not otherwise be present in biological environments and would be unlikely to cause adverse effects to neighboring cells or tissues.
This investigation builds upon the results of past studies demonstrating that the application of electrochemical potentials can be used to disrupt or deconstruct PEMs. In general, these approaches fall into one of two categories. The first approach involves the fabrication of multilayers using electro-active molecules as film components; the application of oxidation or reduction potentials to these films changes the redox-states of these components and, as a result, leads to changes in intramolecular interactions in the films.42,45,57,58 The second approach is based on the creation of local changes in pH near the surfaces of film-coated electrodes by (i) electrochemical oxidation of water (which results in the production of hydronium ions)43,44,59-62 or (ii) electrochemical reduction of water or dissolved oxygen (which results in the production of hydroxide ions).63 This second approach does not require the use of electro-active film components, and it is useful for promoting the electrochemically-triggered dissolution of PEMs with components that respond to increases or decreases in pH. For example, electrochemically-induced decreases in local pH near electrodes have been used to promote the dissolution or disruption of PEMs fabricated from poly(lysine)/heparin,43 as well as films fabricated from poly(lysine) and fish sperm DNA,60 and avidin and an imminobiotin-labeled polymer.44 Other studies have demonstrated that electrochemically-induced increases in local pH can be used to dissolve or deconstruct hydrogen-bonded multilayers fabricated from poly(vinylpyrrolidone) and tannic acid.63
Here, we report that electrochemically induced changes in pH can be used to accelerate dramatically the erosion of PEMs fabricated using transcriptionally active plasmid DNA. We demonstrate that the application of reduction potentials to stainless steel electrodes coated with polymer 1/DNA films results in the complete release of DNA over periods ranging from seconds to several minutes (as opposed to several days in the absence of applied potentials). We demonstrate further that the rate of release can be tuned by adjusting the magnitude of the reduction potential applied to the electrode, and that the DNA that is released under these conditions remains transcriptionally active and able to promote transgene expression in mammalian cells. Our results are consistent with a mechanism of release that involves the localized generation of elevated pH near electrode surfaces. We note that a recent study reported the electron transfer-mediated release of DNA from multilayers fabricated from redox-active zirconium ions.45 Our current approach provides a method for the release of DNA that (i) does not involve electron-transfer to the film itself (or, ultimately, even direct contact of the film with an electrode), and (ii) exploits multilayer structures that promote the simultaneous release64 of DNA and a cationic polymer (polymer 1) used in several past studies to deliver DNA to cells.46,52,65,66 With further development, this approach could therefore lead to new methods for the rapid transfer or patterned delivery of DNA to cells and tissues of interest in a range of fundamental and applied contexts.
Materials and Methods
Materials
Linear poly(ethylene imine) (LPEI, MW = 25000) was purchased from Polysciences, Inc. (Warrington, PA). Poly(sodium 4-styrenesulfonate) (SPS, MW = 70000) was purchased from Aldrich Chemical Company (Milwaukee, WI). All commercial polyelectrolytes were used as received without further purification. Polymer 1 (Mn ∼10,000) was synthesized as previously described.65 Stainless steel sheets (SS 304, 75 μm thick) were obtained from Trinity Brand Industries (Atlanta, GA). Gold-coated glass substrates were prepared by depositing thin layers of titanium (10 nm) and gold (200 nm) sequentially onto clean glass substrates using an electron-beam evaporator (CHA Industries, Fremont, CA). Plasmid DNA encoding green fluorescent protein [gwiz-GFP (5757bp), >95% supercoiled] was purchased from Aldevron (Fargo, ND). Solutions of sodium acetate buffer (Accugene, Rockland, ME) and phosphate-buffered saline (PBS) (EM Science, Gibbstown, NJ) were prepared by diluting commercially available concentrate. Deionized water (18 MΩ) was used for all rinsing steps during film fabrication, the preparation of polymer and DNA solutions, and dilution of concentrated buffer solutions.
General Considerations
The surfaces of stainless steel and gold-coated glass substrates (∼2.5 cm × 0.7 cm) used for the fabrication of multilayered films were prepared by rinsing with acetone, ethanol, methanol, and deionized water and then drying under a stream of filtered air. All buffers and polymer solutions (with the exception of solutions of DNA) were filtered through a 0.2 μm membrane syringe filter prior to use. The amount of DNA released from PEMs during incubation in PBS was quantified by recording UV/vis absorbance values at a wavelength of 260 nm (corresponding to the absorbance maximum of double-stranded DNA) using a DU 520 UV/vis spectrophotometer (Beckman Coulter, Fullerton, CA). COS-7 cells used for in vitro transfection experiments were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Ellipsometric thicknesses of films deposited on silicon and gold-coated glass substrates were determined using a Gaertner LSE Stokes ellipsometer (632.8 nm, incident angle = 70°). Thicknesses were measured in at least five locations and data were processed using the Gaertner Ellipsometer Measurement Program software package to calculate relative thicknesses by assuming an average refractive index of 1.58 for the multilayered films. Prior to ellipsometeric measurements, the films were dried with filtered, compressed air using a 0.4 μm membrane syringe filter. Atomic force microscopy images were obtained in tapping mode using a Nanoscope Multimode atomic force microscope (Digital Instruments, Santa Barbara, CA), using scan rates 5-7 of μm/s to obtain 256 × 256 pixel images. Silicon cantilevers with a spring constant of 40 N/m and a radius of curvature of less than 10 nm were used (model NSC15/NoA1 MicroMasch, USA, Inc., Portland, OR) and images were processed using the NanoScope® III version 5.31R1 software package (Digital Instruments, Santa Barbara, CA). Height data were flattened using a 1st-order fit. Fluorescence microscopy images were acquired using an Olympus IX70 microscope and Metavue version 7.1.2.0 software package (Molecular Devices, Toronto, Canada). Images were analyzed using ImageJ 1.43u (National Institutes of Health; Washington, DC) and Photoshop CS5 (Adobe Systems; San Jose, CA). Experiments involving linear scan voltammetry were performed at a scan rate of 50 mV/sec in a three-electrode cell (i.e., planar stainless steel substrate, platinum wire, and Ag/AgCl reference electrode) to determine the electrochemical responses of stainless steel substrates. Potential scans were performed from 0.2 V down to -1.4 V.
Preparation of Polyelectrolyte Solutions
Solutions of LPEI and SPS used for the fabrication of LPEI/SPS base layers (20 mM with respect to molecular weight of the polymer repeat unit) were prepared using a 13 mM NaCl solution in deionized water (18 MΩ). Solutions of LPEI contained 10 mM HCl to aid polymer solubility. Solutions of polymer 1 (5 mM with respect to molecular weight of the polymer repeat unit) were prepared in sodium acetate buffer (100 mM, pH = 4.9). Solutions of plasmid DNA were prepared at a concentration of 1 mg/mL in sodium acetate buffer (100 mM, pH = 4.9).
Fabrication of Multilayered Films
Prior to the fabrication of polymer 1/DNA films, substrates were pre-coated with a multilayered film composed of 10 bilayers of LPEI/SPS, as previously described.33 Polymer 1/DNA layers were then fabricated on these precursor layers using the following general protocol: (1) substrates were submerged in a solution of polymer 1 for 5 min, (2) substrates were removed and immersed in a wash bath of 100 mM sodium acetate buffer for 1 min followed by a second wash bath for 1 min, (3) substrates were submerged in a solution of plasmid DNA for 5 min, and (4) substrates were rinsed as described for step (2). This cycle was repeated until 8 bilayers of polymer 1/DNA were deposited. Following the final rinse step, the substrates were dried under a stream of filtered air.
Characterization of Film Erosion
Electrochemical experiments were performed using a bipotentiostat (Pine Instruments, Grove City, PA) and a three-electrode cell to maintain a constant potential between the working electrode and a Ag/AgCl reference electrode. For most experiments, planar stainless steel substrates coated with polymer 1/DNA films were used as a working electrode and a platinum wire was used as the counter electrode. The electrolyte solution used was PBS (pH = 7.4, 137 mM NaCl, 2.7 mM KCl, and 10 mM phosphate buffer) maintained at 37 °C. Experiments designed to promote erosion of the films in response to the application of electrochemical potentials were performed in the following general manner: film-coated electrodes were placed in a beaker containing 3.5 mL of PBS. A reduction potential was then applied to the film-coated working electrode for multiple different pre-determined periods of time. At each time point, a 200 μL aliquot of solution was extracted and saved for: (i) analysis of the concentration of DNA released into solution (by measuring the absorbance of the buffer solution at 260 nm), (ii) characterization of the structure of released DNA (using agarose gel electrophoresis), and (iii) investigation of the transcriptional viability of released DNA (in cell-based transfection assays). At the end of the electrochemical dissolution experiments, electrodes were further incubated in PBS (pH = 7.4, at 37 °C) for 48 hours in the absence of potential to exhaustively release any remaining DNA and determine the total amount of DNA released from the films. Values for the percentage of DNA released reported in the text were calculated by dividing the amount of DNA released at each time point during electrochemical erosion experiments by these total amounts of DNA.
Agarose Gel Electrophoresis Assays
Samples of plasmid DNA collected during film erosion experiments were characterized by loading 20 μL of plasmid solution into a 1% agarose gel (HEPES, 20 mM, pH = 7.2; 70 V, 2 h). Samples of DNA were loaded in the gel using a loading solution consisting of 50:50 glycerol:water (v/v). DNA bands were stained using ethidium bromide and visualized using a transluminator.
Cell Transfection Assays
COS-7 cells (American Type Culture Collection, Manassas, VA) were seeded into 96-well plates at a density of 15,000 cells/well in 200 μL of Dulbecco's modified Eagle medium supplemented with 10% fetal serum, 100 units/mL of penicillin, and 100 μg/mL of streptomycin. Cells were grown for 24 h, at which time the medium was aspirated and replaced with fresh medium, and 50 μL of a Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and plasmid DNA mixture was added directly to the cells according to the general protocol provided by the manufacturer. The Lipofectamine 2000/plasmid DNA mixture was prepared by mixing 25 μL of the plasmid DNA solution collected at each time point during release experiments (arbitrary concentrations but constant volumes) with 25 μL of diluted Lipofectamine 2000 (24 μL stock diluted into 976 μL of Opti-MEM I Reduced Serum Medium). Fluorescence microscopy images used to characterize the expression of EGFP were acquired 48 h after the addition of lipoplexes.
Results and Discussion
Layer-by-Layer Assembly of DNA-Containing Films on Planar Stainless Steel Substrates
All studies described below were conducted using PEMs composed of eight bilayers (or ‘layer pairs’) of polymer 1 and a plasmid DNA construct encoding enhanced green fluorescent protein (EGFP). Films were fabricated on electrically conducting planar stainless steel substrates using a previously established alternating dipping procedure.33 We selected stainless steel substrates for this study (as opposed to other commonly used conducting materials such as indium-tin oxide (ITO) glass) in view of a recent study by our group on the design of PEM-coated stainless steel microneedle arrays for the delivery of DNA to skin.51 To provide surfaces suitable for fabrication of DNA-containing films, substrates were precoated with 10 bilayers of linear poly(ethylene imine) (LPEI) and poly(sodium 4-styrenesulfonate) (SPS).33,47 Plasmid DNA encoding EGFP was selected to facilitate characterization of released DNA in cell-based reporter gene assays (described below). Finally, we also fabricated otherwise identical films on the surfaces of silicon and gold-coated glass substrates for use as controls. Characterization of films fabricated on these reflective surfaces using ellipsometry revealed the thicknesses of polymer 1/DNA films eight bilayers thick to be ∼100 nm.33,67
Characterization of DNA Release Profiles Upon Application of Electrochemical Potentials
As described above, PEMs fabricated using polymer 1 and plasmid DNA erode physically and release DNA gradually (e.g., over a period of ∼2-4 days) upon incubation in PBS at 37 °C (pH 7.4).33,47 We conducted a series of experiments to investigate the influence of applied electrochemical potentials on the release profiles of films fabricated on stainless steel substrates (used in these studies as the working electrode). For this study, we chose to investigate reducing potentials, which should generate OH− ions and increase the pH at the working electrode, as opposed to oxidizing potentials (which should generate H+ ions). This decision was based on the results of initial screening experiments investigating the influence of pH on the erosion of polymer 1/DNA films (performed by incubating film-coated substrates in solutions of differing pH). These studies revealed that the release of DNA occurred much more rapidly (e.g., over ∼5 h) under alkaline conditions than in acidic media (e.g., see Figure S1 of the Supporting Information). In a more general context, we note that approaches based on elevated pH are also potentially more attractive because while exposure to alkaline media can result in reversible denaturation of double stranded DNA, exposure to strongly acidic media is associated with other forms of irreversible degradation.68
For the experiments reported in this paper, substrates coated with polymer 1/DNA films were submerged in PBS (pH = 7.4, 137 mM NaCl, 2.7 mM KCl, and 10 mM phosphate buffer) maintained at 37 °C, and a constant reduction potential was applied for a pre-determined period of time to generate OH− ions and increase the pH at the film/electrode interface. The electrochemical response of the stainless steel substrate in PBS was determined using linear sweep voltammetry (LSV). These measurements revealed the reduction of dissolved oxygen to take place at a potential interval between -0.3 V and -1 V, and the reduction of water to occur at potentials lower than -1.0 V (see Figure S2 of the Supporting Information for additional details). Based on these results, we chose to investigate the voltage-dependent rates of erosion of our films at three different reduction potentials that result in the generation of OH− ions: at -0.7 V and -0.9 V (both in the oxygen reduction region) and at -1.1 V (in the water reduction region). As control experiments for many of the studies outlined below, we also analyzed the influence of potentials of lower magnitude (e.g., -0.2 V) at which no oxygen or water reduction takes place (and, thus, no pH increase should occur).
We visually confirmed the reduction reactions and accompanying formation of OH− ions at the selected voltages (i.e., -0.7, -0.9 and -1.1 V) by performing experiments in PBS containing the pH indicator phenolphthalein. Observation of the formation of a pink color near the surfaces of bare stainless steel substrates confirmed the generation of a high pH environment (e.g., pH >8) in the vicinity of the electrode surface (see Figure S3 of the Supporting Information). The application of a potential of -0.2 V did not result in the formation of pink color near the film-coated surface, confirming that a high pH environment is not generated at this potential. We note here that the application of reduction potentials to stainless steel substrates can, under certain conditions, result in reduction of the thin passivating oxide films present on the surface.69 Control experiments revealed that application of potentials of -0.7, -0.9 and -1.1 V to bare stainless steel substrates for short periods of time (e.g., up to 3 min, the longest time scale used in experiments described below) did not result in significant increases in the absorbance of the PBS solutions at wavelengths ranging from 200 to 400 nm.
Figure 1 shows DNA release profiles for stainless steel substrates coated with polymer 1/DNA films incubated in PBS in the absence of an applied potential (Figure 1A) or in the presence of an applied potential (Figure 1B) as a function of time. Inspection of the data in Figure 1A reveals that, in the absence of an applied potential, polymer 1/DNA films coated on these substrates release DNA gradually over a period of ∼24-48 h. These results are consistent with those reported in our past studies for polymer 1/DNA films on a variety of different substrates.33,47,51,52 Inspection of the data in Figure 1B, however, reveals that the release of DNA occurred much more rapidly (e.g., in approximately 1-2 minutes) when reduction potentials were applied to the substrates.
Figure 1.

Plots showing the release of DNA from polymer 1/DNA films (A) in the absence of an applied electrochemical potential, and (B) in the presence of applied electrochemical potentials of -1.1 V (●), -0.9 V (○), -0.7 V (▲), and -0.2 V (■). Error bars represent the standard deviation of the calculated percentages of DNA released obtained from multiple samples collected from experiments using two different film-coated electrodes for each condition. In some cases, error bars are smaller than the symbols used to represent the data.
Inspection of the data in Figure 1B also reveals that release rates varied with the magnitude of the applied potential. For example, whereas the application of a potential of -1.1 V resulted in complete (∼100%) release of DNA after only 1 minute (Figure 1B, closed circles), complete release of DNA from films incubated at a potential of -0.9 V (open circles) required 2 minutes. In contrast, the release of DNA from films incubated at a potential of -0.7 V (closed triangles) was only ∼60% complete after 3 minutes. These results suggest that film erosion and DNA release profiles can be tuned by modulation of the electrochemical potentials applied to the substrates on which the films are deposited. Otherwise identical experiments in which a potential of -0.2 V was applied resulted in only negligible amounts of DNA release over this time period (Figure 1B, closed squares). As described above, neither oxygen nor water reduction takes place at this potential. This result thus suggests that changes in pH play an important role in promoting the rapid release at potentials of higher magnitude (i.e., -0.7, -0.9 and -1.1 V).
To provide additional visual evidence of the complete release of DNA from these film-coated surfaces, we soaked the films in ethidium bromide (a small-molecule DNA-intercalator used for fluorescence-based characterization of DNA).70,71 Figure 2 shows representative fluorescence microscopy images of an ethidium bromide-soaked film before incubation and erosion (Figure 2A) and after the application of a potential of -1.1 V for 1 minute (Figure 2B). The absence of significant fluorescence in the image in Figure 2B is consistent with the exhaustive release of DNA from the surface of this substrate.
Figure 2.
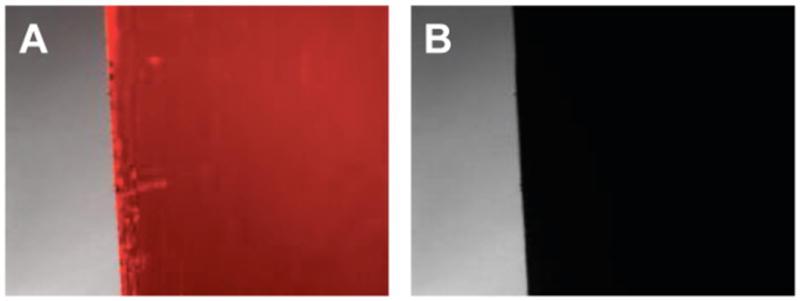
Fluorescence microscopy images (40× magnification; 730 μm × 510 μm) of the edge of a stainless steel substrate coated with a polymer 1/DNA film 8-bilayers thick, incubated in an aqueous ethidium bromide solution that intercalates DNA and labels it with a red color (A) before electrochemically-induced erosion, and (B) after applying a potential of -1.1 V for 1 min in PBS.
Past reports have demonstrated that PEMs fabricated using other materials can be disrupted by the application of electrochemical potentials that lead to local changes in pH (and the influx of counterions) sufficient to disrupt ionic interactions and promote film disassembly.43,44,59-63 Other studies have demonstrated that, at certain potentials, the generation of hydrogen gas can produce bubbles at PEM-coated electrodes that can also contribute to physical film disruption,63 and that, in certain cases, complete delamination of intact films can also occur.62 Our pH indicator-based results, discussed above, demonstrate that the range of reduction potentials used in this study is sufficient to generate OH− ions near our film-coated electrodes. In general, the extent to which OH− ions are generated should vary with the magnitude of the applied reduction potential, with the lowest potential used here (-1.1 V; at which the reduction of water takes place) leading to a higher concentration of OH− ions than those generated at -0.7 or -0.9 V (at which reduction of oxygen occurs). We note, however, that the application of -1.1 V also resulted in the evolution of bubbles of hydrogen gas (observed visually) that could also contribute to film disruption. The faster rates of DNA release at potentials of -1.1 V observed in Figure 1B could therefore be a consequence of both a higher concentration of OH− ions (and the attendant influx of counterions) and the evolution of hydrogen gas at the electrode/film surface. However, the formation of hydrogen should not occur (and was not observed) at the potentials of lower magnitude (e.g., at -0.7 or -0.9 V) used to generate the results in Figure 1B. The observation of rapid DNA release at these less-negative potentials suggests that the formation of gas bubbles is not solely responsible for faster film erosion (i.e., that changes in pH and resulting ion flux – in the absence of the generation of gas bubbles – are sufficient to promote rapid film erosion). Finally, we note also that we did not observe large-scale disruption or the physical delamination of films under any of the conditions used in this study.
Control experiments using otherwise identical films fabricated on gold-coated glass slides also eroded rapidly in the presence of applied potentials (e.g., release was ∼95% complete after 30 sec when a potential of -1.1 V was applied; this potential generates approximately the same current density on gold-coated glass as it does on our stainless steel substrates; see Figure S4 of the Supporting Information for additional details). These results demonstrate that any possible reduction of the passivating oxide film on the surface of the stainless steel substrates under the conditions used here (as discussed above) does not play a critical role in promoting film erosion or the release of DNA. More broadly, these results also demonstrate that this approach can be used to promote the release of DNA from films on electrodes fabricated from other types of conducting materials. The application of oxidizing potentials of +1.1 V to films on these gold-coated substrates (i.e., to promote the generation of acidic pH) did not result in rapid and complete release of DNA (only ∼35 % of DNA was released over three minutes). These results are consistent with those of our solution-based experiments (see Figure S1 and additional discussion above) demonstrating that alkaline pH environments generally lead to much faster film erosion than acidic environments. These control experiments were performed on inert gold-coated substrates, rather than on stainless steel substrates, because stainless steel electrodes oxidize rapidly upon the application of a potential of +1.1 V.
Finally, we mention that the majority of past studies on electrochemically-induced disruption of PEMs have been conducted using films fabricated using polyelectrolytes that either (i) do not degrade (e.g., hydrolyze) in aqueous media, or (ii) are presumed to hydrolyze slowly enough that film erosion can be understood broadly in terms of disruptions of internal ionic crosslinks within the films.43-45,59-63 In contrast to these past studies, however, it is also important to note that the cationic polymer used in this study (polymer 1; a cationic polyester) is also hydrolytically degradable.65 Past studies have demonstrated that hydrolysis plays an important role in promoting the erosion of PEMs fabricated using this polymer (e.g., in PBS, in the absence of applied potential).72,73 In addition to the potential for pH-induced disruption of ionic interactions in these films, it is therefore also possible that the generation of high-pH environments near film-coated electrodes could promote more rapid film disassembly through a mechanism that also involves more rapid polymer backbone hydrolysis.
Physical Characterization of Changes in Film Morphology
We used atomic force microscopy (AFM) to characterize changes in the surface morphologies of polymer 1/DNA films and provide additional insight into factors that could influence film erosion upon the application of electrochemical potentials. Figure 3 shows representative AFM images (5 μm × 5 μm) of polymer 1/DNA films eight bilayers thick before erosion (A) and after incubation in PBS at a potential of -1.1 V for 5 seconds (B) or a potential of -0.7 V for 30 seconds (C). The image in part A reveals the presence of a film that is rough (RMS roughness generally varied from ∼20 nm to ∼100 nm) but continuous over the surface of the substrate prior to incubation. This result is generally consistent with those of our past studies;46,73 increased film roughness observed here likely results from the increased roughness of the underlying stainless steel substrates (e.g., relative to the flatter silicon substrates used in our past studies).
Figure 3.
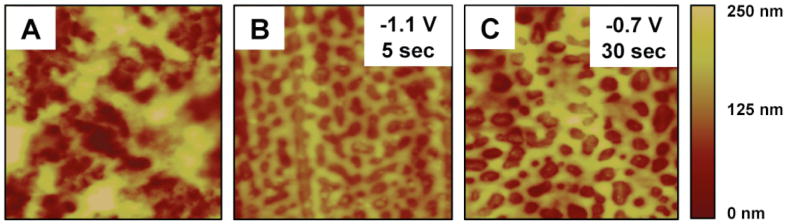
AFM images of polymer 1/DNA films (5 μm × 5 μm) (A) before erosion, (B) after 5 sec of erosion at -1.1 V in PBS, and (C) after 30 sec of erosion at -0.7 V in PBS. The scale in the z-direction, shown at the far right, is 250 nm.
Inspection of the images in parts B and C of Figure 3, however, reveals significant changes in surface structure and a film morphology characterized by the presence of a more topographically complex pattern of valleys and ridges (∼30-60 nm high). This film morphology is similar in many respects to those observed during the erosion and decomposition of polymer 1/DNA films in PBS in the absence of applied potentials, which we have characterized extensively in past studies using AFM, scanning electron microscopy, and other analytical methods.73,74 That past work provided a framework for understanding these changes in film structure as arising from changes in pH (as opposed to changes that occur as a result of polymer hydrolysis, etc.) that reduce ionic crosslinking and permit the transport of film components over larger distances.73,74 The observation of these changes in morphology in this current study upon the application of potentials that increase the pH near film-coated electrodes is also consistent with this physical picture. More generally, these observations reveal that although DNA release occurs much more rapidly upon the application of reduction potentials (e.g., Figure 1B), film erosion nevertheless appears to occur through an overall mechanism that is similar to that observed on the surfaces of other non-conducting substrates. (In this context, we also note that these AFM results also provide additional evidence that the release of DNA measured in Figure 1B is not the result of large-scale delamination or physical disruption, as discussed above.)
Characterization of Released DNA
We performed agarose gel electrophoresis assays to characterize DNA released from polymer 1/DNA films and determine the extent to which the application of reduction potentials could potentially damage DNA. Figure 4A shows the results of an experiment using DNA collected at four different time points (after 30 sec, 1 min, 2 min, and 3 min; the lane labeled C shows a sample of supercoiled DNA used as a control) during the erosion of a film using a potential of -1.1 V. Qualitative inspection of the intensities of the bands suggests that DNA release is essentially complete after a period of 1 minute (consistent with the quantitative data shown in Figure 1B, above). These data also reveal that the majority of the DNA released from these films is present as open-circular DNA, rather than supercoiled DNA (and that a small fraction is present as linear DNA). We note that this distribution of plasmid topologies is similar to that observed for films incubated in the absence of potential (which also release DNA in a predominantly open-circular form)33,48,52 and thus does not appear to result from the potentials used in these current experiments.
Figure 4.

(A) Agarose gel electrophoresis characterization of samples of DNA released from stainless steel substrates coated with polymer 1/DNA films and incubated with an applied potential of -1.1 V. Lane labels indicate the time at which samples were collected for analysis. The lane labeled ‘C’ corresponds to a pEGFP control. (B) Representative fluorescence micrographs (100× magnification; 1194 μm × 895 μm) of confluent monolayers of COS-7 cells showing levels of EGFP expression mediated by samples of released DNA collected after the electrochemically promoted erosion of polymer 1/DNA films (at an applied potential of -1.1 V) for 30 sec, 1 min, 2 min, and 3 min.
Additional characterization of PBS solutions of supercoiled DNA exposed to a potential of -1.1 V (using a bare stainless steel substrate as the working electrode) revealed no large changes in plasmid topology when potentials were applied for up to one minute (although samples incubated for longer times did exhibit significantly higher concentrations of open-circular DNA; see Figure S5 of the Supporting Information). While the reduction potential used above does not appear to result in degradation of released DNA (as revealed by gel electrophoresis), the potential for other forms of degradation must be borne in mind with respect to the future development of this approach (for example, prolonged exposure to high pH can result in reversible denaturation of DNA, and electron transfer processes resulting from the physical contact with electrodes could result in the reduction of DNA bases). In this latter context, we note that our current results (e.g., Figure 1B) demonstrate that it should be possible to optimize or adapt this approach further by the use of potentials of lower magnitude. Further, the development of mechanisms of film disassembly that are based on changes in pH in the vicinity of an electrode – as opposed to electron transfer processes promoted by direct contact of the film with an electrode – should also render it feasible to increase the distance between these films and the electrode (e.g., by increasing the number of LPEI/SPS precursor layers between the electrode and the DNA-containing layers) if electron transfer processes are ultimately found to lead to other forms of degradation. Finally, in the context of this current study, we note that open-circular plasmid DNA remains transcriptionally active. The results of additional studies described below demonstrate that DNA released upon the application of a reduction potential is capable of promoting transgene expression in mammalian cells.
Figure 4B shows representative low-magnification fluorescence microscopy images of mammalian fibroblast cells (COS-7 cells) treated with lipoplexes formed using a commercially-available transfection agent and samples of DNA released from a film incubated at a potential of -1.1 V. The images correspond to cells treated with lipoplexes formed using DNA collected at 30 sec, 1 min, 2 min, and 3 min (generated using a procedure identical to that used to perform the gel electrophoresis experiments, above). These images reveal significant numbers of cells expressing EGFP (visible as cells exhibiting green fluorescence) and demonstrate that the DNA released from these films remains transcriptionally viable. The results of similar experiments performed using DNA collected from films eroded completely in the absence of an applied potential (e.g., over a period of 48 hours) are included in Figure S6 of the Supporting Information and were similar to those reported in our past studies.33,51
Influence of the Application of Electrochemical Potentials for Short Periods
The results of the experiments described above demonstrate that the continuous application of reduction potentials to film-coated electrodes can be used to accelerate film erosion dramatically. We note, however, that some applications for which rapid release of DNA may be useful may not be tolerant of electrochemical potentials (or such potentials may, for various practical reasons, be difficult or impossible to apply). We conducted a final series of experiments to determine whether initial short-term (rather than continuous) exposure of film-coated substrates to electrochemical potentials could be used to influence the rate of release of DNA after the potential was removed. Such an approach could potentially be used to ‘pre-treat’ or prepare film-coated substrates that erode more rapidly than ‘conventional’ polymer 1/DNA films when they are used in the absence of applied potentials.
Figure 5 shows a plot of the release of DNA from a film-coated stainless steel substrate identical to those used in experiments described above that was (i) incubated in PBS at an applied potential of -1.1 V for only 5 seconds, and then (ii) transferred to and incubated further in fresh buffer in the absence of an applied potential. Inspection of these data reveals an initial burst of release (corresponding to ∼30% of the total amount of DNA in the film) over the 5-second period during which the potential was applied. Further inspection, however, also reveals that the release of the remaining DNA occurs much more rapidly (e.g., over a period of ∼2.5 h) after the potential is removed than the release of DNA films that were never exposed to an initial reduction potential (e.g., Figure 1A; release over ∼1-2 days). Additional characterization will be necessary to understand the origins of this behavior more completely. However, this more accelerated release could result from the persistence of a high pH environment inside these films after the potential is removed (and is likely a consequence of the rapid changes in film structure that occur upon the initial application of reduction potentials; e.g., Figure 3B-C). We performed a final set of experiments in which film-coated electrodes were ‘pre-treated’ by the application of potential for 5 seconds, removed immediately from PBS, rinsed, and then dried. In contrast to the results shown in Figure 5, the reintroduction of these pre-treated films to solutions of PBS, in the absence of applied potentials, did not result in rapid release of DNA (e.g., DNA was released completely over a period of ∼18 hours; see Figure S7 of the Supporting Information).
Figure 5.
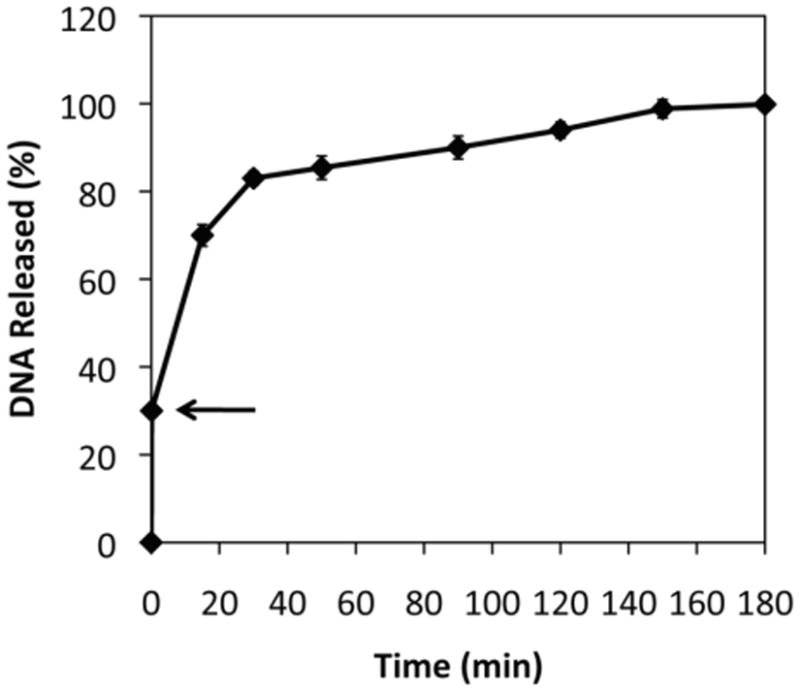
Plot showing the release of DNA from polymer 1/DNA films incubated at an applied potential of -1.1 V for 5 sec and then subsequently incubated in PBS in the absence of an applied potential (see text). The arrow indicates the point at which the potential was removed. Error bars show the standard deviation of the percentage of DNA released using multiple measurements of samples during experiments using two different film-coated electrodes.
Additional optimization of these approaches could lead to practical methods for the in situ treatment of film-coated surfaces to promote rapid or pulsatile-type release in environments for which it is difficult (or undesirable) to apply potentials continuously for prolonged periods. In this context, however, we note that while the approaches reported here lead to rapid and accelerated film erosion, our results do not constitute ideal examples of ‘triggered’ release (that is, these polymer 1/DNA films are not completely stable in aqueous media, and do erode, albeit much more slowly, in the absence of applied reduction potentials). The results of other studies on the electrochemical disruption of otherwise ‘stable’ PEMs,43-45,57-63 however, suggest that it should be possible to use this approach to trigger the onset of disruption of films fabricated using plasmid DNA and other, non-degradable cationic polymers (such as poly(ethylene imine) (PEI), etc.). Studies to this end are currently underway.
Summary and Conclusions
We have reported an electrochemical approach to the rapid release of plasmid DNA from surfaces coated with DNA-containing thin films. Our results demonstrate that applied potentials in the range of -1.1 to -0.7 V (vs. a Ag/AgCl electrode) that generate local high-pH environments near the surfaces of electrodes can be used to disrupt PEMs fabricated from plasmid DNA and a model hydrolytically degradable cationic polymer. This approach leads to the release of DNA from film-coated electrodes over periods ranging from 1-2 minutes (as opposed to 1-2 days for films incubated in the absence of applied potentials). Our results demonstrate that the magnitudes of continuously applied potentials can be used to influence the rate at which DNA is released, and that DNA is released in a form that remains able to promote transgene expression when administered to mammalian cells in vitro. The results of our release studies, and those of additional physical characterization experiments, are consistent with a mechanism of film disruption that involves the rapid, pH-induced disruption of ionic interactions in these materials. Finally, this work demonstrates that films exposed to reduction potentials for short periods of time as a pretreatment, as opposed to continuous application, also erode rapidly (e.g., over 1-2 h rather than 1-2 days) once the potential is removed. With further development, these electrochemical approaches could provide new methods for the rapid, triggered, or spatially patterned transfer of DNA (or other agents) from film-coated objects to cells and tissues of interest in a variety of fundamental studies and applied contexts.
Supplementary Material
Acknowledgments
Financial support was provided in part by the National Institutes of Health (R01 EB006820) and the Alfred P. Sloan Foundation. We thank Adam H. Broderick for assistance with AFM experiments and for many helpful discussions.
Footnotes
Supporting Information Available: Results of additional experiments showing pH-dependent DNA release profiles, a linear scan voltammogram of a stainless steel substrate, DNA release profiles of films fabricated on gold-coated glass substrates, additional gel electrophoresis data, and other results as identified in the main text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Takahashi H, Letourneur D, Grainger DW. Biomacromolecules. 2007;8:3281–3293. doi: 10.1021/bm700540p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Laporte L, Shea LD. Adv Drug Delivery Rev. 2007;59:292–307. doi: 10.1016/j.addr.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jewell CM, Lynn DM. Adv Drug Delivery Rev. 2008;60:979–999. doi: 10.1016/j.addr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zelikin AN. ACS Nano. 2010;4:2494–2509. doi: 10.1021/nn100634r. [DOI] [PubMed] [Google Scholar]
- 5.Segura T, Shea LD. Bioconjugate Chem. 2002;13:621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 6.Fishbein I, Stachelek SJ, Connolly JM, Wilensky RL, Alferiev I, Levy RJ. J Controlled Release. 2005;109:37–48. doi: 10.1016/j.jconrel.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 7.Wang CHK, Jiang S, Pun SH. Langmuir. 2010;26:15445–15452. doi: 10.1021/la1025203. [DOI] [PubMed] [Google Scholar]
- 8.Blocker KM, Kiick KL, Sullivan MO. Langmuir. 2011;27:2739–2746. doi: 10.1021/la104313z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shea LD, Smiley E, Bonadio J, Mooney DJ. Nat Biotechnol. 1999;17:551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- 10.Klugherz BD, Jones PL, Cui XM, Chen WL, Meneveau NF, DeFelice S, Connolly J, Wilensky RL, Levy RJ. Nat Biotechnol. 2000;18:1181–1184. doi: 10.1038/81176. [DOI] [PubMed] [Google Scholar]
- 11.Shen H, Tan J, Saltzman WM. Nat Mater. 2004;3:569–574. doi: 10.1038/nmat1179. [DOI] [PubMed] [Google Scholar]
- 12.Pannier AK, Anderson BC, Shea LD. Acta Biomater. 2005;1:511–522. doi: 10.1016/j.actbio.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Putnam D. Nat Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 14.Decher G. Science. 1997;277:1232–1237. [Google Scholar]
- 15.Bertrand P, Jonas A, Laschewsky A, Legras R. Macromol Rapid Commun. 2000;21:319–348. [Google Scholar]
- 16.Ai H, Jones SA, Lvov YM. Cell Biochem Biophys. 2003;39:23–43. doi: 10.1385/CBB:39:1:23. [DOI] [PubMed] [Google Scholar]
- 17.Peyratout CS, Dahne L. Angew Chem Int Ed. 2004;43:3762–3783. doi: 10.1002/anie.200300568. [DOI] [PubMed] [Google Scholar]
- 18.Tang Z, Wang Y, Podsiadlo P, Kotov NA. Adv Mater. 2006;18:3203–3224. [Google Scholar]
- 19.Angelatos AS, Katagiri K, Caruso F. Soft Matter. 2006;2:18–23. doi: 10.1039/b511930h. [DOI] [PubMed] [Google Scholar]
- 20.De Geest BG, Sanders NN, Sukhorukov GB, Demeester J, De Smedt SC. Chem Soc Rev. 2007;36:636–649. doi: 10.1039/b600460c. [DOI] [PubMed] [Google Scholar]
- 21.Lynn DM. Adv Mater. 2007;19:4118–4130. [Google Scholar]
- 22.Boudou T, Crouzier T, Ren K, Blin G, Picart C. Adv Mater. 2010;22:441–467. doi: 10.1002/adma.200901327. [DOI] [PubMed] [Google Scholar]
- 23.Wood KC, Chuang HF, Batten RD, Lynn DM, Hammond PT. Proc Natl Acad Sci U S A. 2006;103:10207–10212. doi: 10.1073/pnas.0602884103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jessel N, Oulad-Abdeighani M, Meyer F, Lavalle P, Haikel Y, Schaaf P, Voegel JC. Proc Natl Acad Sci U S A. 2006;103:8618–8621. doi: 10.1073/pnas.0508246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Montañez SI, Jewell CM, Lynn DM. Langmuir. 2007;23:11139–11146. doi: 10.1021/la702021s. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Zhang J, Lynn DM. Adv Mater. 2008;20:4148–4153. doi: 10.1002/adma.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer F, Dimitrova M, Jedrzejenska J, Arntz Y, Schaaf P, Frisch B, Voegel JC, Ogier J. Biomaterials. 2008;29:618–624. doi: 10.1016/j.biomaterials.2007.10.027. [DOI] [PubMed] [Google Scholar]
- 28.Dubas ST, Schlenoff JB. Macromolecules. 2001;34:3736–3740. [Google Scholar]
- 29.Schuler C, Caruso F. Biomacromolecules. 2001;2:921–926. doi: 10.1021/bm010052w. [DOI] [PubMed] [Google Scholar]
- 30.Sukhishvili SA, Granick S. Macromolecules. 2002;35:301–310. [Google Scholar]
- 31.Sukhishvili SA. Curr Opin Colloid Interface Sci. 2005;10:37–44. [Google Scholar]
- 32.Vázquez E, Dewitt DM, Hammond PT, Lynn DM. J Am Chem Soc. 2002;124:13992–13993. doi: 10.1021/ja026405w. [DOI] [PubMed] [Google Scholar]
- 33.Zhang JT, Chua LS, Lynn DM. Langmuir. 2004;20:8015–8021. doi: 10.1021/la048888i. [DOI] [PubMed] [Google Scholar]
- 34.Serizawa T, Yamaguchi M, Akashi M. Angew Chem Int Ed. 2003;42:1115–1118. doi: 10.1002/anie.200390293. [DOI] [PubMed] [Google Scholar]
- 35.Picart C, Schneider A, Etienne O, Mutterer J, Schaaf P, Egles C, Jessel N, Voegel JC. Adv Funct Mater. 2005;15:1771–1780. [Google Scholar]
- 36.Ren KF, Ji J, Shen JC. Bioconjugate Chem. 2006;17:77–83. doi: 10.1021/bc050264g. [DOI] [PubMed] [Google Scholar]
- 37.Zelikin AN, Quinn JF, Caruso F. Biomacromolecules. 2006;7:27–30. doi: 10.1021/bm050832v. [DOI] [PubMed] [Google Scholar]
- 38.Blacklock J, Handa H, Manickam DS, Mao G, Mukhopadhyay A, Oupicky D. Biomaterials. 2007;28:117–124. doi: 10.1016/j.biomaterials.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Huang SW, Lin WH, Zhuo RX. Small. 2007;3:636–643. doi: 10.1002/smll.200600301. [DOI] [PubMed] [Google Scholar]
- 40.Radt B, Smith TA, Caruso F. Adv Mater. 2004;16:2184–2189. [Google Scholar]
- 41.Skirtach AG, Antipov AA, Shchukin DG, Sukhorukov GB. Langmuir. 2004;20:6988–6992. doi: 10.1021/la048873k. [DOI] [PubMed] [Google Scholar]
- 42.DeLongchamp DM, Hammond PT. Adv Funct Mater. 2004;14:224–232. [Google Scholar]
- 43.Boulmedais F, Tang CS, Keller B, Vörös J. Adv Funct Mater. 2006;16:63–70. [Google Scholar]
- 44.Sato K, Kodama D, Naka Y, Anzai Ji. Biomacromolecules. 2006;7:3302–3305. doi: 10.1021/bm060819q. [DOI] [PubMed] [Google Scholar]
- 45.Wang F, Li D, Li G, Liu X, Dong S. Biomacromolecules. 2008;9:2645–2652. doi: 10.1021/bm800766t. [DOI] [PubMed] [Google Scholar]
- 46.Jewell CM, Zhang JT, Fredin NJ, Lynn DM. J Controlled Release. 2005;106:214–223. doi: 10.1016/j.jconrel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 47.Jewell CM, Zhang J, Fredin NJ, Wolff MR, Hacker TA, Lynn DM. Biomacromolecules. 2006;7:2483–2491. doi: 10.1021/bm0604808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saurer EM, Jewell CM, Kuchenreuther JM, Lynn DM. Acta Biomater. 2009;5:913–924. doi: 10.1016/j.actbio.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su X, Kim BS, Kim SR, Hammond PT, Irvine DJ. ACS Nano. 2009;3:3719–3729. doi: 10.1021/nn900928u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeMuth PC, Su X, Samuel RE, Hammond PT, Irvine DJ. Adv Mater. 2010;22:4851–4856. doi: 10.1002/adma.201001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saurer EM, Flessner RM, Sullivan SP, Prausnitz MR, Lynn DM. Biomacromolecules. 2010;11:3136–3143. doi: 10.1021/bm1009443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saurer EM, Yamanouchi D, Liu B, Lynn DM. Biomaterials. 2011;32:610–618. doi: 10.1016/j.biomaterials.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Lynn DM. Adv Mater. 2007;19:4218–4223. [Google Scholar]
- 54.Sun B, Lynn DM. J Controlled Release. 2010;148:91–100. doi: 10.1016/j.jconrel.2010.07.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flessner RM, Yu Y, Lynn DM. Chem Commun. 2011;47:550–552. doi: 10.1039/c0cc02926b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zelikin AN, Li Q, Caruso F. Angew Chem Int Ed. 2006;45:7743–7745. doi: 10.1002/anie.200602779. [DOI] [PubMed] [Google Scholar]
- 57.Wood KC, Zacharia NS, Schmidt DJ, Wrightman SN, Andaya BJ, Hammond PT. Proc Natl Acad Sci U S A. 2008;105:2280–2285. doi: 10.1073/pnas.0706994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt DJ, Moskowitz JS, Hammond PT. Chem Mater. 2010;22:6416–6425. doi: 10.1021/cm102578j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guillaume-Gentil O, Akiyama Y, Schuler M, Tang C, Textor M, Yamato M, Okano T, Vörös J. Adv Mater. 2008;20:560–565. [Google Scholar]
- 60.Dieguez L, Darwish N, Graf N, Vörös J, Zambelli T. Soft Matter. 2009;5:2415–2421. [Google Scholar]
- 61.Guillaume-Gentil O, Abbruzzese D, Thomasson E, Vörös J, Zambelli T. ACS Appl Mater Interfaces. 2010;2:3525–3531. doi: 10.1021/am1007062. [DOI] [PubMed] [Google Scholar]
- 62.Guillaume-Gentil O, Graf N, Boulmedais F, Schaaf P, Vörös J, Zambelli T. Soft Matter. 2010;6:4246–4254. [Google Scholar]
- 63.Schmidt DJ, Hammond PT. Chem Commun. 2010;46:7358–7360. doi: 10.1039/c0cc02346a. [DOI] [PubMed] [Google Scholar]
- 64.Bechler SL, Lynn DM. Biomacromolecules. 2012;13:542–552. doi: 10.1021/bm2016338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynn DM, Langer R. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 66.Lynn DM, Anderson DG, Akinc AB, Langer R. In: Polymeric Gene Delivery: Principles and Applications. Amiji M, editor. CRC Press; New York: 2004. pp. 227–241. [Google Scholar]
- 67.Fredin NJ, Zhang JT, Lynn DM. Langmuir. 2005;21:5803–5811. doi: 10.1021/la050596+. [DOI] [PubMed] [Google Scholar]
- 68.Jelen F, Fojta M, Paleček E. J Electroanal Chem. 1997;427:49–56. [Google Scholar]
- 69.Piao T, Park SM. J Electrochem Soc. 1997;144:3371–3377. [Google Scholar]
- 70.Lvov Y, Decher G, Sukhorukov G. Macromolecules. 1993;26:5396–5399. [Google Scholar]
- 71.Lang J, Lin MH. J Phys Chem B. 1999;103:11393–11397. [Google Scholar]
- 72.Zhang J, Fredin NJ, Lynn DM. J Polym Sci Part A: Polym Chem. 2006;44:5161–5173. [Google Scholar]
- 73.Fredin NJ, Zhang J, Lynn DM. Langmuir. 2007;23:2273–2276. doi: 10.1021/la0624182. [DOI] [PubMed] [Google Scholar]
- 74.Fredin NJ, Flessner RM, Jewell CM, Bechler SL, Buck ME, Lynn DM. Microsc Res Tech. 2010;73:834–844. doi: 10.1002/jemt.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


