Introduction
Gap junctions (GJ) are comprised of clusters of intercellular channels that electrotonically and metabolically couple apposing cells. Each channel is formed from the pairing of two connexons (or hemi-channels), which in turn are formed by the assembly of six connexin monomers. The mammalian genome comprises almost two dozen individual connexin genes, and a surprising number of these genes are actively transcribed in the developing or mature heart. The evolutionary advantage for cardiac gap junction diversity remains uncertain. Here, we consider the nature of this diversity and speculate on potential explanations.
Connexin Diversity in the Heart
There are at least five different connexins expressed in the adult mammalian heart (Table I). These include members of the alpha (Cx43 and Cx40), beta (Cx37, Cx30.2 in mouse or its human orthologue Cx31.9) and gamma (Cx45) families. All encode proteins with high degrees of structural homology, including four membrane-spanning helices and two extracellular domains. There is significant divergence in their intracellular segments, particularly in their carboxy terminal domains, which may contribute to the distinct biophysical properties of gap junction channels formed from each connexin.
Table 1. The murine connexin gene family.
Displayed for each connexin is its chromosomal location, number of amino acids, cardiac expression pattern, channel characteristics and cardiac phenotype of murine knockout.
| Connexin | Gene | Chr | AA | Expression | gj (pS) | Knockout phenotype |
|---|---|---|---|---|---|---|
| Cx30.2 | Gjd3 | 11 | 278 | SAN, AVN | 92 | impaired delay in AV nodal conduction14 |
| Cx37 | Gja4 | 4 | 333 | EC | 30022 | female sterility23 |
| Cx40 | Gja5 | 3 | 358 | A, VCS, EC | 16224 | arrhythmias, BBB, congenital defects7 |
| Cx43 | Gja1 | 10 | 382 | A, V | 60–10025 | RV outflow tract malformations26 |
| Cx45 | Gja7 | 11 | 395 | SAN, AVN, VCS | 3227 | endocardial cushion defects6 |
Chr, chromosome; AA, amino acid residues; A, atrial cardiomyocytes; V, ventricular cardiomyocytes; SAN, SA nodal cells; AVN, AV nodal cells; VCS, ventricular conduction system; EC, vascular endothelial cells; gj, unitary channel conductance in picosiemens; BBB, bundle branch block; RV, right ventricle.
Why such diversity? The primary function of the heart is to support the circulation as a dynamic pump – one that is dependable yet highly responsive to acute and chronic changes in metabolic demand. The four-chamber heart and the specialized nodal cells and rapidly conducting His-Purkinje network embedded within the heart have evolved to meet this challenge. Given the functional specialization of individual subsystems that are integrated into the organ as a whole (rate regulation by the nodes, blood collection in the atria; rapid myocardial activation through the His-Purkinje system; blood ejection in the ventricles), one might expect that each component would express connexins with appropriate gap junction channel properties.
Is the nature of connexin diversity consistent with these disparate functional requirements, indicative of strong evolutionary pressure? Indeed, connexin isotype expression in the heart appears well designed to support not only the unique functional attributes of each compartment, but also, the particular complexities that arise at interfaces between individual compartments (see Figure 1).
Figure 1.
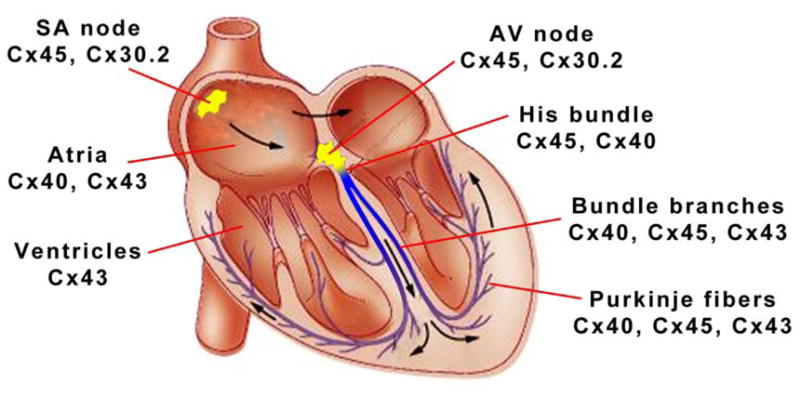
Connexin expression profiles in the adult murine heart.
We begin with the SA node, where both theoretical and experimental studies have demonstrated that pacemaker function requires a population of weakly coupled intrinsically rhythmic cells apposed to a second population of well-coupled cardiomyocytes. Not unexpectedly, we find that the SA node includes a core of pacemaker cells expressing low levels of Cx45 and Cx30.2, each of which encode highly voltage-sensitive, small conductance gap junctional channels1. In contrast, as one moves toward the periphery of the SA node into the so-called “paranodal area”, there is a heterogeneous mixture of myocytes, some of which express Cx431. In the atrium proper, both Cx43 and Cx40 are found. Within the AV node, Cx45 and Cx30.2 (at least in the murine heart) are arranged to create low-conductance GJ2. This expression pattern contributes to the slowing of conduction that is responsible for the delay between atrial and ventricular activation. In contrast, maximal pressure development within the pumping chambers of the heart is dependent upon rapid and highly synchronized triggering of the ventricular myocardium by strongly coupled cells within the His-Purkinje network, and in fact, high conductance Cx40 channels are robustly expressed in the specialized ventricular conduction system. Finally, the ventricular myocardium itself is highly coupled through abundant expression of Cx43. These channels provide an intermediate gating sensitivity and conductance that is best suited for widespread local propagation of the cardiac action potential.
The interface between the Purkinje fiber network and the ventricular myocardium presents a particularly intriguing example of structural and functional specialization. Here, it is believed that heterotypic channels are formed by Cx40 connexons in the Purkinje cells and Cx43 connexons in the ventricular cardiomyocytes. The differential voltage-dependence of Cx40 and Cx43 connexons provides for rectification, supporting antegrade flow of current down the specialized cardiac conduction system (CCS) across the Purkinje-ventricular (PV) junction into the ventricular myocardium, while at the same time providing a degree of protection against retrograde conduction and reentrant arrhythmias3. The difficulty associated with this balancing act may explain why the PV junction is thought to be a common element in various arrhythmic syndromes4.
Connexin Diversity During Heart Development
Expression of different connexin isotypes varies not only within distinct compartments of the adult heart, but also as a function of cardiac developmental stage. The major spatiotemporal expression patterns appear relatively conserved across mammalian species, as the embryonic mouse heart closely parallels that found in the embryonic human heart5. Cx45 is the earliest expressed connexin (E8.5 in mice), initially found in all cardiac compartments but ultimately restricted to primarily cells of the specialized CCS by E15.5. Cx40 is also expressed throughout the early developing heart, most prominently in the trabecular myocardium, but is downregulated in late fetal stages5. Cx43, while present throughout early cardiac development, increases in abundance during late gestation in both atrial and especially in working ventricular cardiomyocytes5.
Targeted mutagenesis studies in the mouse have revealed that each of these connexins are required for normal heart formation and function. Cx45−/− mice have endocardial cushion defects6. Cx40 knockout mice have abnormalities in cardiac conduction and arrhythmias and an increased incidence of congenital defects including pathologic hypertrophy, common atrioventricular junction or ventricular septal defects7. Germline Cx43 knockout mice die perinatally with heart defects involving the right ventricular outflow tract8. While cardiac-specific Cx43 knockout mice circumvent this congenital defect, they develop spontaneous lethal ventricular arrhythmias beginning around one month after birth9. The mechanisms supporting conduction in the absence of Cx43 are uncertain. It is conceivable that low level expression of alternative isoforms such as Cx45 provides adequate electrotonic coupling10. Alternatively, computational modeling and recent studies of sub-cellular localization of voltage-gated sodium channels suggest the intriguing possibility of a role for ephaptic propagation11–13. Mice lacking Cx30.2 do not display structural heart defects, but do have a shortened PR interval on the surface EKG and therefore lack the physiologic delay between atrial and ventricular contraction14. While these knockout studies demonstrate the requirement for multiple connexins during cardiac development, they do not provide much insight into underlying mechanism. Conceivably, channels comprised of specific connexins regulate the flow of critical morphogens within or between different compartments of the developing heart. In addition, non-channel properties of connexins, i.e., interaction with β-catenin or other binding partners may regulate key developmental processes such as cellular proliferation, apoptosis or differentiation15.
Regulatory Diversity
While each of the cardiac connexins form channels with distinct biophysical properties, the multitude of connexin genes provides for several additional levels of control, including transcriptional and post-translational regulation. For example, by virtue of sequence variation, individual connexins differ in their sensitivity to various kinases, acetylases, or other post-translational modifying enzymes, facilitating cell-type specific regulation by various signaling pathways. This form of regulation may provide a mechanism to rapidly and potently modulate coupling of populations of cells in response to physiologic cues or pathologic stimuli.
The surprisingly larger number of connexin genes may also provide a mechanism for fine-tuning connexin expression in stage- and lineage-specific manners through transcriptional control. The different connexin genes vary in their cis-acting regulatory regions, and consequently each can respond individually to the cell autonomous transcriptional pathways. For example, the T-box family transcription factor Tbx5 has been shown to activate the mouse Cx40 promoter, either alone or in concert with the homeoprotein Nkx2-516. Conversely, mouse hearts that overexpress Nkx2-5 have reduced expression of Cx4317. Further, Tbx2 (similar to Tbx5) has been shown to be a negative regulator of Cx43 expression18. Recently, the Iroquois homeobox protein Irx3 has been shown to repress Cx43 expression and conversely activate Cx40. Irx3 mutant mice display QRS widening and ectopic Cx43 expression in the proximal bundle branches19. Such findings offer a glimpse into the complex mechanisms responsible for the heterogeneity of connexin expression.
Connexins and Disease
Connexin diversity may also provide some protective advantage for the heart when challenged by pathologic stressors, although evidence supporting this hypothesis is currently lacking. Certainly “modern” diseases such as coronary artery disease that present after reproductive age would not impart evolutionary pressure, however it is conceivable that differential gating of connexins in response to adrenergic activation (i.e., as part of the fight-or-flight response or with acute hemorrhage), might provide some benefit. For example, expression of multiple connexins with differential sensitivity to protein kinase A dependent phosphorylation might allow for preservation of electrotonic coupling and successful impulse propagation while at the same time minimize diffusion of toxic metabolites from ischemic regions of the heart.
Looking to the future, it is conceivable that “designer” connexins with potenti anti-arrhythmic activity could be expressed in diseased hearts. Our laboratory has already demonstrated that a Cx43 mutant resistant to dephosphorylation in the carboxy terminus diminishes pathologic gap junction remodeling20. One can imagine fine tuning the structure of connexins to optimize their anti-arrhythmic efficacy.
Conclusion
The large number of connexin genes actively transcribed in the mammalian genome suggests that significant evolutionary pressure exists to maintain this high degree of complexity. Connexin diversity in the heart appears well designed to coordinate the initiation and propagation of the action potential from the sinus node to the ventricular myocardium. Moreover, the multitude of connexins expressed in the developing heart may facilitate the remarkable structural and functional plasticity that occurs as the heart matures from a simple linear peristaltic tube to a highly complex four-chambered pump. Finally, connexin diversity may provide some protection against pathologic stimuli.
What does the future hold for gap junction biologists? Despite significant progress, our understanding of the molecular determinants of the cardiac connexin life-cycle are still relatively immature. Further studies in this area will be essential to identify novel targets to manipulate connexin expression and function in the heart for therapeutic effect. In addition, the list of connexin-interacting proteins is expanding, but the biological significance of this interactome is in its infancy21. A multi-disciplinary strategy, channeling the efforts of a diverse assembly of investigators, with expertise in molecular, cellular and systems biology will be essential to unravel the nature of these interactions.
Acknowledgments
Research in the Fishman laboratory is supported by grants from the NIH (R01 HL82727, R01 HL105983, T32 HL098129) and the New York State Department of Health (NYSTEM N08G-132). We apologize to the many outstanding investigators whose work was not cited due to constraints of the Viewpoint format.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Monfredi O, Dobrzynski H, Mondal T, Boyett MR, Morris GM. The anatomy and physiology of the sinoatrial node--a contemporary review. Pacing Clin Electrophysiol. 2010 Nov;33:1392–1406. doi: 10.1111/j.1540-8159.2010.02838.x. [DOI] [PubMed] [Google Scholar]
- 2.Kreuzberg MM, Sohl G, Kim JS, Verselis VK, Willecke K, Bukauskas FF. Functional properties of mouse connexin30.2 expressed in the conduction system of the heart. Circulation research. 2005 Jun 10;96:1169–1177. doi: 10.1161/01.RES.0000169271.33675.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valiunas V, Weingart R, Brink PR. Formation of heterotypic gap junction channels by connexins 40 and 43. Circulation research. 2000 Feb 4;86:E42–49. doi: 10.1161/01.res.86.2.e42. [DOI] [PubMed] [Google Scholar]
- 4.Morley GE, Danik SB, Bernstein S, et al. Reduced intercellular coupling leads to paradoxical propagation across the Purkinje-ventricular junction and aberrant myocardial activation. Proceedings of the National Academy of Sciences of the United States of America. 2005 Mar 15;102:4126–4129. doi: 10.1073/pnas.0500881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coppen SR, Kaba RA, Halliday D, et al. Comparison of connexin expression patterns in the developing mouse heart and human foetal heart. Mol Cell Biochem. 2003 Jan;242:121–127. [PubMed] [Google Scholar]
- 6.Kumai M, Nishii K, Nakamura K, Takeda N, Suzuki M, Shibata Y. Loss of connexin45 causes a cushion defect in early cardiogenesis. Development. 2000 Aug;127:3501–3512. doi: 10.1242/dev.127.16.3501. [DOI] [PubMed] [Google Scholar]
- 7.Kirchhoff S, Kim JS, Hagendorff A, et al. Abnormal cardiac conduction and morphogenesis in connexin40 and connexin43 double-deficient mice. Circulation research. 2000 Sep 1;87:399–405. doi: 10.1161/01.res.87.5.399. [DOI] [PubMed] [Google Scholar]
- 8.Ewart JL, Cohen MF, Meyer RA, et al. Heart and neural tube defects in transgenic mice overexpressing the Cx43 gap junction gene. Development. 1997 Apr;124:1281–1292. doi: 10.1242/dev.124.7.1281. [DOI] [PubMed] [Google Scholar]
- 9.Gutstein DE, Morley GE, Tamaddon H, et al. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circulation research. 2001 Feb 16;88:333–339. doi: 10.1161/01.res.88.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada KA, Kanter EM, Green KG, Saffitz JE. Transmural distribution of connexins in rodent hearts. Journal of cardiovascular electrophysiology. 2004 Jun;15:710–715. doi: 10.1046/j.1540-8167.2004.03514.x. [DOI] [PubMed] [Google Scholar]
- 11.Kucera JP, Rohr S, Rudy Y. Localization of sodium channels in intercalated disks modulates cardiac conduction. Circulation research. 2002 Dec 13;91:1176–1182. doi: 10.1161/01.res.0000046237.54156.0a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori Y, Fishman GI, Peskin CS. Ephaptic conduction in a cardiac strand model with 3D electrodiffusion. Proceedings of the National Academy of Sciences of the United States of America. 2008 Apr 29;105:6463–6468. doi: 10.1073/pnas.0801089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin X, Liu N, Lu J, et al. Subcellular heterogeneity of sodium current properties in adult cardiac ventricular myocytes. Heart rhythm: the official journal of the Heart Rhythm Society. 2011 Jul 20; doi: 10.1016/j.hrthm.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreuzberg MM, Schrickel JW, Ghanem A, et al. Connexin30.2 containing gap junction channels decelerate impulse propagation through the atrioventricular node. Proceedings of the National Academy of Sciences of the United States of America. 2006 Apr 11;103:5959–5964. doi: 10.1073/pnas.0508512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin M. Isolation and community: a review of the role of gap-junctional communication in embryonic patterning. J Membr Biol. 2002 Feb 1;185:177–192. doi: 10.1007/s00232-001-0129-7. [DOI] [PubMed] [Google Scholar]
- 16.Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circulation research. 2002 Mar 22;90:509–519. doi: 10.1161/01.res.0000013072.51957.b7. [DOI] [PubMed] [Google Scholar]
- 17.Kasahara H, Ueyama T, Wakimoto H, et al. Nkx2.5 homeoprotein regulates expression of gap junction protein connexin 43 and sarcomere organization in postnatal cardiomyocytes. J Mol Cell Cardiol. 2003 Mar;35:243–256. doi: 10.1016/s0022-2828(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 18.Borke JL, Chen JR, Yu JC, Bollag RJ, Orellana MF, Isales CM. Negative transcriptional regulation of connexin 43 by Tbx2 in rat immature coronal sutures and ROS 17/2.8 cells in culture. Cleft Palate Craniofac J. 2003 May;40:284–290. doi: 10.1597/1545-1569_2003_040_0284_ntrocb_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 19.Zhang SS, Kim KH, Rosen A, et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proceedings of the National Academy of Sciences of the United States of America. 2011 Aug 16;108:13576–13581. doi: 10.1073/pnas.1106911108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remo BF, Qu J, Volpicelli FM, et al. Phosphatase-resistant gap junctions inhibit pathological remodeling and prevent arrhythmias. Circulation research. 2011 Jun 10;108:1459–1466. doi: 10.1161/CIRCRESAHA.111.244046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herve JC, Bourmeyster N, Sarrouilhe D, Duffy HS. Gap junctional complexes: from partners to functions. Progress in biophysics and molecular biology. 2007 May-Jun;94:29–65. doi: 10.1016/j.pbiomolbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Traub O, Hertlein B, Kasper M, et al. Characterization of the gap junction protein connexin37 in murine endothelium, respiratory epithelium, and after transfection in human HeLa cells. Eur J Cell Biol. 1998 Dec;77:313–322. doi: 10.1016/S0171-9335(98)80090-3. [DOI] [PubMed] [Google Scholar]
- 23.Simon AM, Goodenough DA, Li E, Paul DL. Female infertility in mice lacking connexin 37. Nature. 1997 Feb 6;385:525–529. doi: 10.1038/385525a0. [DOI] [PubMed] [Google Scholar]
- 24.Desplantez T, Dupont E, Severs NJ, Weingart R. Gap junction channels and cardiac impulse propagation. J Membr Biol. 2007 Aug;218:13–28. doi: 10.1007/s00232-007-9046-8. [DOI] [PubMed] [Google Scholar]
- 25.Fishman GI, Moreno AP, Spray DC, Leinwand LA. Functional analysis of human cardiac gap junction channel mutants. Proceedings of the National Academy of Sciences of the United States of America. 1991 May 1;88:3525–3529. doi: 10.1073/pnas.88.9.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reaume AG, de Sousa PA, Kulkarni S, et al. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995 Mar 24;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- 27.Desplantez T, Halliday D, Dupont E, Weingart R. Cardiac connexins Cx43 and Cx45: formation of diverse gap junction channels with diverse electrical properties. Pflugers Arch. 2004 Jul;448:363–375. doi: 10.1007/s00424-004-1250-0. [DOI] [PubMed] [Google Scholar]


