Summary
Agrobacterium tumefaciens incites plant tumors that produce nutrients called opines, which are utilized by the bacteria during host colonization. Various opines provide sources of carbon, nitrogen, and phosphorous, but virtually nothing was previously known about how A. tumefaciens acquires sulfur during colonization. Some strains encode an operon required for the catabolism of the opine octopine. This operon contains a gene, msh, that is predicted to direct the conversion of S-methylmethionine (SMM) and homocysteine (HCys) to two equivalents of methionine. Purified Msh carried out this reaction, suggesting that SMM could be an intermediate in opine catabolism. Purified octopine synthase (Ocs, normally expressed in plant tumors) utilized SMM and pyruvate to produce a novel opine, designated sulfonopine, whose catabolism by the bacteria would regenerate SMM. Sulfonopine was produced by tobacco and Arabidopsis when colonized by A. tumefaciens and was utilized as sole source of sulfur by A. tumefaciens. Purified Ocs also used thirteen other proteogenic and nonproteogenic amino acids as substrates, including three that contain sulfur. Sulfonopine and eleven other opines were tested for induction of octopine catabolic operon and all were able to do so. This is the first study of the acquisition of sulfur, an essential element, by this pathogen.
Keywords: S-methylmethionine, Sulfonopine, Agrobacterium, Opines
Introduction
Agrobacterium tumefaciens provides fascinating examples of the chemical ecology that underlies the interactions between plant-associated bacteria and their hosts (Brencic & Winans, 2005, Zhu et al., 2000, Ranocha et al., 2001). This bacterium uses a conjugation-like machinery to transfer discrete fragments of oncogenic DNA from the tumor-inducing (Ti) plasmid to the nuclei of host plants (Alvarez-Martinez & Christie, 2009, Gelvin, 2009, Tzfira & Citovsky, 2006). These DNA fragments are integrated into the host genome, and genes encoded within them are expressed in the plant nuclei. Several transferred genes direct the production of phytohormones such as auxin and cytokinin, which can lead to uncontrolled cell proliferation, producing a crown gall tumor (Kamada-Nobusada & Sakakibara, 2009, Tzfira & Citovsky, 2006). Other transferred genes direct the production of novel compounds called opines, which serve as nutrients for the colonizing bacteria (Savka et al., 2002, White & Winans, 2007).
Different strains of Agrobacterium collectively can direct plant tumors to synthesize a bewildering array of opines that can serve as sources of nitrogen, carbon or phosphorous (Dessaux et al., 1998). First, different strains transfer diverse types of opine biosynthetic genes. Second, each strain generally transfers more than one opine biosynthetic gene. Third, particular opine biosynthetic enzymes can sometimes utilize a variety of substrates. For example, A. tumefaciens strains that carry a so-called octopine type Ti plasmid, such as pTiA6, pTiB6, pTi15955, pTiR10, or pTiAch5, transfer the opine biosynthetic genes ocs, ags, mas1, and mas2. Ocs can reductively conjugate pyruvate with several different amino acids, including arginine, lysine, ornithine, histidine, methionine, and glutamine, synthesizing the corresponding opines (Menage & Morel, 1964, Menage & Morel, 1965, Biemann et al., 1960, Hack & Kemp, 1980, Otten et al., 1977). Five of these are utilized as sources of carbon and nitrogen, while the compound containing methionine was described as a pseudo-opine, as it was not utilized by the bacterium (Firmin et al., 1985). Mas1 and Mas2 conjugate mannose with glutamine, forming mannopine, which can be lactonized by Ags to form agropine or can spontaneously lactonize to form agropinic acid (Hong et al., 1997, Dessaux et al., 1988, Dessaux et al., 1986).
Opine catabolic operons are generally located on non-transferred portions of the Ti plasmids, and are transcriptionally induced by the opines whose catabolism they direct (Dessaux et al., 1998). These operons invariably include genes that encode ABC-type uptake systems and one or more genes that direct opine catabolism. For example, the octopine catabolism (occ) operon of octopine-type Ti plasmids contains 15 genes (Fig. 1A), and is activated by the product of the divergent occR gene in response to octopine and similar opines (Wang et al., 1992, Habeeb et al., 1991, von Lintig et al., 1991). This operon contains one confirmed (Valdivia et al., 1991) and one putative ABC-type permease (Fig. 1A) and six genes known or suspected to be involved in opine catabolism. One such catabolic gene, msh, is described below. The last gene in the operon is traR, which encodes a LuxR-type transcription factor that directs the transcription of Ti plasmid vegetative replication and conjugative transfer genes (Fuqua & Winans, 1996a, Piper et al., 1999). TraR functions only in the presence of 3-oxooctanoylhomoserine lactone (OOHL), whose synthesis is directed by TraI, also encoded on Ti plasmids (Fuqua & Winans, 1994, Piper et al., 1993). TraR expression requires expression of the occ operon (Fuqua & Winans, 1996b), and quorum sensing therefore occurs only within or near colonized plants, as they are the only natural source of these opines in the rhizosphere.
Fig. 1.
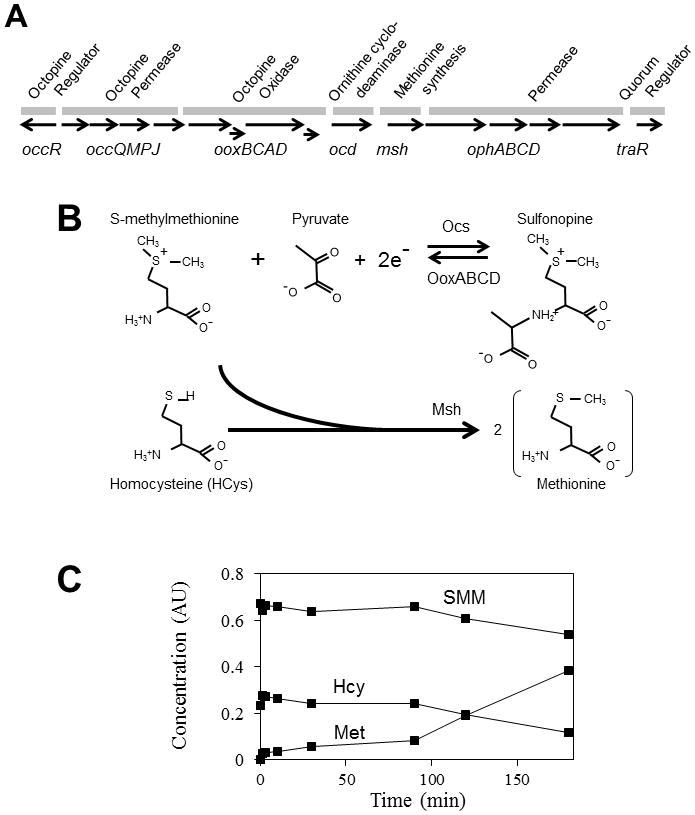
Proposed synthesis and catabolism of sulfonopine. (A) Genetic map of the octopine catabolic operon. The occQMPJ genes direct the uptake of octopine and probably related opines, while ooxBCAD direct the oxidative hydrolysis of octopine-type opines, yielding pyruvate and the corresponding amino acid. The ooxC and ooxD genes are provisional and based upon our DNA sequence analysis (data not shown). The ocd gene directs the conversion of ornithine to proline (Schindler et al., 1989). The msh resembles S-methylmethionine-homocysteine methyltransferase of E. coli. The remaining five genes constitute an ABC-type permease for an unknown substrate, and the traR gene, which directs quorum-dependent Ti plasmid conjugation and vegetative replication. OccR positively regulates the operon in the presence of octopine-type opines. (B) Proposed synthesis of sulfonopine in plant cells by Ocs and its metabolism in bacterial cells by OoxABCD (top). SMM and HCys, undergo transmethylation by Msh, yielding Met (bottom). (C) Purified Msh can convert SMM and HCys to two equivalents of Met via a methyltransferase activity.
The msh gene encodes a protein having a limited similarity to proteins involved in methionine biosynthesis, hence the mnemonic methionine synthase homolog (Fuqua & Winans, 1996b). However, this homology appeared to be limited to the amino and carboxyl termini. In this study, we describe an end-to-end homolog of Msh, and confirm that Msh, like its homolog, is a methyltransferase that converts S-methylmethionine (SMM) and homocysteine (HCys) to methionine. We also show that a T-DNA encoded protein, octopine synthase (Ocs), can provide a substrate for Msh by synthesizing a novel opine containing SMM, and can also utilize a broad variety of other amino acids, including three that contain sulfur. The ability of Ocs to derivatize one or more sulfur-containing amino acids provides a new perspective on nutrient acquisition during plant colonization.
Results
Msh converts SMM and HCys to methionine
As described above, the predicted Msh protein was previously found to have a limited sequence similarity to a family of proteins involved in methionine biosynthesis. Simply by decreasing the gap penalty of the BLAST algorithm (Altschul et al., 1990), we detected an end-to-end sequence similarity between Msh and the MmuM protein of E. coli (Fig. S1). MmuM transfers a methyl group from SMM to HCys to yield two molecules of methionine (Met) (Thanbichler et al., 1999, Neuhierl et al., 1999). If Msh were to carry out a similar reaction, it would suggest that SMM and/or HCys might be an intermediate in the catabolism of a previously undescribed opine. We hypothesized that one or both of these hypothetical new opines could be synthesized in crown gall tumors by Ocs (Fig. 1B), and could be converted back to the corresponding amino acids by octopine oxidase (Fig. 1B). These amino acids could then be converted by Msh to Met.
To test these ideas, we wanted first to determine whether Msh could convert SMM and HCys to two equivalents of Met. We purified recombinant His6-Msh and quantitated reactants and products by electrospray ta ndem mass spectrometry (ESI-MS/MS). As predicted, Msh used SMM and HCys as substrates to produce methionine (Fig. 1C). Msh also carried out similar reactions with lower efficiency with three other methyl donors in place of SMM (Table 1). These properties are similar to those of MmuM (Thanbichler et al., 1999, Neuhierl et al., 1999), suggesting that these two enzymes are functionally similar.
Table 1.
Msh catalytic specificity for methyl donor substratesa.
| Methyl Donor | km (mM) | kcat (min−1) | Enzymatic efficiency (kcat/km) | Relative activity (%) |
|---|---|---|---|---|
| SMM | 3.7 ± 2b | 114 | 31 | (100) |
| Methylcobalamin | 17 ± 8 | 14 | 0.8 | 2.7 |
| Dimethylglycine | 21 ± 8 | 2.4 | 0.114 | 0.4 |
| SAM | 18 ± 4 | 0.7 | 0.04 | 0.12 |
| Betaine | >1.3 ± 0.6c | <0.04 c | >0.03 c | >0.9 c |
Production of Met from homocysteine (10 mM) and the indicated methyl donors was carried out by using 0.61 μM His6-Msh in 20 mM Tris buffer (pH 7.9). Met production was analyzed by ESI-MS/MS.
Mean ± SD of n=3 enzymatic reactions.
Detection limit.
Octopine synthase utilizes SMM and HCys as substrates
The model proposed above predicts that SMM and/or HCys could be derived from hypothetical opines, possibly synthesized by the Ocs protein that is expressed within crown gall tumors. In order to purify Ocs, we constructed a gene encoding an MBP-His6-Ocs fusion protein that contains a cleavage site for TVMV protease between MBP and the His6 tag. This fusion was co-expressed with TVMV protease to remove the MBP portion of the fusion (Donnelly et al., 2006). We added an additional plasmid that overexpresses the chaperone proteins GroESL, as this caused a significant increase in His6-Ocs solubility (data not shown). His6-Ocs protein was purified to apparent homogeneity by sequential immobilized nickel affinity and gel filtration chromatography.
Enzymatic reactions were prepared containing Ocs, pyruvate, NADPH, and either SMM or arginine (Arg), and the reaction was monitored by using UV spectroscopy to detect the conversion of NADPH to NADP+. Both amino acids stimulated NADPH oxidation (Table 2). We confirmed using ESI-MS/MS and NMR that reactions containing arginine produced octopine (N-(R-1-carboxyethyl)-arginine), while reactions containing SMM produced a novel compound, N-(R-1-carboxyethyl)-S-methyl-S-methionine (Fig. S2, S3B, S4) which we designate sulfonopine. The enzymatic efficiency (kcat/km) for SMM was similar to that for arginine, indicating that Ocs utilized SMM with high efficiency. Similar reactions containing homocysteine produced yet another novel compound, (N-(R-1-carboxyethyl)-S-homocysteine, which in principle could be converted by octopine oxidase to homocysteine and pyruvate. The homocysteine thus produced could provide the second substrate for Msh.
Table 2.
Substrate specificity of octopine synthase for amino acids and α-keto acids.
| Amino Acid | α-keto acid | km (mM) | kcat (min-1) | Enzymatic Efficiency (kcat/km) | Relative activity (%) |
|---|---|---|---|---|---|
| L-Arginine | Pyruvate | 3.5 ± 0.3 | 720 | 210 | (100) |
| L-SMM | Pyruvate | 0.31 ± 0.04 | 49 | 160 | 76 |
| L-HCys | Pyruvate | 8.9 ± 1.1 | 1600 | 180 | 86 |
| L-Methionine | Pyruvate | 27 ± 6 | 4700 | 170 | 81 |
| L-Cysteine | Pyruvate | 11 ± 3 | 670 | 61 | 29 |
| L-Histidine | Pyruvate | 2.0 ± 0.21 | 1200 | 600 | 286 |
| L-Canavanine | Pyruvate | 2.8 ± 0.5 | 1000 | 360 | 171 |
| L-Ornithine | Pyruvate | 3.0 ± 0.4 | 540 | 180 | 86 |
| L-Lysine | Pyruvate | 0.74 ± 0.08 | 130 | 178 | 85 |
| L-Glutamine | Pyruvate | 5.6 ± 0.6 | 780 | 140 | 67 |
| L-Homoserine | Pyruvate | 15 ± 3 | 1800 | 120 | 57 |
| L-Glycine | Pyruvate | 30 ± 6 | 1300 | 43 | 20 |
| L-Homoarginine | Pyruvate | 11 ± 2 | 410 | 37 | 18 |
| L-Leucine | Pyruvate | 11 ± 2 | 410 | 37 | 18 |
| L-Serine | Pyruvate | 12 ± 2 | 220 | 18 | 9 |
| L-Alanine | Pyruvate | 47 ± 14 | 340 | 7.2 | 3.4 |
| L-Asparagine | Pyruvate | 41 ± 13 | 220 | 5.4 | 2.6 |
| L-Threonine | Pyruvate | 976 ± 30 | 4600 | 4.7 | 2.2 |
| L-Valine | Pyruvate | 36 ± 5 | 170 | 4.7 | 2.2 |
| L-Isoleucine | Pyruvate | 70 ± 30 | 180 | 2.6 | 1.2 |
| L-Glutamic acid | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| L-Aspartic acid | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| L-Tryptophan | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| L-Tyrosine | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| L-Phenylalanine | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| L-Proline | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| L-Carnosine | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| D-Arginine | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| D-Methionine | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| D-SMM | Pyruvate | >1.5 × 106 | <6.9 × 104 | <0.6 | <0.09 |
| Arginine | Pyruvate | 0.44 ± 0.07 | 561 | 1275 | (100) |
| Arginine | α-ketobutyrate | 6 ± 1 | 573 | 96 | 7.5 |
| Arginine | Oxaloacetate | 28 ± 4.5 | 1073 | 38 | 3 |
| Arginine | Glyoxylate | 22 ± 5 | 354 | 16 | 1.3 |
| Arginine | α-ketoglutarate | >1.5 e+6 α | <6.9 e+4 α | >0.6 α | >0.09 α |
Mean ± SD of n=3 enzymatic reactions.
Detection limit
To better understand Ocs substrate specificity, we compared all 20 proteinogenic amino acids as well as several nonproteinogenic ones. We also tested several α-keto acids for the ability to replace pyruvate in this reaction, and compared the utilization of NADPH and NADH. We confirmed that the enzyme can utilize lysine, ornithine, histidine, methionine, and glutamine. It also efficiently utilized canavanine, homoserine, cysteine, glycine, and homoarginine. Of these, histidine and canavanine were utilized even more efficiently than arginine. Ocs can also utilize other amino acids at reduced efficiencies (Table 2). The enzyme did not detectably utilize aromatic or acidic amino acids, nor did it utilize proline, carnosine, or any of three tested D-amino acids. The enzyme utilized α-ketobutyrate, oxaloacetate, and glyoxylate in place of pyruvate, although significantly less efficiently (Table 2). It did not detectably utilize α-ketoglutarate as a substrate. We confirmed an earlier report (Otten et al., 1977) that Ocs utilizes NADPH slightly more efficiently than NADH (data not shown).
Homogenates and exudates of plants colonized by A. tumefaciens contain sulfonopine
We set out to determine whether tobacco seedlings can synthesize and exude sulfur-containing opines when colonized by A. tumefaciens. To address this, tobacco seedlings were cultured hydroponically and inoculated using four different strains: R10 (wild type), KYC55 (which lacks a Ti plasmid), R10 ΔvirD4 (which cannot transfer T-DNA to plant cells), and R10 Δocs (which fails to produce octopine-type opines). After three days of cocultivation, the bacteria were killed by addition of carbenicillin and the seedlings were suspended in water. Two weeks later the water and seedling tissues were assayed by ESI-MS/MS for sulfur-containing amino acids and sulfur-containing opines.
The amino acid SMM and the corresponding opine, sulfonopine, were readily detected in tobacco exudates and homogenates (Table 3, Fig. S3D–E). We also looked for the presence of three other sulfur-containing amino acids, homocysteine, methionine, and cysteine, and their corresponding opines, but did not detect them (data not shown). Sulfonopine was not detected in mock-infected seedlings, nor in seedlings infected with a strain lacking a Ti plasmid, nor in seedlings infected with a strain lacking virD4 or ocs (Table 3). These results show that Ocs, when expressed in plant cells, can direct sulfonopine production, and that this opine is released to the rhizosphere, where it could be consumed by Agrobacteria (Savka et al., 1996). It is plausible that transformed plants can also synthesize some or all of the opines that we detected with the purified enzyme, but that the substrates and products were not sufficiently abundant to be detected using our methods.
Table 3.
SMM and sulfonopine content in tobacco tissue extracts and exudate of infected seedlings with A. tumefaciens R10 and mutants strains.
| Compound | Location | R10 | KYC55 | ΔvirD4 | Δocs | Mock infected |
|---|---|---|---|---|---|---|
| SMMa | Homogenate | 21±16a,c | 18±14 | 31 ±25 | 26 ±19 | 40±25 |
| Sulfonopinea | Homogenate | 5±0.6a,c | n. d.d | n. d. | n. d. | n. d. |
| Sulfonopineb | Exudate | 2 ±0.7b,c | n. d. | n. d. | n. d. | n. d. |
: nmolmg−1 of plant tissue
: nmol ml−1
: Mean ± SD of triplicate samples.
: ND. Not detected
We wanted to know whether the production of sulfonopine by plants is dependent on SMM. We therefore used Arabidopsis thaliana wild type (Col-0), and two mutant lines, one of which (hmt2-2) overexpresses SMM, while the other (mmt) does not synthesize SMM (Lee et al., 2008). Sulfonopine was produced by infected Col-0 and was produced at higher levels by infected hmt2-2 plants, but was not produced by infected mmt plants (Table 4, Fig. S3C). The concentration of SMM in the host therefore influences the production and accumulation of sulfonopine, consistent with SMM being a direct substrate for Ocs.
Table 4.
Sulfonopine is detected only in plants that produce SMM.
| Arabidopsis Strain | Sulfonopine Contect in Floral Tissue | ||
|---|---|---|---|
| Wild typea | mmtb | hmt2-2c | |
| Infected | 7 ± 1e,f | n. d. | 140 ± 90 |
| Noninfected | n. d.d | n. d. | n. d. |
: Wild type (Columbia ecotype).
: does not produce SMM.
: SMM hyperaccumulation.
: Not detected
: (nmol mg−1)
: Mean ± SD of triplicate samples.
Sulfonopine induces transcription of the octopine catabolism operon
At least some octopine-type opines induce the transcription of the occ operon via the LysR-type regulator OccR (Habeeb et al., 1991). To investigate whether the octopine-type opines found in exudates and extracts from infected-tobacco seedlings were able to induce transcription of the occ operon, we used the strain KYC16, which harbors an octopine-inducible ooxA-uidA fusion made by transposition of Tn5gusA7 (Cho et al., 1996). KYC16 was cultured in the presence of exudates or extracts from the infected-tobacco seedlings. Gus activity was elevated by exudates and extracts of tobacco seedlings infected with A. tumefaciens R10 (Fig. S5A–B).
We tested sulfonopine and ten other enzymatically synthesized opines (Table 2), including two that contain sulfur (derived from methionine or cysteine) for induction of the ooxA-uidA fusion. We used a commercial source of octopine as a positive control and the corresponding amino acids as negative controls. Each of these opines activated the fusion (Fig. 2A), while the corresponding amino acids did not (Fig. S5C).
Fig. 2.
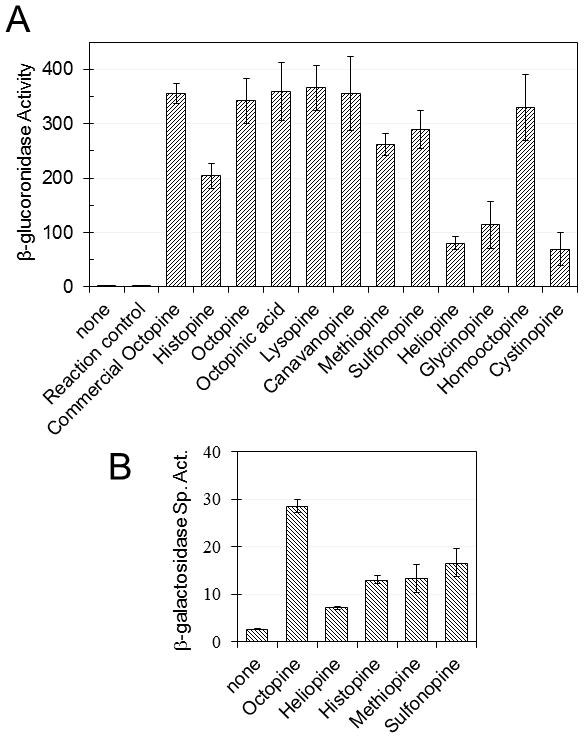
Induction of occ operon of strain KYC16 (R10 ooxA-uidA) by octopine-type opines. (A) Enzymatically produced opines induce expression of an ooxA-uidA fusion on the Ti plasmid. “Reaction control” refers to an enzymatic reaction mixture lacking any amino acid. “Commercial octopine” was purchased from Aldrich. (B) β-galactosidase activity of A. tumefaciens KYC1203(pKYC148)(pRM101) cultured in the presence of different opines. pKYC148 contains an occQ-lacZ translational fusion, while pRM101 contains a Plac-occR fusion. Histopine, N-(R-1-carboxyethyl)-Histidine; Octopinic acid, N-(R-1-carboxyethyl)-Ornithine; Lysopine, N-(R-1-carboxyethyl)-Lysine; Canavanopine, N-(R-1-carboxyethyl)-Canavanine; Methiopine, N-(R-1-carboxyethyl)-Methionine; Heliopine, N-(R-1-carboxyethyl)-Glutamine; Glycinopine, N-(R-1-carboxyethyl)-Glycine; Homooctopine, N-(R-1-carboxyethyl)-Homoarginine; Cystinopine, N-(R-1-carboxyethyl)-Cysteine. Mean ± SD of n=3.
To examine whether OccR is required for induction, we cultured two congenic strains in the presence or absence of sulfonopine, and four other opines. Strains KYC1203(pKY148) (pRJM101) and KYC1203(pKY148) have a plasmid-borne occQ-lacZ fusion, and the former strain expresses OccR, while the latter strain does not. The former strain was induced by all five opines (Fig. 2B), while the latter strain was uninduced (Fig. S6A), indicating that OccR is required for detection of these opines.
A. tumefaciens can utilize sulfonopine as sole source of sulfur
We next determined whether A. tumefaciens is able to utilize sulfonopine as a source of sulfur. For this experiment, A. tumefaciens strains R10 and KYC55 (Ti-plasmid less) were cultured in the presence or absence of sulfonopine, SMM, or methionine as sole sulfur sources. All other essential nutrients were provided. Strain R10 utilized all three compounds (Fig. 3A and 3B). Unexpectedly, strain KYC55 also grew at the expense of any of these three compounds. This indicates that the occ operon is dispensable for the use of sulfonopine as a sulfur source.
Fig. 3.
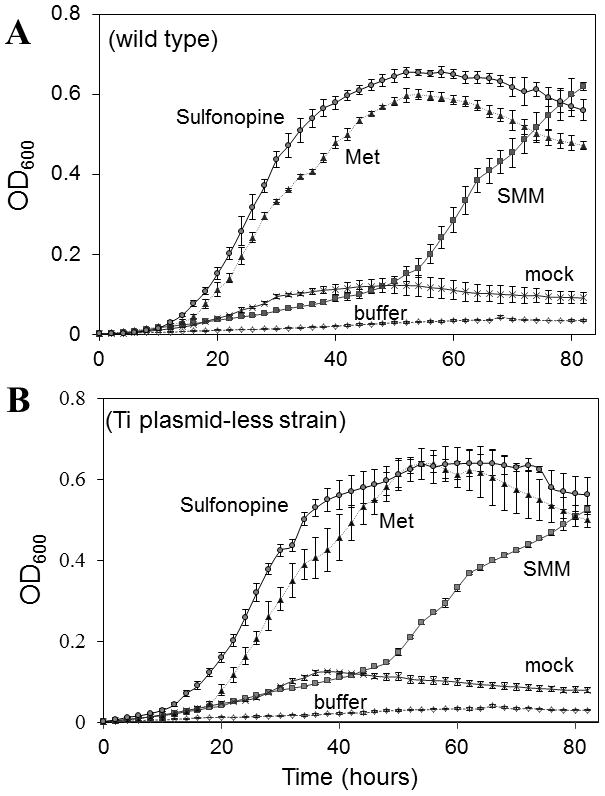
Utilization of sulfonopine as a sulfur source by strain R10 (wild type, A) and KYC55 (lacking a Ti plasmid, B). Strains were cultured in modified AB minimal lacking sulfur supplemented with 1 mM of the tested compounds. Sulfonopine was synthesized enzymatically, and “mock” refers to a control reaction conducted in the absence of any amino acid. “Buffer” refers to growth medium lacking any additions. Mean ± SD of n=3.
Discussion
In E. coli, there are three known pathways for methionine biosynthesis; all involve methylation of homocysteine, but differ in the source of the methyl group. The MetE protein transfers the methyl group directly from N5-methyl-tetrahydrofolate (Gonzalez et al., 1996). MetH transfers the methyl group from N5-methyl-tetrahydrofolate to a cobalamine coenzyme and from there to homocysteine (Frasca et al., 1988). The third methionine synthase, MmuM, transfers the methyl group from SMM, converting it to a second molecule of methionine (Thanbichler et al., 1999). The Msh protein of A. tumefaciens has limited sequence and structural homology with MetE and MetH, restricted to their homocysteine binding domains (Evans et al., 2004). In contrast, Msh has end-to-end sequence similarity with MmuM of E. coli. Msh showed a strong preference for SMM over three other methyl donors (Table 1), much like five other SMM-homocysteine methyltransferases found in E. coli, Arabidopsis, S. cerevisiae, and humans (Thanbichler et al., 1999, Ranocha et al., 2000, Thomas et al., 2000, Szegedi et al., 2008).
SMM is a ubiquitous metabolite in many or possibly all plants, although it is not essential in Arabidopsis (Lee et al., 2008). The methylation of methionine creates a positive charge on the sulfur atom, causing a net positive charge and enhancing its solubility. Perhaps for this reason, SMM is the transported form of methionine, and is concentrated in floral tissues (Ranocha et al., 2001). It should perhaps not be surprising that SMM could be a substrate for Ocs, as Ocs can utilize a rather broad variety of amino acids, especially the positively charged amino acids arginine, lysine, ornithine and histidine, as well as the non-charged amino acids glutamine and methionine (Chilton et al., 2001, Bates et al., 1984, Menage & Morel, 1964, Hack & Kemp, 1977, Biemann et al., 1960).
All previous studies of Ocs were done using enzyme that was purified from crown gall tumors (Hack & Kemp, 1980, Otten et al., 1977). In the present study, we produced a highly purified recombinant Ocs, which was stable and provided highly reproducible kinetic data for different substrates. This enzyme utilizes a surprising variety of proteogenic and nonproteogenic substrates, and several different alpha-keto acids, as well as NADH or NADPH. Many of the nonproteinogenic substrates are found in plants, including canavanine, homocysteine, homoserine, homoarginine, and selenomethionine (Bell, 2003). From an evolutionary perspective, it would seem advantageous for A. tumefaciens to utilize such a wide array of compounds as substrates, using either NADH or NADPH, and to derivatize them in such a way that they are unavailable to most other bacteria or presumably to plants.
Among the new sulfur-containing compounds synthesized by Ocs, only sulfonopine was detected in plant exudates and homogenates, probably because its precursor, SMM, was far more abundant than homocysteine, methionine, and cysteine. However, Ocs efficiently utilized all four amino acids in vitro, suggesting that all four could be potential sources of sulfur. Of these, the methionine-containing compound was previously described as a pseudo-opine, in that it was found to be produced by crown gall tumors but not imported by A. tumefaciens (Firmin et al., 1985). However, we showed that this compound induces expression of the occ operon via OccR. OccR is a cytoplasmic opine-responsive transcription factor of the LysR family. This indicates that the methionine-containing molecule can be internalized. Of the eleven compounds tested for induction of the occ operon, all were able to do so, suggesting that all can be internalized.
Production of sulfonopine by tobacco required transfer of ocs from the bacterium into plant cells, as a bacterial vir mutant or ocs mutant blocked sulfonopine production. Sulfonopine was exuded from tobacco, however, we do not know how sulfonopine or other opines are released from plant cells. An older report suggested that the ons gene facilitated opine export (Messens et al., 1985), but Ons is now generally thought to be a member of the RolB-RolC family, whose members regulate the abundance or activity of phytohormones (Bulgakov, 2008). Therefore, any effect of Ons on opine export would probably be indirect. The export of opines from transformed plant cells is therefore an unanswered question. However, the roots of many nontransformed plants exude surprisingly high levels of amino acids and carbohydrates (Phillips et al., 2004), suggesting that opine secretion may not require T-DNA encoded proteins.
Given the abundance of SMM in plants and the fact that it contains sulfur, an essential nutrient, it should not be surprising that at least some strains of A. tumefaciens can utilize it to form a sulfur-containing opine. We have not tested whether nopaline-type Ti plasmids can direct plant tumors to make a similar opine, but it seems quite plausible that nopaline synthase could conjugate SMM with α-ketoglutarate (rather than pyruvate) to create a compound similar to sulfonopine.
Opines have long been known to induce genes required for their uptake and catabolism (von Lintig et al., 1991, Klapwijk et al., 1978, Montoya et al., 1977). Octopine has been reported to activate the transcription of the occ operon by binding to OccR. (Habeeb et al., 1991). In the present study, we found that all tested opines can activate the transcription of the occ operon and that for all five opines tested, OccR was required for induction. These findings suggest that OccR detects primarily the amino acid backbone and pyruvate moieties of these opines, and has little or no specificity for their amino acid side chains.
Our finding that the utilization of sulfonopine did not require the Ti plasmid was somewhat counterintuitive. Apparently the uptake and utilization of these compounds can be directed by other genes of the rather large (5.6 megabase) genome of this organism. Previous reports have shown that the first four genes of the occ operon direct octopine uptake into E. coli (Valdivia et al., 1991), but are not essential for its uptake into A. tumefaciens (Cho et al., 1996). Evidently, octopine uptake is encoded redundantly. The present study indicates that this is also true of sulfonopine uptake. However, utilization of octopine as a carbon or nitrogen source requires at least some genes of the occ operon (Cho et al., 1996). Our observations that sulfonopine can provide sulfur in the absence of occ indicates that its catabolism is encoded redundantly.
A. tumefaciens spp. are well known to have evolved an efficient mechanism to obtain nutrients from the plants they infect. Previously, opines have been described that can be used as a source of carbon, nitrogen or phosphorous. Here we show that also sulfur can be obtained by opine catabolism. This new opine, sulfonopine, represents a novel way to derivatize a source of sulfur, SMM, from the infected plant and make it available for colonizing Agrobacteria.
Experimental Procedures
Bacterial strains, plasmids, and oligonucleotides
The bacterial strains, plasmids and oligonucleotides used in this study are listed in Table S1 and S2, respectively. Luria Broth (LB) and Terrific Broth (TB) were used as complex media, while AT and AB media were used as defined media (Tartoff & Hobbs, 1987, Cangelosi et al., 1991). AB medium. modified to lack sulfur, was used to test the ability of various opines and amino acids to provide sulfur. E. coli strains were grown at 37°C and Agrobacterium tumefaciens strains at 27°C. Octopine, amino acids, NADPH, NADH, and α-keto acids were purchased from Sigma-Aldrich. Sulfonopine was produced either by chemical synthesis or enzymatic synthesis and other opines were enzymatically synthesized (described below).
Chemical synthesis of SMM
L-SMM and D-SMM were synthesized using 1.5 g of L-methionine or D -methionine dissolved in 16 ml of 89% formic acid and 5 ml of acetic acid and combined with 2.5 ml of methyl iodide (Tuennies & Kolb, 1945). The reaction was incubated for 3 days at room temperature in the dark, and then evaporated to a syrup. Methanol (10 ml) was added to obtain granular particles, which were filtered and washed with methanol and acetone, and dissolved in 8 ml of warm 50% ethanol, and 25 ml of 100% ethanol was then added to allow crystallization in the dark. Crystals were filtered, washed and dried with acetone. Purity of L-SMM and D-SMM was determined using ESI-MS/MS and NMR.
Chemical synthesis of sulfonopine
The chemical synthesis of sulfonopine was done in two steps: 1) reductive condensation of methionine with pyruvate to yield N-(R-1-carboxyethyl)-S-methionine (methiopine), and 2) methylation of methiopine to yield sulfonopine. In the first step, 2 g of methionine methyl ester-HCl (Sigma-Aldrich) was combined with 10 ml of water and 1 M NaOH was added to reach pH 10.0 (~10 ml). The free base was extracted using chloroform and dried using Na2SO4. A solution of 35 ml of chloroform containing 1.5 g of methyl pyruvate and 4 g of sodium triacetoxyborohydride was added and incubated overnight under slight N2 pressure at room temperature. Ten ml of NaHCO3 solution (pH ~7.9), was then added and mixed until bubble formation ceased. The mixture was transferred to a separatory funnel, 20 ml of 1M NaOH was added, and extracted three times using 50 ml of CH2Cl2. The organic phases were combined and dried using MgSO4. The dried extract was dissolved in 3 ml of MeCN and then treated with 2 ml of water. After centrifugation, the supernatant was subjected to preparative HPLC using a 1×25 cm C-18 column (Phenomenex) and eluted with an acetonitrile-water gradient. Testing with ESI-MS/MS showed two major peaks to correspond to the two expected diastereomers of methiopine dimethyl ester. The pooled fractions of each diastereomer were evaporated to dryness, redissolved in 10 ml of 1M HCl, hydrolyzed by heating for 15 hours using an autoclave (120°C), and dried in vacuo to remove the HCl. The purity of each diastereomer was checked by ESI-MS/MS.
Conversion of the methiopine diastereomers to those of sulfonopine was carried out as for the methylation of methiopine to yield SMM (above). The purity of the sulfonopine diastereomers was checked by ESI-MS/MS (Fig. S3A) and NMR (Fig. S4). Only one of the diastereomers was found to have biological activity (Fig. S6B).
Enzymatic synthesis of opines
Octopine-type opines were produced using purified Ocs and glucose dehydrogenase, which regenerates NADPH from NADP+ at the expense of glucose (Weckbecker & Hummel, 2005). Enzymatic assays were performed using 150 mM sodium pyruvate, 90 mM of the amino acid substrate, 10 mM NADPH, 150 mM glucose, 150 mM sodium phosphate (pH 6.6), 0.08 μM of His6-Ocs and 2 units of glucose dehydrogenase. Reactions were incubated at 28°C for 24 hours. Enzymes were removed by methanol precipitation and filtration (Amicon Ultra, Millipore; molecular mass cut-off 3,000). The concentrations of reactants and products were determined using ESI-MS/MS.
DNA manipulation
Recombinant DNA techniques were performed using established procedures (Sambrook et al., 1989). PCR amplification of genes was done using Platinum Taq DNA Polymerase High Fidelity (Invitrogen). Plasmid DNA was isolated using QIAprep spin miniprep kits (Qiagen). DNA fragments generated by PCR or restriction digestion were gel purified using QIAquick Gel Extraction Kit (Qiagen). Restriction enzymes and other DNA modification reagents were purchased from New England Biolabs and used according to the methods described by the manufacturers. Plasmid DNA was introduced into E. coli and A. tumefaciens by electroporation (Cangelosi et al., 1991). Plasmids and bacterial strains are listed in Table S1, and primers are described in Table S2.
Plasmid construction
The ocs gene was PCR amplified by using primers ALFM28 and ALFM29, and inserted into pMCSG19 using ligation-independent cloning (LIC) (Donnelly et al., 2006), resulting in pAFM04, which encodes MBP-His6-Ocs fusion. Between MBP and His6- tag, there is a recognition site for TVMV protease. pAFM04 and pT7-groESL were electroporated into E. coli BL21/DE3(pRK1037) (pRK1037 encodes TVMV protease), so the MBP portion of the tripartite fusion was removed immediately after protein synthesis. Plasmid pT7-groESL was provided to enhance accumulation of soluble protein (Choi & Greenberg, 1991). The msh gene was subcloned by PCR amplification using primers ALFM21 and ALFM27 (Table S2) and pMCSGG19, resulting in pAFM11, which encodes a MBP-His6-Msh fusion. This plasmid was electroporated into BL21/DE3(pRK1037)(pT7-groESL).
Mutagenesis of ocs and virD4 by Campbell-type integration
Plasmids pAFM110 and pAFM111 are suicide plasmids containing 0.5 kb internal fragments of ocs and virD4, respectively. These fragments were PCR amplified using ALFM218 and ALFM219 (for ocs) and ALFM220 and ALFM221 (for virD4), and inserted between the KpnI and BamHI sites of the suicide plasmid pVIK107 (Kalogeraki & Winans, 1997). The resulting plasmids, pAFM110 and pAFM111, respectively, were transferred into strain R10 by conjugation (Kalogeraki & Winans, 1997), and transconjugants were selected using AB minimal agar plates containing 200 μg/ml of kanamycin. Since these plasmids cannot replicate in A. tumefaciens, they conferred antibiotic resistance by Campbell-type integration into ocs and virD4, respectively, creating null mutations in each gene. Campbell-type integration was confirmed by PCR amplification.
Overproduction and purification of Ocs and Msh
To overproduce Ocs, E. coli strain BL21/DE3(pAFM04)(pRK1037)(pT7-groESL) was cultured at 37°C in 1 L of TB containing 400 μg/ml of ampicillin, 400 μg/ml of kanamycin, and 35 μg/ml of chloramphenicol until OD600 0.6 was reached. The culture was cooled on ice to 28°C and overexpression of Ocs was induced by adding 0.3 mM IPTG. Incubation was continued at 28°C for 5 additional hours. Cells were concentrated by centrifugation for 10 min at 4°C. The pellet was suspended in lysis buffer (20 mM sodium phosphate buffer (pH 7.4), 200 mM NaCl, 20% glycerol, and 10 mM imidazole) and disrupted by passage through a French pressure cell (20,000 psi). The lysate was cleared by ultracentrifugation (25,000 × g at 4°C for 30 min). The supernatant was applied to Ni Sepharose™ 6 Fast Flow (GE Lifescience) chromatography resin. The column was washed extensively using lysis buffer, and Ocs was eluted using lysis buffer supplemented with 250 mM imidazole. Fractions containing Ocs were combined and concentrated by using an Amicon Ultra cell with YM-30 filter membrane (30,000 MWCO; Millipore, Eschborn, Germany). During concentration the buffer was changed to 20 mM sodium phosphate buffer (pH 7.4), 200 mM NaCl, 20% glycerol and 1 mM DTT. Ocs was further purified by gel filtration chromatography using a Superdex 200 column (GE Lifescience). Peak fractions were pooled and concentrated as described above and dialyzed using a buffer containing 150 mM sodium phosphate buffer (pH 6.6), 50% glycerol, and 1 mM DTT. Protein purity was analyzed using 12% SDS-PAGE gels and visualized by Coomassie staining. Msh was purified according to the same techniques using strain BL21/DE3(pAFM11)(pRK1037)(pT7-groESL). After purification, Msh was dialyzed using a buffer containing 20 mM Tris (pH 7.9), 50% glycerol and 1 mM DTT.
Msh Activity Assays
Msh enzymatic reactions were carried out by using 0.61 μM His6-Msh in 20 mM Tris buffer (pH 7.9), 10 mM homocysteine, and different concentrations of a variety of methyl donors such as SMM, SAM, methylcobalamine, betaine and, dimethylglycine. The reactions were stopped by adding an equal volume of 75% methanol and 0.25% formic acid solution and centrifuged for 30 min to precipitate Msh. The supernatants were analyzed for methionine production by ESI-MS/MS. Enzyme kinetics were calculated using the initial velocities for various concentrations of methyl donor.
Ocs Activity Assays
Enzymatic assays were performed by using 10 mM sodium pyruvate, 0.3 mM NADPH and different concentrations of amino acids in 150 mM PIPES (pH 6.6) and 0.08 μM of His6-Ocs. The reaction was started by the addition of the amino acid to be tested and followed by measuring the oxidation of NADPH spectrophotometrically at 340 nm at room temperature by using a Synergy™ HT multi-detection microplate reader (Biotek Intruments). The concentrations of NADPH in solution were calculated on the basis of the molar absorption coefficient 6200 M−1 cm−1 at 340 nm. Production of opines in the enzymatic mixture was confirmed by ESI-MS/MS. Enzyme kinetics were calculated using the initial velocities. Initial velocities obtained from Msh and Ocs enzymatic reactions were used to calculate the kinetics constants by using nonlinear least square analysis of the data fitted to the Michaelis-Menten and Lineweaver-Burk rate equation using SIGMA PLOT 9.0 (Systat Software, Ekrath, Germany) and the enzyme kinetic module 2.0.
Plant cultivation and inoculation
Arabidopsis thaliana wild-type Col-O, and mutants hmt2-2 and mmt-2 (Lee et al., 2008) were obtained as seeds from G. Jander (Boyce Thompson Institute). Nicotiana tabacum seeds were obtained from A. Colmer (Cornell University). Seeds were surface-sterilized by soaking in 50% bleach and 0.1% SDS for 10 min, followed by extensive washing in sterile water. Seeds were transferred to sterile water in Petri dishes and incubated for 48 h at 4°C in the dark. A. thaliana sterilized seeds were transferred to trays containing sterilized soil. Plants were maintained at room temperature with exposure to natural and artificial lighting.
Prior to plant infection, A. tumefaciens strains were cultured overnight in AT medium. Cells were then washed with sterile water and suspended to OD600 of 0.5 in a solution containing 5% sucrose and 0.005% of Triton-X100. A. thaliana plants cultivated in soil were infected by the floral dip method (Zhang et al., 2006). Plants were then cultivated for 2 weeks. Floral bolts were collected and homogenized using a Potter-Elvehjem homogenizer. The homogenate was centrifuged for 5 min at 10,000× g at 4°C and the supernatant was evaporated to dryness under a vacuum. The resulting solid was resuspended in 3:1 (v/v) methanol: 1% formic acid in water and centrifuged. The clear supernatant was analyzed by ESI-MS/MS.
Approximately 200 N. tabacum seeds were germinated in water supplemented with MS salts (Murashige, 1962). When the seedlings were 1–2 cm in length (approximately 2 weeks), they were submerged in the bacterial suspensions for 5 min, washed gently in 5% sucrose, and transferred to MS salts medium. Three days later, the infected seedlings were extensively washed with medium containing 300 μg per ml of carbenicillin, and incubated for 1 day in medium containing carbenicillin, then transferred to water and incubated for 2 weeks. Seedlings and water were collected for ESI-MS/MS analysis. Seedling extracts were obtained following the same protocol as for A. thaliana described above. Exudates were obtained by evaporating the water to dryness and resuspending in 3:1 (v/v) methanol: 1% formic acid in water for further ESI MS/MS analysis.
Electrospray mass-spectroscopy and nuclear magnetic resonance
Mass spectroscopy analysis was carried out using a Micromass Quattro II tandem MS operated in positive ion electrospray mode. Samples were injected directly using a syringe pump at a rate of 4 μl per min. Data acquisition and processing for the MS scans was controlled by the MassLynx software (Waters Corporation, Milford, MA). Twenty or more scans were averaged for each sample. When possible, spectra of all compounds were compared to those of commercial preparations of the same or similar compounds. NMR analysis was done by using a JEOL ECX-400 NMR spectrometer (Peabody, MA)
Utilization of sulfonopine and other sulfur source compounds
Sulfonopine, SMM, and methionine were tested as sources of sulfur by culturing strains in modified AB broth containing all essential nutrients except sulfur and supplemented with 1 mM of the tested compound as a sole sulfur source. Cultures were incubated at 28°C with vigorous aeration. Experiments were performed in triplicate using three different isolates for each strain.
Induction of the occ operon by opines
Opines were tested for induction of the occ operon using strain KYC16 (ooxA-uidA) cultured in AT medium containing 100 μg/ml of kanamycin at 27°C. β-glucuronidase specific activity was measured as described previously (Gallagher, 1992).
To determine whether OccR is responsible for induction, KYC1203(pKY148) and KYC1203(pKY148)(pRJM101) were cultured in AT medium supplemented with 1 mM of opines under vigorous aeration at 27°C to an OD600 of 0.3–0.4, and assayed for β-galactosidase specific activity (Miller, 1972).
Supplementary Material
Acknowledgments
We thank the members of our lab for helpful discussions and critical review of this manuscript. Special thanks to Drs. Frank Schroeder and Georg Jander for help with mass spectroscopy. We thank Dr. Chun Li of Ithaca College for the NMR work. This study was supported by grants from the NIGMS (GM042893) to S.C.W., from the TRIAD Foundation to F. Schroeder, and from the National Science Foundation (NSF0416567) to G. Jander.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates HA, Kaushal A, Deng PN, Sciaky D. Structure and synthesis of histopine, a histidine derivative produced by crown gall tumors. Biochemistry. 1984;23:3287–3290. doi: 10.1021/bi00309a026. [DOI] [PubMed] [Google Scholar]
- Bell EA. Nonprotein amino acids of plants: significance in medicine, nutrition, and agriculture. J Agric Food Chem. 2003;51:2854–2865. doi: 10.1021/jf020880w. [DOI] [PubMed] [Google Scholar]
- Biemann K, Lioret C, Asselineau J, Lederer E, Polonsky J. On the structure of lysopine, a new amino acid isolated from crown gall tissue. Biochim Biophys Acta. 1960;40:369–370. doi: 10.1016/0006-3002(60)91370-6. [DOI] [PubMed] [Google Scholar]
- Brencic A, Winans SC. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol Mol Biol Rev. 2005;69:155–194. doi: 10.1128/MMBR.69.1.155-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakov VP. Functions of rol genes in plant secondary metabolism. Biotechnol Adv. 2008;26:318–324. doi: 10.1016/j.biotechadv.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Cangelosi GA, Best EA, Martinetti G, Nester EW. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- Chilton WS, Petit A, Chilton MD, Dessaux Y. Structure and characterization of the crown gall opines heliopine, vitopine and rideopine. Phytochemistry. 2001;58:137–142. doi: 10.1016/s0031-9422(01)00166-2. [DOI] [PubMed] [Google Scholar]
- Cho K, Fuqua C, Martin BS, Winans SC. Identification of Agrobacterium tumefaciens genes that direct the complete catabolism of octopine. J Bacteriol. 1996;178:1872–1880. doi: 10.1128/jb.178.7.1872-1880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Greenberg EP. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci U S A. 1991;88:11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y, Guyon P, Farrand SK, Petit A, Tempe J. Agrobacterium Ti and Ri plasmids specify enzymic lactonization of mannopine to agropine. J Gen Microbiol. 1986;132:2549–2559. doi: 10.1099/00221287-132-9-2549. [DOI] [PubMed] [Google Scholar]
- Dessaux Y, Guyon P, Petit A, Tempe J, Demarez M, Legrain C, Tate ME, Farrand SK. Opine utilization by Agrobacterium spp.: octopine-type Ti plasmids encode two pathways for mannopinic acid degradation. J Bacteriol. 1988;170:2939–2946. doi: 10.1128/jb.170.7.2939-2946.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y, Petit A, Farrand SK, Murphy PJ. Opines and opine-like molecules involved in plant-Rhizobiaceae interactions. In: Spaink HP, Kondorosk A, Hooykaas PJ, editors. The Rhizobiaceae. Dordrecht: Kluwer Academic Publishers; 1998. pp. 173–197. [Google Scholar]
- Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc Natl Acad Sci U S A. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmin JL, Stewart IM, Wilson KE. N2-(1-carboxyethyl)methionine. A ‘pseudo-opine’ in octopine-type crown-gall tumours. Biochem J. 1985;232:431–434. doi: 10.1042/bj2320431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca V, Banerjee RV, Dunham WR, Sands RH, Matthews RG. Cobalamin-dependent methionine synthase from Escherichia coli B: electron paramagnetic resonance spectra of the inactive form and the active methylated form of the enzyme. Biochemistry. 1988;27:8458–8465. doi: 10.1021/bi00422a025. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Winans SC. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996a;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Winans SC. Localization of OccR-activated and TraR-activated promoters that express two ABC-type permeases and the traR gene of Ti plasmid pTiR10. Mol Microbiol. 1996b;20:1199–1210. doi: 10.1111/j.1365-2958.1996.tb02640.x. [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. Gus Protocols: Using the Gus geneas a reporter of gene expression. Academic Press; San Diego: 1992. [Google Scholar]
- Gelvin SB. Agrobacterium in the genomics age. Plant Physiol. 2009;150:1665–1676. doi: 10.1104/pp.109.139873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JC, Peariso K, Penner-Hahn JE, Matthews RG. Cobalamin-independent methionine synthase from Escherichia coli: a zinc metalloenzyme. Biochemistry. 1996;35:12228–12234. doi: 10.1021/bi9615452. [DOI] [PubMed] [Google Scholar]
- Habeeb LF, Wang L, Winans SC. Transcription of the octopine catabolism operon of the Agrobacterium tumor-inducing plasmid pTiA6 is activated by a LysR-type regulatory protein. Mol Plant Microbe Interact. 1991;4:379–385. doi: 10.1094/mpmi-4-379. [DOI] [PubMed] [Google Scholar]
- Hack E, Kemp JD. Comparison of octopine, histopine, lysopine, and octopinic acid synthesizing activities in sunflower crown gall tissues. Biochem Biophys Res Commun. 1977;78:785–791. doi: 10.1016/0006-291x(77)90248-0. [DOI] [PubMed] [Google Scholar]
- Hack E, Kemp JD. Purification and Characterization of the Crown Gall-specific Enzyme, Octopine Synthase. Plant Physiol. 1980;65:949–955. doi: 10.1104/pp.65.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Hwang I, Dessaux Y, Guyon P, Kim KS, Farrand SK. A T-DNA gene required for agropine biosynthesis by transformed plants is functionally and evolutionarily related to a Ti plasmid gene required for catabolism of agropine by Agrobacterium strains. J Bacteriol. 1997;179:4831–4840. doi: 10.1128/jb.179.15.4831-4840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeraki VS, Winans SC. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/s0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- Kamada-Nobusada T, Sakakibara H. Molecular basis for cytokinin biosynthesis. Phytochemistry. 2009;70:444–449. doi: 10.1016/j.phytochem.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Klapwijk PM, Scheulderman T, Schilperoort RA. Coordinated regulation of octopine degradation and conjugative transfer of Ti plasmids in Agrobacterium tumefaciens: evidence for a common regulatory gene and separate operons. J Bacteriol. 1978;136:775–785. doi: 10.1128/jb.136.2.775-785.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Huang T, Toro-Ramos T, Fraga M, Last RL, Jander G. Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J. 2008;54:310–320. doi: 10.1111/j.1365-313X.2008.03419.x. [DOI] [PubMed] [Google Scholar]
- Menage A, Morel G. Sur la presence d’octopine dans les tissus de crown-gall. C R Acad Sci Paris. 1964;259:4795–4796. [PubMed] [Google Scholar]
- Menage A, Morel G. Sur la presence d’un acide amine nouveau dans les tissus de crown-gall. C R Acad Sci Paris. 1965;261:2001–2002. [Google Scholar]
- Messens EALMV, WHR Genetic basis for opine secretion from crown gall tumor cells. Mol Gen Genet. 1985;199:344–348. [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1972. [Google Scholar]
- Montoya AL, Chilton MD, Gordon MP, Sciaky D, Nester EW. Octopine and nopaline metabolism in Agrobacterium tumefaciens and crown gall tumor cells: role of plasmid genes. J Bacteriol. 1977;129:101–107. doi: 10.1128/jb.129.1.101-107.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neuhierl B, Thanbichler M, Lottspeich F, Bock A. A family of S-methylmethionine-dependent thiol/selenol methyltransferases. Role in selenium tolerance and evolutionary relation. J Biol Chem. 1999;274:5407–5414. doi: 10.1074/jbc.274.9.5407. [DOI] [PubMed] [Google Scholar]
- Otten LA, Vreugdenhil D, Schilperoort RA. Properties of D(+)-lysopine dehydrogenase from crown gall tumour tissue. Biochim Biophys Acta. 1977;485:268–277. doi: 10.1016/0005-2744(77)90163-2. [DOI] [PubMed] [Google Scholar]
- Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR. Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 2004;136:2887–2894. doi: 10.1104/pp.104.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper KR, Beck von Bodman S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- Piper KR, Beck Von Bodman S, Hwang I, Farrand SK. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol Microbiol. 1999;32:1077–1089. doi: 10.1046/j.1365-2958.1999.01422.x. [DOI] [PubMed] [Google Scholar]
- Ranocha P, Bourgis F, Ziemak MJ, Rhodes D, Gage DA, Hanson AD. Characterization and functional expression of cDNAs encoding methionine-sensitive and -insensitive homocysteine S-methyltransferases from Arabidopsis. J Biol Chem. 2000;275:15962–15968. doi: 10.1074/jbc.M001116200. [DOI] [PubMed] [Google Scholar]
- Ranocha P, McNeil SD, Ziemak MJ, Li C, Tarczynski MC, Hanson AD. The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 2001;25:575–584. doi: 10.1046/j.1365-313x.2001.00988.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Savka MA, Black RC, Binns AN, Farrand SK. Translocation and exudation of tumor metabolitesin crown galled plants. Mol Plant Microbe Interact. 1996;9:310–313. doi: 10.1094/mpmi-9-0310. [DOI] [PubMed] [Google Scholar]
- Savka MA, Dessaux Y, Oger P, Rossbach S. Engineering bacterial competitiveness and persistence in the phytosphere. Mol Plant Microbe Interact. 2002;15:866–874. doi: 10.1094/MPMI.2002.15.9.866. [DOI] [PubMed] [Google Scholar]
- Schindler U, Sans N, Schroder J. Ornithine cyclodeaminase from octopine Ti plasmid Ach5: identification, DNA sequence, enzyme properties, and comparison with gene and enzyme from nopaline Ti plasmid C58. J Bacteriol. 1989;171:847–854. doi: 10.1128/jb.171.2.847-854.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedi SS, Castro CC, Koutmos M, Garrow TA. Betaine-homocysteine S-methyltransferase-2 is an S-methylmethionine-homocysteine methyltransferase. J Biol Chem. 2008;283:8939–8945. doi: 10.1074/jbc.M710449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartoff KD, Hobbs CA. Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus. 1987;9:12. [Google Scholar]
- Thanbichler M, Neuhierl B, Bock A. S-methylmethionine metabolism in Escherichia coli. J Bacteriol. 1999;181:662–665. doi: 10.1128/jb.181.2.662-665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Becker A, Surdin-Kerjan Y. Reverse methionine biosynthesis from S-adenosylmethionine in eukaryotic cells. J Biol Chem. 2000;275:40718–40724. doi: 10.1074/jbc.M005967200. [DOI] [PubMed] [Google Scholar]
- Tuennies G, Kolb JJ. Methionine studies VII. Sulfonium derivatives. J Am Chem Soc. 1945;67:1141–1144. [Google Scholar]
- Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol. 2006;17:147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Wang L, Winans SC. Characterization of a putative periplasmic transport system for octopine accumulation encoded by Agrobacterium tumefaciens Ti plasmidpTiA6. J Bacteriol. 1991;173:6398–6405. doi: 10.1128/jb.173.20.6398-6405.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Zanker H, Schroder J. Positive regulators of opine-inducible promoters in the nopaline and octopine catabolism regions of Ti plasmids. Mol Plant Microbe Interact. 1991;4:370–378. doi: 10.1094/mpmi-4-370. [DOI] [PubMed] [Google Scholar]
- Wang L, Helmann JD, Winans SC. The A. tumefaciens transcriptional activator OccR causes a bend at a target promoter, which is partially relaxed by a plant tumor metabolite. Cell. 1992;69:659–667. doi: 10.1016/0092-8674(92)90229-6. [DOI] [PubMed] [Google Scholar]
- Weckbecker A, Hummel W. Glucose Dehydrogenase for the Regeneration fo NADPH and NADH. In: Barredo JL, editor. Microbial Enzymes and Biotransformations. SpringerLink; 2005. [Google Scholar]
- White CE, Winans SC. Cell-cell communication in the plant pathogen Agrobacterium tumefaciens. Philos Trans R Soc Lond B BiolSci. 2007;362:1135–1148. doi: 10.1098/rstb.2007.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zhu J, Oger PM, Schrammeijer B, Hooykaas PJ, Farrand SK, Winans SC. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


