Understanding the mechanisms of gene expression requires a thorough knowledge of the balance between the biogenesis of an mRNA and its degradation. While advances in transcriptome profiling methods have enabled the large-scale analysis of RNA steady-state levels, monitoring the synthesis, processing and decay rates of specific transcripts remains technically challenging. The application of nucleoside analogs such as 4-thiouridine (4-sU) has recently enabled metabolic labeling and enrichment of nascent transcripts to monitor RNA dynamics in mammalian cells.[1] Thiol-selective labeling allows the capture and enrichment of RNA but is incompatible with cellular imaging due to competing free thiols in cells. Alternatively, metabolic labeling with 5-ethynyluridine (EU) is compatible with fluorescence imaging of nascent transcripts in cells, tissues and model organisms following copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC or “click chemistry”) with azide-functionalized fluorescent dyes.[2] Here we report N6-propargyl adenosine[3] (N6pA; Figure 1a) is efficiently incorporated into RNA in mammalian cells by polyadenylate (poly(A)) polymerase and all three mammalian RNA polymerases: pol I, which transcribes ribosomal RNA in the nucleolus, pol II which transcribes mRNA and some noncoding RNAs, and pol III which transcribes specific small RNAs (tRNAs, et al.). We show that N6pA can be used for CuAAC-mediated fluorescence imaging and affinity enrichment of nascent transcripts. Finally, we demonstrate how “clickable” nucleosides can be employed to monitor poly(A) tail dynamics in cells.
Figure 1.
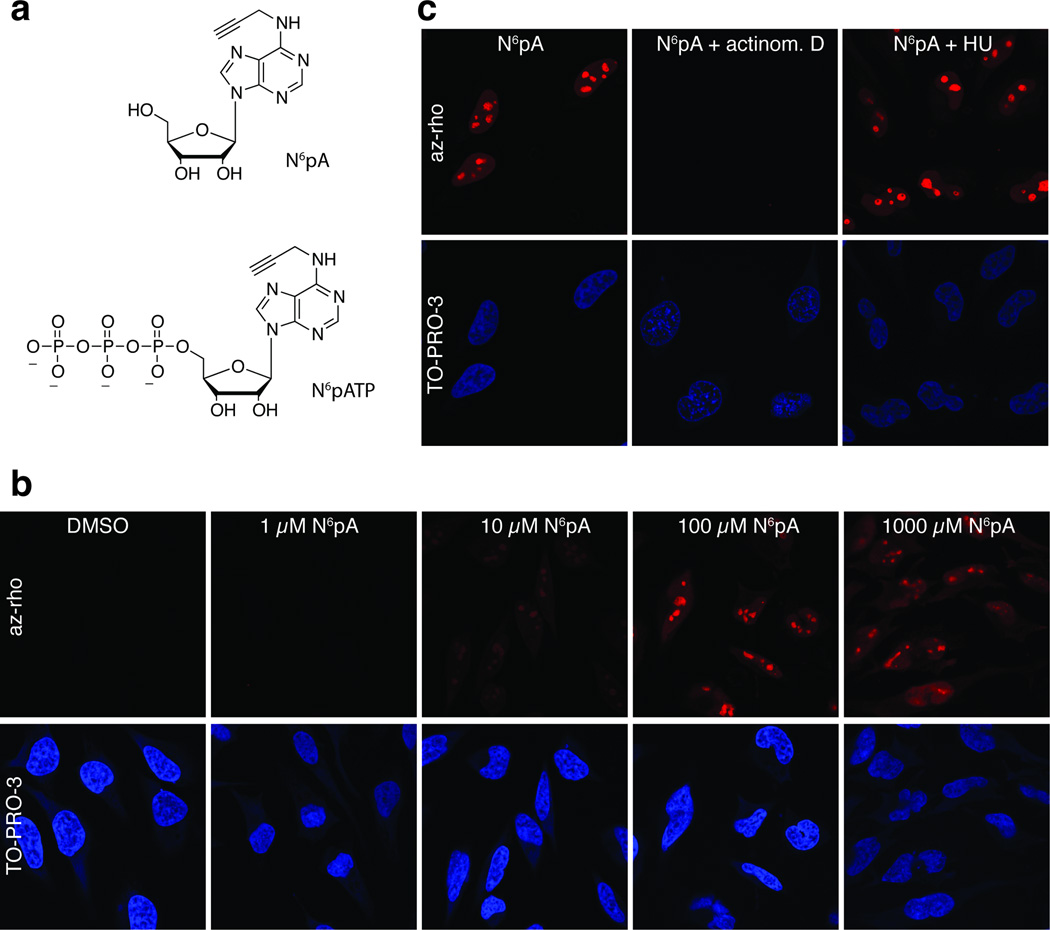
N6pA is incorporated by pol I in cells. (a) Structure of N6pA and N6pATP. (b) Cells were incubated for 12 hours in media containing the indicated concentrations of N6pA. After fixation and permeabilization, rhodoamine-azide was conjugated via click chemistry. Top panels are rhodamine signal and the bottom panels are nuclei counterstained with TO-PRO-3. (c) Cells were incubated with 1 mM N6pA in the presence of actinomycin D or hydroxyurea as indicated; fluorescent assay performed as in (b).
To determine whether N6pA labels cellular RNA, we analyzed metabolic incorporation of N6pA into mammalian cells by fluorescence microscopy.[2] HeLa cells were incubated in complete medium containing a range of N6pA concentrations (1.0 µM - 1 mM). After 12 hours, the cells were washed, permeabilized, fixed, reacted with azido-rhodamine via CuAAC and visualized by fluorescence microscopy (Figure 1b). The majority of N6pA-specific fluorescence signal localized to distinct substructures of the nucleus, most likely nucleoli, in accordance with the dominant transcriptional activity of pol I. Incorporation of N6pA was observed with as little as 10 µM of N6pA and appeared to reach its maximum between 10 µM and 100 µM. To distinguish between DNA and RNA labeling, we examined N6pA incorporation in the presence of hydroxyurea, a ribonucleotide reductase inhibitor, or actinomycin D, a transcription inhibitor (Figure 1c). While the addition of hydroxyurea had no effect, actinomycin D completely abolished fluorescence, demonstrating that N6pA is incorporated into RNA, but not DNA. We conclude that N6pA is efficiently incorporated into rRNA by pol I in cells.
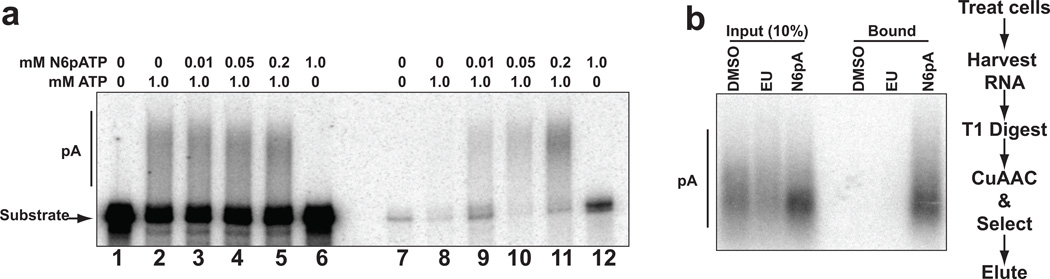
In contrast to EU, N6pA is a potential substrate for incorporation into poly(A) tails by cellular poly(A) polymerases. To test this, we performed in vitro polyadenylation reactions with HeLa nuclear extracts, a uniformly 32P-labeled RNA substrate and N6-propargyl adenosine-5′-triphosphate (N6pATP; Figure 1a). Incubation of the substrate in nuclear extract under polyadenylation conditions leads to the addition of a poly(A) tail of ~150–250 nt, as previously observed[4] (Figure 2a, lanes 1 and 2). Inclusion of N6pATP in the reaction at various concentrations (lanes 3–5) had no apparent effect on polyadenylation efficiency, as judged by the intensity and lengths of the polyadenylated products (lanes 2–5). However, when ATP was replaced completely with N6pATP, no poly(A) extension was observed (lane 6). To show incorporation of N6pA into poly(A) tails, polyadenylation products were biotinylated using CuAAC with biotin-azide and captured on streptavidin beads. Polyadenylated RNAs generated in the presence of N6pATP were retained by streptavidin beads, whereas those generated in its absence were not (compare lanes 9–11 to lane 8). In principle, incorporation of only one N6pATP and its subsequent biotinylation should be sufficient for binding. However, the recovery of the polyadenylated substrates correlated with higher N6pATP:ATP ratios. This may indicate that N6pATP is not utilized by poly(A) polymerase as efficiently as ATP or that any one of the downstream steps (i.e. biotinylation, binding or retention on the column) requires more than one modified residue for maximal recovery. Interestingly, while N6pATP alone did not support processive polyadenylation, this substrate was recovered more efficiently than the control (lane 12 versus lane 7). It is therefore likely that a few N6pATP molecules were incorporated into the substrate, but the tail could not be further extended. Perhaps natural adenosines in the poly(A) tail are required to promote processive polyadenylation or ATP may be necessary for the activity of putative ATP-dependent enzymes (e.g. helicases) involved in polyadenylation in extract. In either case, these data clearly show that N6pA is utilized by a poly(A) polymerase in vitro.
Figure 2.
N6pA is incorporated into poly(A) tails in vitro and in vivo. (a) In vitro polyadenylation assays with a 32P-labeled “pre-cleaved” substrate in HeLa nuclear extract.[4] The substrate is a 247-nt in vitro transcribed RNA derived from the 3′ end of PANΔ79 RNA[14] that contains the AAUAAA polyadenylation signal and terminates at the natural cleavage site. As a result, polyadenylation is uncoupled from the endonucleolytic cleavage step of 3′-end formation. The concentrations of N6pATP and ATP in the reaction are shown above each lane. Lanes 1–6 contain the products of the polyadenylation reaction (20% of the total reaction), while lanes 7–12 are the streptavidin-bound transcripts. (b) N6pA is incorporated into poly(A) tails in vivo. 10% of the RNase T1-treated RNA (Input) or 100% of the streptavidin-bound RNA was analyzed by northern blot using an oligo dT probe to detect poly(A) tails. The experimental flow is shown to the right.
Next we tested whether N6pA is incorporated into poly(A) tails in mammalian cells. To do this, we labeled HEK293 cells with 200 µM EU or N6pA for 12 hr, harvested cells, and extracted RNA. To degrade total RNA but leave poly(A) tails intact, we treated the RNA with RNase T1, a G-specific endonuclease, prior to biotinylation and selection. Only the poly(A) tails from cells labeled with N6pA bound the streptavidin column (Figure 2b), while the EU-treated or control (DMSO) poly(A) tails did not. These results demonstrate that N6pA is incorporated into poly(A) tails in cells. While several poly(A) polymerases are expressed in cells, our results are consistent with the conclusion that we are monitoring incorporation by the “canonical” poly(A) polymerase, PAPα.[5] PAPα is responsible for the polyadenylation of cellular mRNAs that contain a typical AAUAAA poly(A) signal. Both our in vitro substrate (Figure 2a) and the majority of the “bulk” poly(A) tails (Figure 2b) fall into this category, so it seems likely that this is the poly(A) polymerase responsible for the activity we are monitoring. Further studies are necessary to determine whether the “noncanonical” poly(A) polymerases involved in a variety of alternative polyadenylation pathways[6] can also utilize N6pA.
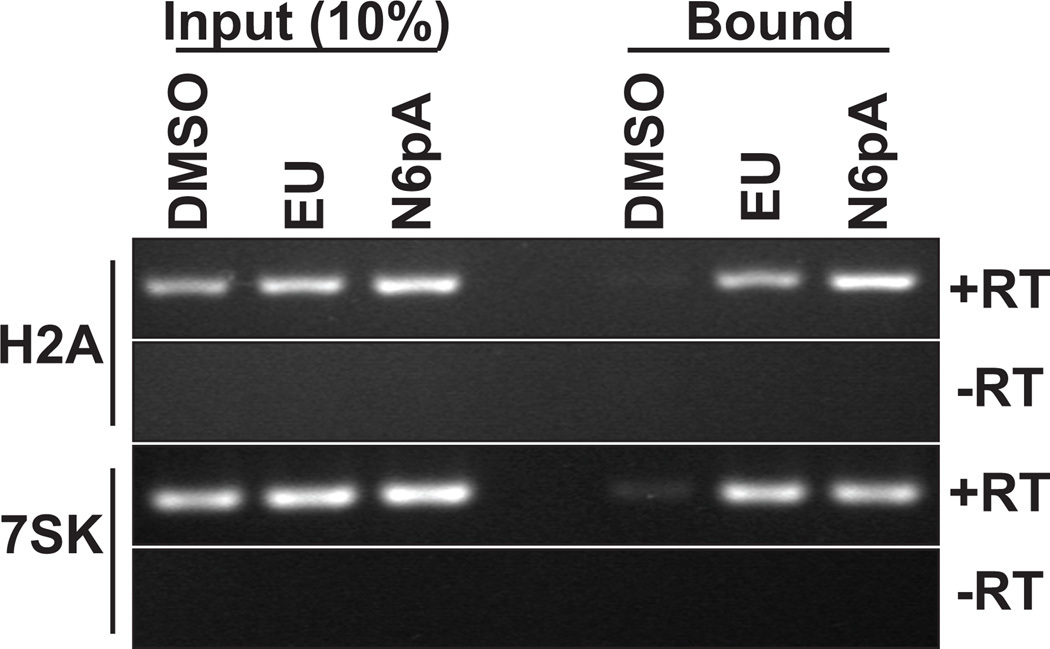
We next examined whether pol II and pol III utilize N6pA. For pol II, we needed to experimentally distinguish between pol II and PAPα activity in cells. To do so, we tested whether the histone mRNAs, which are non-polyadenylated pol II transcripts[7], were labeled with N6pA. Total RNA from N6pA-treated cells was biotinylated, selected, and recovered transcripts were analyzed by semi-quantitative RT-PCR (Figure 3, top). H2A mRNA was efficiently recovered from N6pA-treated cells as well as from positive control (EU) cells. In contrast, minimal H2A mRNA was observed in the negative control (DMSO). Similar results were obtained for histone HIST3H2BB mRNA (data not shown). Additionally, the pol III-transcribed 7SK RNA was detected among the selected transcripts from EU and N6pA-treated cells (Figure 3, bottom). From these results and those presented above, we conclude that N6pA is incorporated into RNA by pol I, II, III and PAPα.
Figure 3.
N6pA is incorporated into pol II and pol III transcripts in vivo. RNA harvested from cells treated with DMSO, 200 µM EU, or 200 µM N6pA for 12 hr was subject to click chemistry with biotin-azide and selected on a streptavidin column. 10% of the input and 100% of the bound RNA was analyzed by semi-quantitative RT-PCR using primers specific for the histone H2A mRNA or the 7SK small noncoding RNA. Negative controls lacking reverse transcriptase are also shown (-RT).
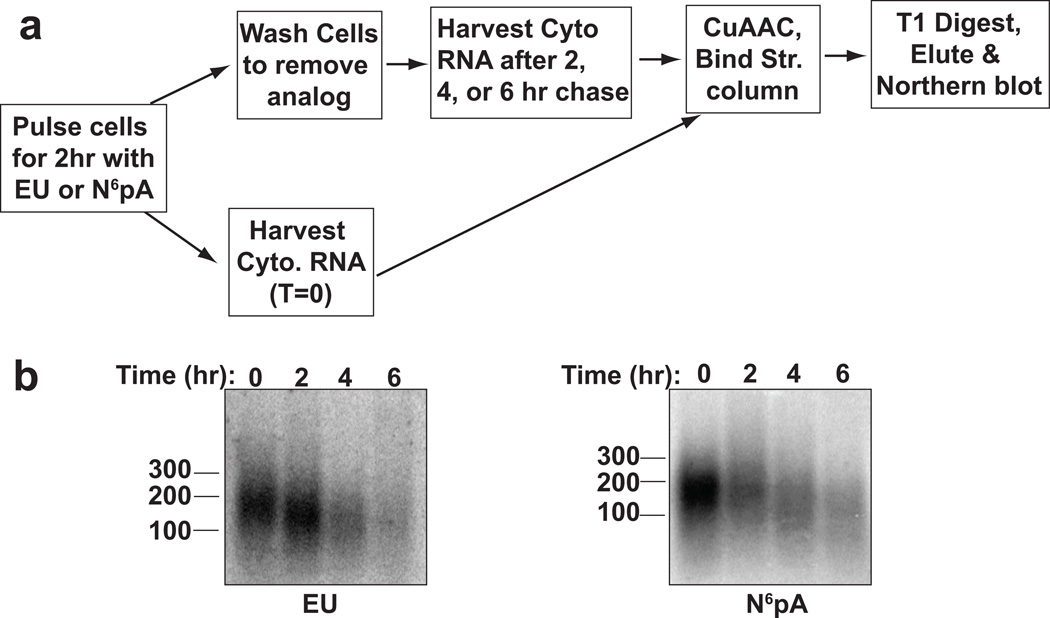
The general pathway for destruction of cytoplasmic mRNAs begins with a slow distributive deadenylation of the transcript, which is followed by nearly coincident decapping, processive deadenylation, and transcript decay[8]. In most cases, distributive deadenylation has been observed using specific reporter transcripts driven from inducible promoters, but has not been demonstrated at the bulk level. Changes in bulk poly(A) lengths have been limited to steady-state observations primarily due to the lack of simple approaches for global pulse-chase analysis of poly(A) tail lengths. While steady-state analysis can be informative (e.g.[9]), interpreting poly(A) tail length differences in a pool of transcripts that contains both newly synthesized transcripts and older transcripts can be difficult. We therefore developed a pulse-chase strategy using EU or N6pA to study bulk poly(A) tail dynamics in cells. HEK293 cells were pulse-labeled with 200 µM EU or N6pA for 2 hr, washed, and chased with media lacking modified nucleoside. Cytoplasmic RNA was extracted from cells at given time intervals following the chase. Metabolically labeled transcripts were biotinylated by CuAAC, captured on streptavidin columns, and digested with RNase T1. Northern blot analysis of the resulting poly(A) tails showed that after a 2-hr pulse (t=0), the cytoplasmic poly(A) tails were ~150–200 nt long (Figure 4). Importantly, the tails shortened over time and decreased in their abundance, consistent with the model that most cytoplasmic mRNAs are gradually deadenylated prior to their destruction. This assay has the advantage of observing RNAs transcribed from endogenous promoters in the absence of general transcription inhibitors, which often have pleiotropic effects.[10] The ability to detect bulk poly(A) tails after a short labeling pulse allows us to examine poly(A) dynamics of transcripts that were synthesized within a 2-hr window, thereby limiting the heterogeneity of poly(A) tail length that arises over time. We conclude that alkyne-nucleosides and click chemistry can be employed to examine poly(A) tail dynamics in cells. In principle, this approach could be extended to examine poly(A) tail dynamics of specific cellular mRNAs using previously described RT-PCR techniques to detect poly(A) tail length.[11]
Figure 4.
Pulse-chase assay to analyze poly(A) tail dynamics. (A) Experimental flow. Cells were labeled for two hours with EU or N6pA and chased for 2, 4, or 6 hr prior to cytoplasmic RNA extraction. Poly(A) tails of captured transcripts were then analyzed by northern blot using an oligo dT probe. (B) Data from an in vivo poly(A) tail dynamics assay.
In summary, we report N6pA is incorporated by all three cellular RNA polymerases (pol I, pol II, pol III) in mammalian cells. Moreover, N6pA is added to poly(A) tails in vitro and in vivo, demonstrating that this chemical reporter is utilized by poly(A) polymerase. Furthermore, we showed that clickable nucleosides can be employed to monitor cellular poly(A) tail dynamics. These alkyne-adenosine reporters should provide new tools to monitor RNA metabolism in vitro and in vivo using bioorthogonal chemistry [12] and vibrational spectroscopy.[13]
Methods
Detailed methods are given in the supplementary information.
Acknowledgments
We thank Olga Hunter for technical assistance and Ivan D’Orso for reagents. N.K.C. was funded by the NIH/NIAID AI081710 and by the Cancer Prevention and Research Institute of Texas grant RP110132. N.K.C. is a Southwestern Medical Foundation Scholar in Biomedical Research. H.C.H was funded by Irma T. Hirschl/Monique Weill-Caulier Trust, Lerner Trust and NIH/NIGMS (1R01GM087544).
References
- 1.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. Nat Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jao CY, Salic A. Proc Natl Acad Sci U S A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grammel M, Luong P, Orth K, Hang HC. J Am Chem Soc. 2011 doi: 10.1021/ja205137d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilusz J, Shenk T. Cell. 1988;52:221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- 5.Eckmann CR, Rammelt C, Wahle E. Wiley Interdiscip Rev RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt MJ, Norbury CJ. Wiley Interdiscip Rev RNA. 2010;1:142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- 7.Marzluff WF, Wagner EJ, Duronio RJ. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a Parker R, Song H. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]; b Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 9.Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. Hum Mol Genet. 19:1058–1065. doi: 10.1093/hmg/ddp569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loflin PT, Chen CY, Xu N, Shyu AB. Methods. 1999;17:11–20. doi: 10.1006/meth.1998.0702. [DOI] [PubMed] [Google Scholar]
- 11.Salles FJ, Richards WG, Strickland S. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- 12.Boyce M, Bertozzi CR. Nat Methods. 2011;8:638–642. doi: 10.1038/nmeth.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamakoshi H, Dodo K, Okada M, Ando J, Palonpon A, Fujita K, Kawata S, Sodeoka M. J Am Chem Soc. 2011;133:6102–6105. doi: 10.1021/ja108404p. [DOI] [PubMed] [Google Scholar]
- 14.Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Proc Natl Acad Sci U S A. 2007;104:10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]