Abstract
The CELLULOSE SYNTHASE (CESA) superfamily of proteins contains several sub-families of closely related CELLULOSE SYNTHASE-LIKE (CSL) sequences. Among these, the CSLA and CSLC families are closely related to each other and are the most evolutionarily divergent from the CESA family. Significant progress has been made with the functional characterization of CSLA and CSLC genes, which have been shown to encode enzymes with 1,4-β-glycan synthase activities involved in the biosynthesis of mannan and possibly xyloglucan backbones, respectively. This review examines recent work on the CSLA and CSLC families from evolutionary, molecular, and biochemical perspectives. We pose a series of questions, whose answers likely will provide further insight about the specific functions of members of the CSLA and CSLC families and about plant polysaccharide biosynthesis is general.
Keywords: CELLULOSE SYNTHASE-LIKE, mannan, xyloglucan, CSLA, CSLC, plant cell wall
INTRODUCTION
Plant cell walls are complex composites that consist mainly of carbohydrates, including cellulose, hemicelluloses, and pectins (Somerville et al., 2004; Lerouxel et al., 2006; Sandhu et al., 2009; Doblin et al., 2010; Liepman et al., 2010; Scheller and Ulvskov, 2010; Carpita, 2011). The carbohydrates present in plant cell walls vary in structure and composition among plants (Carpita, 2011) and even within different cells and tissues of a single plant (Lee et al., 2011). This structural heterogeneity among carbohydrates is a key factor underlying the functional diversity of plant cell walls (Pauly and Keegstra, 2010). Plant cell walls also represent the most abundant source of renewable biomass, and provide materials with a multitude of human uses and other important roles in the biosphere (Pauly and Keegstra, 2010; Lee et al., 2011).
Due to the complexity of monosaccharide composition and glycosidic linkages present in plant cell walls, it is predicted that hundreds of enzymes are involved in cell wall carbohydrate biosynthesis (Keegstra and Raikhel, 2001; Scheible and Pauly, 2004). This estimate substantially increases when including other cell wall-related proteins (Girke et al., 2004; McCann and Carpita, 2008). Spurred by genome sequencing and the availability of powerful comparative and functional genomic tools, the cell wall research community has made significant progress over the last decade in identifying and determining the function of numerous enzymes involved in the synthesis of plant cell wall carbohydrates (Farrokhi et al., 2006; Lerouxel et al., 2006; Penning et al., 2009; Sandhu et al., 2009; Doblin et al., 2010; Liepman et al., 2010). Among these are members of the CELLULOSE SYNTHASE (CESA) superfamily of proteins (CAZy GT2; Cantarel et al., 2009). The CESA superfamily includes bona fide CESA proteins involved in cellulose synthesis (Youngs et al., 2007; Endler and Persson, 2011), as well as CELLULOSE SYNTHASE-LIKE (CSL) proteins (Richmond and Somerville, 2000; Hazen et al., 2002; Fincher, 2009) that have been implicated in the synthesis of various β-glycan polymers.
This minireview focuses upon the CSLA and CSLC subgroups, the most divergent CSL subgroups relative to the CESA proteins (Richmond and Somerville, 2000; Youngs et al., 2007). Because members of the CSLA and CSLC subgroups are thought to have evolved through duplication and diversification from a common ancestral gene (Yin et al., 2009; Del Bem and Vincentz, 2010), they share some structural and physicochemical features (Youngs et al., 2007), however they differ in membrane topology and in enzymatic function (Davis et al., 2010). A number of CSLA genes have been shown to encode mannan synthase enzymes that polymerize the 1,4-β-linked backbone of mannans and glucomannans (Dhugga et al., 2004; Liepman et al., 2005, 2007; Suzuki et al., 2006; Gille et al., 2011). The CSLC proteins have been implicated in the synthesis of 1,4-β-glucan backbone of xyloglucans (Cocuron et al., 2007) and possibly other polysaccharides (Dwivany et al., 2009). Due to space limitations, we are unable to provide a comprehensive review of these topics; instead we will focus upon some important unanswered questions about the CSLA and CSLC families.
ARE ALL CSLA PROTEINS INVOLVED IN MANNAN SYNTHESIS?
CSLA genes appear to be present in all land plants, and ancestral genes with characteristics similar to CSLA and CSLC sequences have been identified in a number of green algal genomes, in which they are thought to represent a homolog of the progenitor gene from which CSLA and CSLC genes evolved (Del Bem and Vincentz, 2010). It has been hypothesized that these CSLA/CSLC-like sequences encode mannan synthases (Yin et al., 2009; Popper et al., 2011), however experimental evidence is needed to test this hypothesis. Heterologous expression of recombinant CSLA proteins has proven particularly effective for determining their enzymatic functions. The involvement of CSLA family members from diverse plant species in the synthesis of 1,4-β-mannan and glucomannan backbones has been demonstrated by a number of studies (Table 1; Dhugga et al., 2004; Liepman et al., 2005, 2007; Suzuki et al., 2006; Goubet et al., 2009; Gille et al., 2011). Studies of recombinant CSLA proteins have further demonstrated that expression of a single CSLA protein in a heterologous host is sufficient to impart enzymatic activity, and that the incorporation of mannose and glucose into glucomannan chains is mediated by a single enzyme (Liepman et al., 2005, 2007; Suzuki et al., 2006; Gille et al., 2011). Since recombinant CSLA proteins from a variety of plants exhibit mannan synthase activity, it is possible that all CSLA proteins are involved in the synthesis of mannans. An alternative possibility is that certain CSLA proteins may catalyze the synthesis of other polysaccharides; in particular a clade of CSLA proteins present only in monocots may have divergent function (Dhugga et al., 2004; Liepman et al., 2007; Del Bem and Vincentz, 2010; Dhugga, 2011). Efforts to characterize members of this clade will provide more information about the biosynthetic capabilities of CSLA proteins. Detailed biochemical studies of CSLA proteins from plants producing mannans of different structures also are needed in order to define whether structural features of these enzymes govern mannan product structure.
Table 1.
Biochemical attributes of recombinant CSLA and CSLC proteins.
| Species*/protein name | Gene identifier [GenBank ID (Phytozome** ID)] | Enzymatic function(s)*** | Reference |
|---|---|---|---|
| AkCSLA3 | HQ833588 | Mannan synthase, glucomannan synthase | Gille et al. 2011 |
| AtCSLA1 | NM_117760 (At4g16590) | Mannan synthase, glucomannan synthase | Liepman et al. 2007 |
| AtCSLA2 | NM_122180 (At5g22740) | Mannan synthase, glucomannan synthase | Liepman et al. 2005 |
| AtCSLA7 | NM_129120 (At2g35650) | Mannan synthase | Liepman et al. 2005 |
| AtCSLA9 | NM_120457 (At5g03760) | Mannan synthase, glucomannan synthase, | Liepman et al. 2005 |
| glucan synthase (GDP) | |||
| CtMANS | AY372247 | Mannan synthase | Dhugga et al. 2004 |
| OsCSLA1 | NM_001052699 (Os02g09930) | Mannan synthase, glucomannan synthase | Liepman et al. 2007 |
| PpCSLA1 | DQ417756 (Pp1s65_194V6) | Mannan synthase, glucomannan synthase | Liepman et al. 2007 |
| PpCSLA2 | DQ417757 (Pp1s36_62V6) | Mannan synthase, glucomannan synthase | Liepman et al. 2007 |
| PtCSLA1 | XM_002311936 (POPTR_0008s02650) | Mannan synthase, glucomannan synthase | Suzuki et al. 2006 |
| PtCSLA3 | XM_002326203 (POPTR_0006s11810) | Mannan synthase | Suzuki et al. 2006 |
| PtaCSLA1 | DQ641986 | Mannan synthase, glucomannan synthase | Liepman et al. 2007 |
| AtCSLC4 | NM_113737 (At3g28180) | Glucan synthase (UDP) | Cocuron et al. (2007) |
| TmCSLC | Not present in databases | Glucan synthase (UDP) | Cocuron et al. (2007) |
Prefixes: Ak, Amorphophallus konjac; At, Arabidopsis thaliana; Ct, Cyamopsis tetragonoloba; Os, Oryza sativa; Pp, Physcomitrella patens; Pt, Populus trichocarpa; Pta, Pinus taeda; Tm, Tropaeolum majus.
Phytozome (www.phytozome.net; Goodstein et al., 2012) identifiers provided, where available.
Mannan synthase, GDP-mannose-dependent 1,4-β-mannan synthase activity; glucomannan synthase, GDP-glucose-dependent 1,4-$β-glucomannan synthase activity; glucan synthase (GDP), GDP-glucose-dependent 1,4-β-glucan synthase activity; glucan synthase (UDP), UDP-glucose-dependent 1,4-β-glucan synthase activity.
WHAT ARE THE PHYSIOLOGICAL FUNCTIONS OF MANNANS?
Within plants and algae, mannans are structurally and functionally diverse, and they serve well-known roles as structural elements and as energy reserves (Moreira and Filho, 2008). In angiosperms, mannans are cross-linking glycans that are present at low levels in primary cell walls (Zablackis et al., 1995; Schroder et al., 2009; Marcus et al., 2010), and in greater abundance in secondary cell walls (Handford et al., 2003; Goubet et al., 2009). In gymnosperms, mannans are the most abundant hemicellulosic polysaccharide present in wood (Maeda et al., 2000; Pauly and Keegstra, 2010). Mannans also are very abundant in the primary cell walls of ferns, where they appear to be the dominant cross-linking glycan of the recently defined Type III cell wall (Silva et al., 2011). A variety of plants store energy in the form of mannans in their endosperm tissue, including members of the Palmae, Liliaceae, Iridaceae, and Leguminosae families (Meier and Reid, 1982; Buckeridge, 2010). Glucomannans also are used for energy storage in corms of plants within the genus Amorphophallus. The AkCSLA3 protein, involved in the synthesis of glucomannan stored in the corms of Konjac, recently has been characterized along with many other sequences encoding proteins involved in other aspects of glucomannan biosynthesis (Gille et al., 2011).
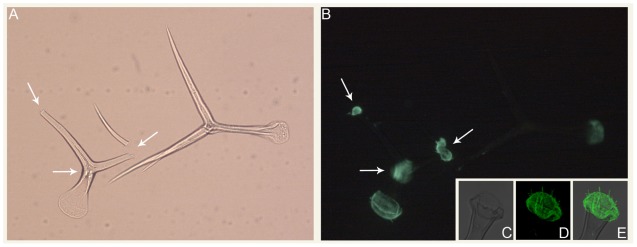
In addition to carbohydrate storage and structure, mannans serve a variety of other functions. In fern roots, mannans are deposited as constituents of cell wall appositions as a defense mechanism to limit microbial ingress (Leroux et al., 2011). Mannans impart hardness to seeds of some plants, such as tomato and lettuce, thereby protecting the embryo and controlling radicle protrusion (Schroder et al., 2009; Buckeridge, 2010). In tomato fruits, mannans also have roles in cell adhesion (Ordaz-Ortiz et al., 2009), and recent immunological studies have revealed a much wider distribution of mannans in cell walls than previously appreciated (Marcus et al., 2010). Pre-treatment of tissue sections with pectate lyase revealed homogalacturonan-masked mannan epitopes (Marcus et al., 2010). Mannose is an abundant constituent of Arabidopsis trichomes and the CSLA9 gene encoding a glucomannan synthase is among the top one hundred most abundant transcripts present in trichomes (Marks et al., 2008). Mannan epitopes appear to be present throughout trichome cell walls and at trichome bases, indicating that mannans may be involved in trichome to leaf adherence (Figure 1). Mannans also are involved in pollen tube growth, as this process is perturbed in Arabidopsis csla7 mutant plants (Goubet et al., 2003).
FIGURE 1.
Immunolocalization of mannans in isolated Arabidopsis trichomes. (A) Bright field and (B) epifluorescence micrographs of intact and broken trichomes isolated using the procedure of Marks et al. (2008). Arrows denote the locations of trichome branch breakpoints. (C–E) Confocal average projections of the base region of an isolated Arabidopsis trichome. For (B, D, E), the LM21 and LM22 antibodies (Marcus et al., 2010) were used for mannan immunolocalization.
A number of studies also implicate mannans within plant developmental signaling pathways. For example, Arabidopsis csla7 mutant embryos exhibit defective embryogenesis, arresting at the globular stage (Goubet et al., 2003), and csla9 mutants have reduced numbers of lateral roots (Zhu et al., 2003). Complementation of the csla7 mutant phenotype has been achieved by overexpression of CSLA9, demonstrating that the CSLA7 and CSLA9 proteins likely make structurally interchangeable mannans in vivo (Goubet et al., 2009). Interestingly, aborted embryos and developmental asynchrony were documented in siliques of transgenic Arabidopsis plants overexpressing various CSLA genes, indicating that mannan abundance influences the progression of embryogenesis (Goubet et al., 2009). A number of other studies have documented growth and developmental responses of plants and cultured plant cells to the application of galactoglucomannan oligosaccharides (GGMOs). For example, GGMOs enhance cell population density and alter the protoxylem:metaxylem ratio of xylogenic cultures of Zinnia (Benova-Kakosova et al., 2006). Treatment of pea stem segments with GGMOs also inhibits auxin-stimulated elongation growth (Auxtova-Samajova et al., 1996), possibly through the action of recently discovered mannan transglycosylases (Schroder et al., 2006, 2009). Additional studies are needed to provide more insight about the biological significance of GGMOs and the mechanism of their action.
Efforts to understand the physiological roles of mannans likely have been hindered by functional redundancy, since CSLA genes are members of multiple-gene families in many plants (Richmond and Somerville, 2001; Hazen et al., 2002; Liepman et al., 2007; Roberts and Bushoven, 2007). In Arabidopsis, csla single mutants have been identified for each of the nine CSLA genes (Table 2). Aside from the csla7 mutant, none of these csla single mutants exhibited notable phenotypic abnormalities (Goubet et al., 2009). It seems likely that at least some of the remaining uncharacterized Arabidopsis CSLA proteins also are mannan synthases, however the significant degree of overlap among the expression patterns of these sequences (Hamann et al., 2004; Liepman et al., 2007) probably masks defects resulting from their loss of function in csla single mutants, necessitating the analysis of higher order mutants. One such mutant, the Arabidopsis csla2/csla3/csla9 triple mutant, lacks detectable glucomannan in stems. This glucomannan deficiency did not impact stem strength, indicating that mannans are not required for stem strength in Arabidopsis, or that a compensatory mechanism may exist in their absence (Goubet et al., 2009).
Table 2.
Analyses of csla mutants.
| Mutant name | Gene identifier | Mutant phenotype | Reference |
|---|---|---|---|
| atcsla1 | At4g16590 | No apparent mutant phenotype | Goubet et al. (2009) |
| atcsla2 | At5g22740 | No apparent mutant phenotype | Goubet et al. (2009) |
| atcsla2 | At5g22740 | Reduction of phase II xylem in secondary thickened hypocotyl tissue | Ubeda-Tomas et al. (2007) |
| atcsla3 | At1g23480 | No apparent mutant phenotype | Goubet et al. (2009) |
| atcsla! | At2g35650 | Defective embryogenesis, impaired pollen tube growth | Goubet et al. (2003) |
| rat4 (atcsla9) | At5g03760 | Reduction of lateral root formation and rate of growth, decreased efficiency of root-mediated transformation by Agrobacterium tumefaciens | Zhu et al. (2003) |
| atcsla9 | At5g03760 | ~81% reduction of mannose quantity in inflorescence stems | Goubet et al. (2009) |
| atcsla10 | At1g24070 | No apparent mutant phenotype | Goubet et al. (2009) |
| atcsla11 | At5g16190 | No apparent mutant phenotype | Goubet et al. (2009) |
| atcsla14 | At3g56000 | No apparent mutant phenotype | Goubet et al. (2009) |
| atcsla15 | At4g13410 | No apparent mutant phenotype | Goubet et al. (2009) |
| atcsla2/3 | At5g22740/At1g23480 | Slight reduction of mannan quantity in inflorescence stems | Goubet et al. (2009) |
| atcsla2/9 | At5g22740/At5g03760 | Reduction of mannose quantity to trace level in inflorescence stems | Goubet et al. (2009) |
| atcsla3/9 | At1g23480/At5g03760 | Reduction of mannose quantity to trace level in inflorescence stems | Goubet et al. (2009) |
| atcsla2/3/9 | At5g22740/At1g23480/At5g03760 | Reduction of mannose quantity below detection level in inflorescence stems | Goubet et al. (2009) |
ARE DIFFERENT FORMS OF MANNANS SYNTHESIZED BY DIFFERENT CSL SUBCLASSES?
Recent studies have shed interesting new light on mannan synthesis, by implicating members of the CSLD family in this process (Verhertbruggen et al., 2011; Yin et al., 2011). Analysis of a collection of single mutants revealed that mutations within the CSLD2 and CSLD3 genes result in abnormal root hair morphology. These observations are consistent with other studies that indicate that CSLD proteins are important in tip-growing cells (Doblin et al., 2001; Favery et al., 2001; Wang et al., 2001; Kim et al., 2007; Bernal et al., 2008; Galway et al., 2011; Park et al., 2011). The csld2 and csld3 single mutants, along with all possible double and triple mutant combinations of csld2, csld3, and csld5 also exhibited abnormal mannan immunolocalization patterns in root hairs. Inflorescence stem development and mannan patterning therein of the csld2/csld3/csld5 triple mutant also was disrupted. To complement these loss of function studies, the CSLD2, CSLD3, and CSLD5 proteins were transiently expressed in tobacco leaves. Elevated mannan synthase activity was observed in microsomal membrane fractions prepared from leaf tissue of tobacco plants expressing either CSLD5 or both CSLD2 and CSLD3, but not in lines expressing CSLD2 or CSLD3 individually (Yin et al., 2011).
The CSLD proteins long have been suspected to be involved in the synthesis of a β-glucan polymer (Doblin et al., 2001), so the implication of CSLD proteins in mannan synthesis comes as a surprise for several reasons. First, among the CSL proteins, sequences of the CSLD proteins are the most similar to the CESA proteins, which synthesize cellulose (Youngs et al., 2007). Furthermore, genetic complementation of the csld3 mutant phenotype has been achieved using a chimaeric protein consisting of the CSLD3 protein containing the CESA6 catalytic domain (Park et al., 2011). Additionally, the csla2/csla3/csla9 triple mutant lacks detectable glucomannan in the inflorescence stem (Goubet et al., 2009), indicating that the quantity of mannan synthesized by CSLD proteins must be small. It also is not clear why the patterns of mannans synthesized by CSLA proteins are aberrant in the csld2/csld3/csld5 triple mutant. In light of the lack of consensus about the function(s) of CSLD proteins, additional studies are needed to clarify their functions.
DO THE CSLCs ENCODE XYLOGLUCAN GLUCAN SYNTHASES?
Like the CSLAs, CSLC sequences have been found in many extant species of Viridiplantae, spanning several divisions, including Magnoliophyta, Lycopodiophyta, Bryophyta, and Charophyta (Del Bem and Vincentz, 2010). Within angiosperms, CSLCs have been found in all species surveyed thus far (Yin et al., 2009; Del Bem and Vincentz, 2010), including three species that have been used as models to investigate CSLC function: nasturtium (TmCSLC), Arabidopsis (AtCSLC4, AtCSLC5, AtCSLC6, AtCSLC8, AtCSLC12) and barley (HvCSLC1, HvCSLC2, HvCSLC3, HvCSLC4, and possibly HvCSLC5).
The first evidence that xyloglucan glucan synthase (XGS) might be encoded by a member of the CSLC family was provided by Cocuron et al. (2007) using a comparative genomics approach. Using nasturtium (Tropaeolum majus) seeds, which utilize xyloglucan as the primary seed storage polysaccharide (Gidley et al., 1991), transcriptional profiling was used to identify genes preferentially expressed during the stage of seed development when xyloglucan deposition occurs. The only CSL gene transcripts detected by this analysis were those of the TmCSLC gene, a homolog of Arabidopsis CSLC4 (AtCSLC4). Transgenic Pichia pastoris cells expressing the TmCSLC or AtCSLC4 protein produced soluble 1,4-β-glucans with a low degree of polymerization (DP4–DP6), indicating that these two proteins have glucan synthase activity (Table 1). While efforts to coexpress, in P. pastoris, the Arabidopsis CSLC4 and a xyloglucan xylosyltransferase (AtXXT1) protein did not result in the synthesis of xyloglucan (likely due, at least in part, to the absence of UDP-xylose metabolism in P. pastoris; De Schutter et al., 2009), several additional lines of evidence support the hypothesis that AtCSLC4 is an XGS. P. pastoris cultures expressing AtCSLC4 and AtXXT1 together produced insoluble 1,4-β-glucans with a higher degree of polymerization than glucans synthesized by expression of AtCSLC4 alone, possibly indicating that the AtCSLC4 and AtXXT1 proteins act in a cooperative manner to produce xyloglucan. Additionally, in Arabidopsis there is a strong correlation between the expression patterns of the AtCSLC4 and AtXXT1 genes, suggesting that these two proteins participate in related processes. Finally, AtCSLC4 is a Golgi-localized protein, and xyloglucan synthesis is known to take place within this organelle (Ray et al., 1969; Ray, 1980; White et al., 1993). Although Cocuron et al. (2007) show that CSLC4 has glucan synthase activity and present a reasonable argument that CSLC4 is an XGS, it is unknown if other CSLC members have glucan synthase activity and whether mutation of one or more CSLC genes would affect xyloglucan content or structure.
In contrast to using an experimental system in which a significant portion of the hemicellulose present is xyloglucan, Dwivany et al. (2009) studied CSLCs of barley (Hordeum vulgare L.), where xyloglucan is a minor cell wall component (Sakurai and Masuda, 1978; Kato et al., 1981; Fincher, 1993). The authors identified and characterized four barley CSLCs, HvCSLC1–4 (a fifth, HvCSLC5 also was identified but not characterized). Phylogenetic analysis of CSLC family members from several eudicots and monocots, Physcomitrella patens, Selaginella moellendorffii, and Chara globularis indicates that the CSLC family contains four clades: HvCSLC1, 2, 4, and AtCSLC12 belonging to clade 1, AtCSLC4, 5, 8, and HvCSLC3 belonging to clade 2, Physcomitrella and Selaginella CSLCs comprise clade 3, and AtCSLC6 belongs to clade 4. In addition to taxonomic relationships having an effect on tree structure (i.e., clade 3), the authors hypothesize that when considered in conjunction with the biochemical and molecular evidence (discussed below), the structure of the phylogenetic tree may show functional specialization, with members of clades 2 and 4 having GS and XGS activities, respectively. Based on results of transcriptional profiling of barley organs and tissues, coexpression analysis of HvCSLCs and putative barley xyloglucan xylosyltransferases (HvGT1–5), and its high sequence identity and similarity to AtCSLC4, Dwivany et al. (2009) concluded that HvCSLC3 is probably involved in xyloglucan biosynthesis. Alternatively, in barley suspension cultured cells, which the authors show have transient, low levels of xyloglucan and barely detectable HvCLSC3 transcripts, the CSLC proteins immunolocalize to the plasma membrane in immuno-EM and membrane fractionation experiments. Based on results from these experiments and the molecular characterization at the gene and transcript levels, Dwivany et al. (2009) conclude that there is insufficient evidence to assign functions to HvCSLC1 and HvCSLC4. However, the authors propose that HvCSLC2 likely is not involved in xyloglucan biosynthesis, and instead suggest that it may be involved in cellulose biosynthesis. Although the conclusions reached by Dwivany et al. (2009) are plausible, additional evidence from heterologous expression and mutant genetic studies would strengthen their arguments.
Key to identifying the function(s) of the CSLC family members is to determine how polysaccharide content and structure is affected in plants with mutant CSLC genes. To this end, there is an abundance of Arabidopsis T-DNA insertion lines available from several sources, and mutants for all members of the CSLC family are present within these mutant collections. Furthermore, reverse genetics resources currently are being developed in the model grass species Brachypodium distachyon (Thole et al., 2010, 2012). Because of the likelihood that genetic redundancy exists among the five members of the Arabidopsis CSLC family, it probably will be necessary to generate mutant lines harboring multiple mutant CSLC genes to determine whether members of the Arabidopsis CSLC family are involved in xyloglucan biosynthesis. While it is possible that mutants with severe reductions of xyloglucan content could prove lethal, the existence of the xxt1/xxt2 mutant, which lacks detectable xyloglucan and grows normally under laboratory conditions (Cavalier et al., 2008), shows that it is possible to develop viable xyloglucan mutants. Therefore, if members of the CSLC family are involved in xyloglucan biosynthesis it should be possible to develop Arabidopsis lines harboring multiple mutant CSLC genes.
In addition to genetic redundancy and lethality, determining the effects upon polysaccharide content in cslc mutants could pose a significant challenge if members of the CSLC family are involved in cellulose biosynthesis. One potential difficulty would be the inability to distinguish between cellulose synthesized by CSLCs versus CESAs. Another would be determining if changes in amorphous or crystalline cellulose are due directly to the mutation or are the result of a secondary response to the mutation. Keeping these challenge in mind, detailed studies of such mutants, coupled with biochemical studies of recombinant CSLC proteins ultimately are expected to provide the evidence needed to conclusively define the function(s) of many CSLC family members.
CONCLUDING REMARKS
Within the last decade, our understanding of the functions of CSLA and CSLC proteins has markedly improved. However, many important questions remain relating to the evolution and functions of these related sequences: What is the architecture of the transcriptional network controlling expression of these genes? What proteins are present in the carbohydrate synthesizing enzyme complexes likely to contain these proteins, and how are these complexes regulated? What factors influence the processes of polysaccharide synthesis (initiation, elongation, and termination)? What are the physiological roles and factors influencing the structures of polysaccharides synthesized by CSLA and CSLC proteins? A body of research shows that the CSLA and CSLC families can be successfully studied using heterologous expression and forward and reverse genetics. By leveraging these powerful tools it should be possible to gain significant insights into the specific functions of the CSLA and CSLC proteins and the synthesis of plant polysaccharides in general.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to acknowledge those authors whose studies we were unable to discuss due to space constraints, and reviewers of this manuscript for their suggestions and constructive criticism. Aaron H. Liepman thanks Gregg Sobocinski (Department of Molecular, Cellular and Developmental Biology, University of Michigan) for assistance with confocal imaging. This work was funded, in part, by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494).
REFERENCES
- Auxtova-Samajova O., Liskova D., Kakoniova D., Kubackova M., Karacsonyi S., Bilisics L. (1996). Inhibition of auxin stimulated short-term elongation growth of pea steam segments by galactoglucomannan-derived oligosaccharides. J. Plant Physiol. 147 611–613 [Google Scholar]
- Benova-Kakosova A., Digonnet C., Goubet F., Ranocha P., Jauneau A., Pesquet E., Barbier O., Zhang Z., Capek P., Dupree P., Liskova D., Goffner D. (2006). Galactoglucomannans increase cell population density and alter the protoxylem/metaxylem tracheary element ratio in xylogenic cultures of zinnia. Plant Physiol. 142 696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A. J., Yoo C. M., Mutwil M., Jensen J. K., Hou G., Blaukopf C., Sorensen I., Blancaflor E. B., Scheller H. V., Willats W. G. (2008). Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 148 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckeridge M. S. (2010). Seed cell wall storage polysaccharides: models to understand cell wall biosynthesis and degradation. Plant Physiol. 154 1017–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier D. M., Lerouxel O., Neumetzler L., Yamauchi K., Reinecke A., Freshour G., Zabotina O. A., Hahn M. G., Burgert I., Pauly M., Raikhel N. V., Keegstra K. (2008). Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell. 20 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37 D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N. C. (2011). Progress in the biological synthesis of the plant cell wall: new ideas for improving biomass for bioenergy. Curr. Opin. Biotechnol. 10.1016/j.copbio.2011.12.003 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Cocuron J. C., Lerouxel O., Drakakaki G., Alonso A. P., Liepman A. H., Keegstra K., Raikhel N., Wilkerson C. G. (2007). A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J., Brandizzi F., Liepman A. H., Keegstra K. (2010). Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 64 1028–1037 [DOI] [PubMed] [Google Scholar]
- Del Bem L. E., Vincentz M. G. (2010). Evolution of xyloglucan-related genes in green plants. BMC Evol. Biol. 10 341 10.1186/1471-2148-10-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K., Lin Y.-C., Tiels P., Van Hecke A., Glinka S., Weber-Lehmann J., Rouze P., Van de Peer Y., Callewaert N. (2009). Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotech. 27 561 [DOI] [PubMed] [Google Scholar]
- Dhugga K. S. (2011). Biosynthesis of non-cellulosic polysaccharides of plant cell walls. Phytochemistry 74 8–19 [DOI] [PubMed] [Google Scholar]
- Dhugga K. S., Barreiro R., Whitten B., Stecca K., Hazebroek J., Randhawa G. S., Dolan M., Kinney A. J., Tomes D., Nichols S., Anderson P. (2004). Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303 363–366 [DOI] [PubMed] [Google Scholar]
- Doblin M., Pettolino F., Bacic A. (2010). Plant cell walls: the skeleton of the plant world. Funct. Plant Biol. 37 357–381 [Google Scholar]
- Doblin M. S., De Melis L., Newbigin E., Bacic A., Read S. M. (2001). Pollen tubes of Nicotiana alata express two genes from different beta-glucan synthase families. Plant Physiol. 125 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivany F. M., Yulia D., Burton R. A., Shirley N. J., Wilson S. M., Fincher G. B., Bacic A., Newbigin E., Doblin M. S. (2009). The CELLULOSE-SYNTHASE LIKE C (CSLC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol. Plant 2 1025–1039 [DOI] [PubMed] [Google Scholar]
- Endler A., Persson S. (2011). Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 4 199–211 [DOI] [PubMed] [Google Scholar]
- Farrokhi N., Burton R. A., Brownfield L., Hrmova M., Wilson S. M., Bacic A., Fincher G. B. (2006). Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 4 145–167 [DOI] [PubMed] [Google Scholar]
- Favery B., Ryan E., Foreman J., Linstead P., Boudonck K., Steer M., Shaw P., Dolan L. (2001). KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher G. B. (1993). “Cell wall metabolism in barley,” in Barley: Genetics, Biochemistry, Molecular Biology and Biotechnology, ed. Shewry P. R. (Wallingford: CAB International; ) 413–437 [Google Scholar]
- Fincher G. B. (2009). Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 149 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway M. E., Eng R. C., Schiefelbein J. W., Wasteneys G. O. (2011). Root hair-specific disruption of cellulose and xyloglucan in AtCSLD3 mutants, and factors affecting the post-rupture resumption of mutant root hair growth. Planta 233 985–999 [DOI] [PubMed] [Google Scholar]
- Gidley M. J., Lillford P. J., Rowlands D. W., Lang P., Dentini M., Crescenzi V., Edwards M., Fanutti C., Reid J. S. (1991). Structure and solution properties of tamarind-seed polysaccharide. Carbohydr. Res. 214 299–314 [DOI] [PubMed] [Google Scholar]
- Gille S., Cheng K., Skinner M. E., Liepman A. H., Wilkerson C. G., Pauly M. (2011). Deep sequencing of voodoo lily (Amorphophallus konjac): an approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 234 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T., Lauricha J., Tran H., Keegstra K., Raikhel N. (2004). The Cell Wall Navigator database. A systems-based approach to organism-unrestricted mining of protein families involved in cell wall metabolism. Plant Physiol. 136 3003–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D. S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40 D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F., Barton C. J., Mortimer J. C., Yu X., Zhang Z., Miles G. P., Richens J., Liepman A. H., Seffen K., Dupree P. (2009). Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 60 527–538 [DOI] [PubMed] [Google Scholar]
- Goubet F., Misrahi A., Park S. K., Zhang Z., Twell D., Dupree P. (2003). AtCslA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 131 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T., Osborne E., Youngs H., Misson J., Nussaume L., Somerville C. (2004). Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11 279–286 [Google Scholar]
- Handford M. G., Baldwin T. C., Goubet F., Prime T. A., Miles J., Yu X., Dupree P. (2003). Localisation and characterisation of cell wall mannan polysaccharides in Arabidopsis thaliana. Planta 218 27–36 [DOI] [PubMed] [Google Scholar]
- Hazen S. P., Scott-Craig J. S., Walton J. D. (2002). Cellulose synthase-like (CSL) genes of rice. Plant Physiol. 128 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Iki K., Matsuda K. (1981). Cell-wall polysaccharides of immature barley plants. II. Characterization of a xyloglucan. Agric. Biol. Chem. 45 2745–2753 [Google Scholar]
- Keegstra K., Raikhel N. V. (2001). Plant Glycosyltransferases. Curr. Opin. Plant Biol. 4 219–224 [DOI] [PubMed] [Google Scholar]
- Kim C. M., Park S. H., Je B. I., Park S. H., Park S. J., Piao H. L., Eun M. Y., Dolan L., Han C. D. (2007). OsCSLD1, a Cellulose Synthase-Like D1 gene, is required for root hair morphogenesis in rice (Oryza sativa L.). Plant Physiol. 143 1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J., Marcus S. E., Knox J. P. (2011). Cell wall biology: perspectives from cell wall imaging. Mol. Plant 4 212–219 [DOI] [PubMed] [Google Scholar]
- Leroux O., Leroux F., Bagniewska-Zadworna A., Knox J. P., Claeys M., Bals S., Viane R. L. (2011). Ultrastructure and composition of cell wall appositions in the roots of Asplenium (Polypodiales). Micron 42 863–870 [DOI] [PubMed] [Google Scholar]
- Lerouxel O., Cavalier D. M., Liepman A. H., Keegstra K. (2006). Biosynthesis of plant cell wall polysaccharides – a complex process. Curr. Opin. Plant Biol. 9 621–630 [DOI] [PubMed] [Google Scholar]
- Liepman A. H., Nairn C. J., Willats W. G., Sorensen I., Roberts A. W., Keegstra K. (2007). Functional genomic analysis supports conservation of function among CELLULOSE SYNTHASE-LIKE A gene family members and suggests diverse roles of mannans in plants. Plant Physiol. 143 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A. H., Wightman R., Geshi N., Turner S. R., Scheller H. V. (2010). Arabidopsis – a powerful model system for plant cell wall research. Plant J. 61 1107–1121 [DOI] [PubMed] [Google Scholar]
- Liepman A. H., Wilkerson C. G., Keegstra K. (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. U.S.A. 102 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Awano T., Takabe K., Fujita M. (2000). Immunolocalization of glucomannans in the cell wall of differentiating tracheids in Chamaecyparis obtusa. Protoplasma 213 148–156 [Google Scholar]
- Marcus S. E., Blake A. W., Benians T. A., Lee K. J., Poyser C., Donaldson L., Leroux O., Rogowski A., Petersen H. L., Boraston A., Gilbert H. J., Willats W. G., Knox J. P. (2010). Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 64 191–203 [DOI] [PubMed] [Google Scholar]
- Marks M. D., Betancur L., Gilding E., Chen F., Bauer S., Wenger J. P., Dixon R. A., Haigler C. H. (2008). A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J. 56 483–492 [DOI] [PubMed] [Google Scholar]
- McCann M. C., Carpita N. C. (2008). Designing the deconstruction of plant cell walls. Curr. Opin. Plant Biol. 11 314–320 [DOI] [PubMed] [Google Scholar]
- Meier H., Reid J. S. G. (1982). “Reserve polysaccharides other than starch in higher plants,” in Encyclopedia of Plant Physiology, eds. Loewus F. A., Tanner W. (Berlin: Springer; ) 418–471 [Google Scholar]
- Moreira L. R., Filho E. X. (2008). An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79 165–178 [DOI] [PubMed] [Google Scholar]
- Ordaz-Ortiz J. J., Marcus S. E., Knox J. P. (2009). Cell wall microstructure analysis implicates hemicellulose polysaccharides in cell adhesion in tomato fruit pericarp parenchyma. Mol. Plant 2 910–921 [DOI] [PubMed] [Google Scholar]
- Park S., Szumlanski A. L., Gu F., Guo F., Nielsen E. (2011). A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 13 973–980 [DOI] [PubMed] [Google Scholar]
- Pauly M., Keegstra K. (2010). Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 13 305–312 [DOI] [PubMed] [Google Scholar]
- Penning B. W., Hunter C. T., III., Tayengwa R., Eveland A. L., Dugard C. K., Olek A. T., Vermerris W., Koch K. E., McCarty D. R., Davis M. F., Thomas S. R., McCann M. C., Carpita N. C. (2009). Genetic resources for maize cell wall biology. Plant Physiol. 151 1703–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper Z. A., Michel G., Herve C., Domozych D. S., Willats W. G., Tuohy M. G., Kloareg B., Stengel D. B. (2011). Evolution and diversity of plant cell walls: from algae to flowering plants. Annu. Rev. Plant Biol. 62 567–590 [DOI] [PubMed] [Google Scholar]
- Ray P. M. (1980). Cooperative action of β-glucan synthetase and UDP-xylose xylosyl transferase of Golgi membranes in the synthesis of xyloglucan-like polysaccharide. Biochim. Biophys. Acta 629 431–444 [DOI] [PubMed] [Google Scholar]
- Ray P. M., Shininger T. L., Ray M. M. (1969). Isolation of β-glucan synthetase particles from plant cells and identification with Golgi membranes. Proc. Natl. Acad. Sci. U.S.A. 64 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T., Somerville C. R. (2001). Integrative approaches to determining Csl function. Plant Mol. Biol. 47 131–143 [PubMed] [Google Scholar]
- Richmond T. A., Somerville C. R. (2000). The cellulose synthase superfamily. Plant Physiol. 124 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. W., Bushoven J. T. (2007). The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol. Biol. 63 207–219 [DOI] [PubMed] [Google Scholar]
- Sakurai N., Masuda Y. (1978). Auxin-induced changes in barley coleoptile cell wall composition. Plant Cell Physiol. 19 1217–1223 [Google Scholar]
- Sandhu A. P., Randhawa G. S., Dhugga K. S. (2009). Plant cell wall matrix polysaccharide biosynthesis. Mol. Plant 2 840–850 [DOI] [PubMed] [Google Scholar]
- Scheible W.-R., Pauly M. (2004). Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 7 285–295 [DOI] [PubMed] [Google Scholar]
- Scheller H. V., Ulvskov P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61 263–289 [DOI] [PubMed] [Google Scholar]
- Schroder R., Atkinson R. G., Redgwell R. J. (2009). Re-interpreting the role of endo-beta-mannanases as mannan endotransglycosylase/hydrolases in the plant cell wall. Ann. Bot. 104 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder R., Wegrzyn T. F., Sharma N. N., Atkinson R. G. (2006). LeMAN4 endo-β-mannanase from ripe tomato fruit can act as a mannan transglycosylase or hydrolase. Planta 224 1091–1102 [DOI] [PubMed] [Google Scholar]
- Silva G. B., Ionashiro M., Carrara T. B., Crivellari A. C., Tine M. A., Prado J., Carpita N. C., Buckeridge M. S. (2011). Cell wall polysaccharides from fern leaves: evidence for a mannan-rich Type III cell wall in Adiantum raddianum. Phytochemistry 72 2352–2360 [DOI] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Perrson S., Raab T., Vorwerk S., Youngs H. (2004). Toward a systems approach to understanding plant cell walls. Science 306 2206–2211 [DOI] [PubMed] [Google Scholar]
- Suzuki S., Li L., Sun Y. H., Chiang V. L. (2006). The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 142 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole V., Peraldi A., Worland B., Nicholson P., Doonan J. H., Vain P. (2012). T-DNA mutagenesis in Brachypodium distachyon. J. Exp. Bot. 63 567–576 [DOI] [PubMed] [Google Scholar]
- Thole V., Worland B., Wright J., Bevan M. W., Vain P. (2010). Distribution and characterization of more than 1000 T-DNA tags in the genome of Brachypodium distachyon community standard line Bd21. Plant Biotechnol. J. 8 734–747 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomas S., Edvardsson E., Eland C., Singh S. K., Zadik D., Aspeborg H., Gorzsas A., Teeri T. T., Sundberg B., Persson P., Bennett M., Marchant A. (2007). Genomic-assisted identification of genes involved in secondary growth in Arabidopsis utilising transcript profiling of poplar wood-forming tissues. Physiol. Plant. 129 415–428 [Google Scholar]
- Verhertbruggen Y., Yin L., Oikawa A., Scheller H. V. (2011). Mannan synthase activity in the CSLD family. Plant Signal. Behav. 6 1620–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cnops G., Vanderhaeghen R., De Block S., Van Montagu M., Van Lijsebettens M. (2001). AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol. 126 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A. R., Xin Y., Pezeshk V. (1993). Xyloglucan glucosyltransferase in Golgi membranes from Pisum sativum (pea). Biochem. J. 294 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Verhertbruggen Y., Oikawa A., Manisseri C., Knierim B., Prak L., Jensen J. K., Knox J. P., Auer M., Willats W. G., Scheller H. V. (2011). The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol. Plant 4 1024–1037 [DOI] [PubMed] [Google Scholar]
- Yin Y., Huang J., Xu Y. (2009). The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9 99 10.1186/1471-2229-9-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngs H. L., Hamann T., Osborne E., Somerville C. R. (2007). “The cellulose synthase superfamily,” in Cellulose: Molecular and Structural Biology, eds Brown M., Saxena I. M. (Berlin: Springer; ) 35–49 [Google Scholar]
- Zablackis E., Huang J., Darvill A. G., Albersheim P. (1995). Characterization of the cell wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 107 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Nam J., Carpita N., Matthysse A. G., Gelvin S. B. (2003). Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol. 133 1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]