Abstract
Although epidemiological data provides evidence that there is an interaction between genetics (nature) and the social and physical environments (nurture) in human development; the main open question remains the mechanism. The pattern of distribution of methyl groups in DNA is different from cell-type to cell type and is conferring cell specific identity on DNA during cellular differentiation and organogenesis. This is an innate and highly programmed process. However, recent data suggests that DNA methylation is not only involved in cellular differentiation but that it is also involved in modulation of genome function in response to signals from the physical, biological and social environments. We propose that modulation of DNA methylation in response to environmental cues early in life serves as a mechanism of life-long genome “adaptation” that molecularly embeds the early experiences of a child (“nurture”) in the genome (“nature”). There is an emerging line of data supporting this hypothesis in rodents, non-human primates and humans that will be reviewed here. However, several critical questions remain including the identification of mechanisms that transmit the signals from the social environment to the DNA methylation/demethylation enzymes.
Key words: DNA methylation, psychiatry, development, epidemiology, environment
Introduction
DNA methylation is a covalent modification of DNA by addition of methyl groups to cytosines at specific positions in the genome.1 The pattern of distribution of methyl groups in cytosines in the genome differs from cell type to cell type creating cell-type identity to DNA in addition to the genetic or ancestral identity, which is encoded in the sequence of nucleotides in DNA. DNA methylation patterns are generated during development and it was believed that they were involved in terminal cell differentiation.2 The main enigma in cellular differentiation in multicellular organisms has been: how could one genome encode multitude of phenotypes? DNA methylation by providing cell-type identity within the chemical entity of DNA provides an attractive mechanism for genomes to acquire differential identities within the same organism. The last three decades of research in DNA methylation focused on the role of DNA methylation in cellular differentiation. Data supporting the role of DNA methylation emerged almost three decades ago3 and was recently confirmed by whole genome methods of mapping DNA methylation including next generation sequencing.4
The delineation of the role of DNA methylation in cellular differentiation illustrates that DNA methylation functions as a mechanism that expands the functionality of the genome by enabling similar genomes to encode diversely stable phenotypes. One attractive hypothesis is that DNA methylation is not limited to cellular differentiation as dictated by endogenous innate programs but could also function as a genome adaptation mechanism to external signals from the environment. Responses to external environments would not necessarily override cell type specific DNA methylation patterns but could modulate these patterns.
Similar to cellular differentiation, DNA methylation variations in response to the environment could explain how certain environments stably alter the phenotype without affecting the genotype. DNA methylation variation in response to environmental exposures might emerge sporadically and then maintained by the DNA methylation maintenance machinery. However, recent data suggests that DNA methylation variations might be driven by organized responses not unlike the processes that drive cellular differentiation. It is proposed here that DNA methylation could play a role in genome adaptation at multiple time scales and in response to multiple signals and be involved in several roles that require expanding the functionality of the genome.
DNA Methylation is a Mechanism for Expanding the Scope of Phenotypes Encoded by a Single Genotype: The Example of Cellular Differentiation
Every multicellular organism is a system whereby one genotype expresses multiple phenotypes. Although all the cells in the body contain the same genetic sequence, the DNA methylation pattern is not identical in different tissues.1,2 Thus a DNA molecule contains two layers of identity. The ancestral information encoded in the sequence and the cell type-specific identity contained in the pattern of distribution of methyl moieties and possibly 5-hydroxymethylcytosine moieties. 5-hydroxymethylcytosine is a downstream enzymatic modification of 5-methylcytosine.5 Different enzymatic processes replicate these two layers of information. While the DNA polymerase enzymes replicate the ancestral genetic information, DNA methylation is copied and maintained by DNA methyltransferases (DNMT).6 These enzymes catalyze the transfer of methyl groups from the methyl donor S-adenosyl methionine to the 5′ position in cytosine in DNA.7 Since cellular differentiation is maintained for a life-time, there must be mechanisms that accurately maintain the DNA methylation pattern as well as mechanisms that prevent a drift in the DNA methylation pattern during the life course. It has been generally accepted that the following particular biochemical elements ensured the accurate maintenance of DNA methylation pattern in differentiated tissues. First, a large fraction of cytosines in DNA are found in the palindrome sequence CG;8 following DNA replication unmethylated CGs in the nascent strand are positioned across methylated CGs in the parental strand that could serve as a template for copying the DNA methylation pattern. Second, DNMT1 preferentially recognizes a newly synthesized CG dinucleotide when the CG on the template sequence is methylated.9 Third, it was believed that there were no enzymatic processes that could add (de novo methylation) or remove methyl groups (demethylation) from DNA in a mature differentiated cell. These processes in combination were believed to be responsible for faithfully maintaining the DNA methylation pattern in differentiated cells and tissues.
This rigid picture of the state of the DNA methylation pattern in differentiated tissues is in accordance with the role that it is hypothesized to play in maintaining the terminally differentiated state of cells and tissues. Indeed, loss of methylation driven by either knock down of DNMT1 with antisense depletion10 or with drugs that inhibit DNA methylation such as 5-azacytidine11 changes the state of differentiation of cells. However, there are reasons to believe that in spite of the perceived consistency of tissue specific differential DNA methylation patterns throughout the life course, there is a measure of plasticity in the DNA methylation pattern that could expand the scope of its roles in defining a more dynamic relationship between environments the genome and the phenotype.
The Possibility of Adaptive Dynamic DNA Methylation Responses after Birth and in Postmitotic Tissues: De Novo DNMTs and Demethylases
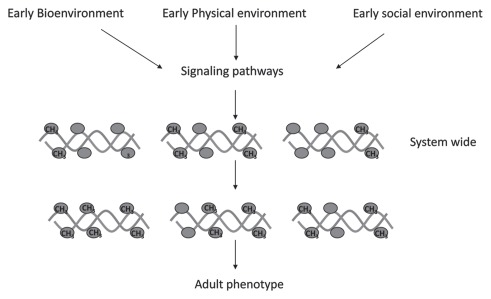
The social and physical environment influences human development after birth and during different life cycle stations. For example, social adversity early in life has a profound impact on life-long physical health and behavior.12–14 Thus, differentiation of the phenotype occurs in response to external signals from the social environment. Similar to cellular differentiation it involves diversification of the phenotype without altering the genotype. An attractive hypothesis is that DNA methylation might play a role in diversification of the phenotypic potential of a single genome in response to external signals during post partum development as much as it plays a role in diversification of genome function in response to innate signals of differentiation (Fig. 1).
Figure 1.
Adaptive response of the genome in early life. Signals triggered by early life environments turn on signaling pathways in brain as well as peripheral tissues that target chromatin and DNA methylation/demethylation enzymes to specific loci in the genome resulting in system-wide differential DNA methylation patterns. These adapt the life long phenotype to the anticipated environments.
The first evidence that early environmental exposures could alter the phenotype through altering DNA methylation patterns came from the Jirtle lab that demonstrated an effect of maternal diet on the agouti color phenotype in agouti mice which was mediated through methylation of a transposable element.15 The impact of methyl-rich diets during gestation or the impact of other chemicals during gestation could be explained just as a stochastic chemical interference in the enzymatic DNA methylation reactions that are actively laying down the DNA methylation pattern during embryogenesis. However, the responsivity of the DNA methylation pattern to social-adversity signals after birth and completion of embryogenesis as discussed below could not be explained just as a stochastic change in DNA methylation reaction kinetics.
If DNA methylation acts as a responsive biological signal even in postmitotic tissues such as neurons, the DNA methylation reaction has to be reversible;16 both demethylation and de novo methylation should be possible in nondividing tissues. It was long understood that during gestation changes in DNA methylation that sculpt the DNA methylation pattern in a tissue specific manner do occur and that these changes must be catalyzed by enzymatic processes that add and remove DNA methylation.1 For decades, DNA methylation enzymology focused on DNA methyl transferases. Several DNA methyl transferases (DNMTs) were characterized. DNMT1 is a hemimethylated DNA methyltransferase that is believed to be responsible for replicating the DNA methylation pattern during cell division and to maintain the fidelity of DNA methylation during cell division.9 DNMT3a and DNMT3b were shown to act as de novo methyltransferases.17 De novo methylation was originally believed to be limited to the early stages of development. This is essential if the DNA methylation pattern is to remain rigid after completion of DNA replication. De novo methylation in a differentiated cell would alter the DNA methylation pattern. It is clear however that DNMT3A is present in adult neurons18 supporting the possibility of change in DNA methylation in postmitotic neurons.
CG is a palindrome sequence and therefore a methylated CG in the parental strand lends itself to template dependent copying during cell division. The discovery of a large number of non-CG methylation in the genome4 raises questions on the mechanisms involved in maintaining these patterns of methylation.19 Although these non-CG methylation sites were discovered mainly in stem cells,4 it is still possible that non-CG methylation is present to a certain extent in mature cells as well.20 Methylation of nonpalindromic sequences cannot be guided by the state of methylation of the template and each round of methylation following DNA replication is essentially de novo methylation. Is there a mechanism that guides the de novo enzymes to specific sites? If there are such mechanisms and indeed there is evidence for targeting of DNMTs to specific sites, then it implies that replication of the DNA methylation pattern is not exclusively an automatic copying process. This is more consistent with a dynamic DNA methylation state.
DNA demethylation is critical for a dynamic DNA methylation pattern. DNA demethylation could occur as a passive process during cell division when DNA methyltransferases are blocked by specific factors.1 Since cell division is obviously abundant during embryogenesis, passive demethylation could theoretically explain DNA demethylation during gestation and cellular and tissue differentiation. This could explain why DNA demethylases didn't attract much attention in the past. The absence of a DNA demethylase is consistent with a rigid DNA methylation pattern post cellular differentiation. A situation where maintenance DNA methyltransferases exclusively maintain the fidelity of DNA methylation in mitotic differentiating cells in the absence of de novo methyltransferases and demethylases would prevent a drift in the DNA methylation pattern and is critical for a rigid “terminal” DNA methylation pattern that guards “terminal” differentiation. However, data that has consistently pointed to replication-independent DNA demethylation has forced us to revisit this issue and a wealth of enzymatic processes that could remove DNA demethylation in the absence of cell division have been defined.21–25 It has been shown that brain extracts are capable of demethylating “naked” DNA substrate in vitro.20,26,27 The strongest evidence for dynamic methylation-demethylation comes from several studies showing active demethylation in postmitotic neurons.18,28–30 Conditional knock out of DNMT1 in postmitotic neurons results in DNA demethylation suggesting the presence of demethylation activity in nondividing neurons which is critical for a dynamic methylation pattern in the brain.31
The main issue in the field remains however whether DNA methylation is truly a reversible reaction that involves removal of the methyl moiety and its release16,32 or whether DNA demethylation requires excision of the methylated base and its replacement by an unmethylated cytosine through a process of DNA repair. The vast majority of the data to date points to a repairbased demethylation process. First, the methylated cytosine could be removed by a glycosylase activity and the abasic site that was created is then repaired and replaced with an unmethylated cytosine.33,34 Second, DNMTs were proposed to deaminate the methyl cytosine to thymidine creating a C/T mismatch, which is then corrected by a mismatch-repair mechanism.35 DNMTs were previously shown to deaminate 5-methylcytosines36,37 under conditions of low SAM. Third, growth arrest and DNA-damage-inducible, a (GADD45A), a DNA repair protein was proposed to participate in catalysis of active DNA demethylation by an unknown DNA repair based mechanism.38 However, this was disputed.39 Other studies have suggested involvement of GADD45B in demethylation in the brain.40 Fourth, a complex sequence of coupled enzymatic reactions of deamination and mismatch repair were shown to be involved in demethylation in zebrafish: activation-induced cytidine deaminase (AID, which converts 5-meC to thymine), a G:T mismatch-specific thymine glycosylase methyl-CpG binding domain protein 4 (MBD4) and repair promoted by GADD45A.41 AID has been implicated in the global demethylation in mouse primordial germ cells as well.42 An open question is the role of the newly discovered modification 5-hydroxymethylcytosine as a potential intermediate in the DNA demethylation reaction.5 5-hydroxymethylcytosine was proposed to serve as a modification of 5-methylcytosine that marks it for base excision repair and demethylation. Recent data suggest that TET1 the enzyme that catalyzes the hydroxylation of 5-methylcytosine is present and required for stem cell maintenance of inner cell mass specification43 and for activity driven demethylation in neurons. 5-hydroxymethylation catalyzed by TET1 is followed by deamination of the 5-hydroxymethylated base by AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA-editing enzyme complex) family of cytidine deaminases and base excision repair enzymes replace the deaminated base with an unmethylated cytosine (BER).44
The main challenge in accepting such a complex multienzyme repair-based mechanisms that involves a sequence of modifications to 5-methylcytosine as a life-long physiological process is that it invokes constant mutagenic stress and damage to the integrity of DNA; constant modification by deamination, breaking and fixing of the DNA seems to be an extremely dangerous way to maintain the DNA methylation equilibrium in both developing embryo and postmitotic neurons. Nevertheless, there is evidence for BER activity during reprogramming of mouse germ cells, a time-point in development that involves extensive DNA demethylation.45 It is unclear however why is there a need for modification of the 5-methylcytosine base first by hydroxylation and then by deamination to target it for base excision especially since glycosylases that could recognize methylated cytosines such as MBD4 and 5-methylcytosine DNA glycosylase do exist.46,47
In contrast to these complicated repair based mechanisms we have previously proposed that demethylation is truly a reversible reaction that involves removal of the methyl moiety rather than modifying and breaking the DNA and then fixing it with an unmethylated cytosine.16 We proposed that the methylated DNA binding protein MBD2 was a bona fide demethylase that removed methyl groups from DNA and truly reversed the DNA methylation reaction. This is to date the only proposed bona fide demethylase. MBD2 has been implicated in the activation of both methylated and unmethylated genes.48,49 Several groups50,51 have contested the demethylase and transcriptional activating properties of MBD2. Studies by Detich et al. have demonstrated however MBD2 demethylase activity in vitro.52 Hamm et al. have proposed an oxidative mechanism of 5-methylcytosine DNA demethylation by MBD2.53 According to this mechanism, oxidation of the methyl moiety generates 5-hydroxymethylcytosine, which is followed by release of the methyl residue as formaldehyde. Although this mechanism implicates 5-hydroxymethylcytosine in demethylation it suggests however that it is an intermediary in the demethylation enzymatic reaction rather than a modification that targets 5-hydroxymethylcytosine for excision repair.
There is no evidence that the recently described complex enzymatic reactions could catalyze demethylation of methylated DNA in vitro. Most of the evidence is based on depletion of the different predicted components of the complex in cells. However, there is evidence from several groups for enzymatic demethylation activity that doesn't require DNA repair in brain cell extracts.26,27 In a very interesting study Fuso et al. showed DNA demethylase activity in brain extracts and that depletion of MBD2 by an antibody blocks this activity supporting a role for MBD2 as a demethylase in the brain.20 Moreover the authors elegantly show that this activity is modulated in vivo by modulating one carbon metabolism. These data suggest that it is possible that DNA demethylation is a true enzymatic reaction that reveres the DNA methylation state rather than a complex of repair and modification activities. Future studies are critical for resolving this central question in DNA methylation.
DNA Methylation Pattern as a Genome Adaptation Mechanism to the External Environment
The possibility that replication of the DNA methylation pattern is not exclusively determined by the template allows for some degrees of freedom in the DNA methylation pattern. This degree of freedom could potentially be utilized to modify the DNA methylation pattern in response to external signals even after the completion of cellular differentiation. The main question that must be addressed is whether these additional changes in DNA methylation are stochastic and result from random interference in enzymes that replicate DNA methylation during cell division or whether these are organized “adaptive” responses similar to the innate processes that delineate tissue specific DNA methylation patterns during gestation. For example, there are signaling pathways that could respond to external signals and the response of neurons to neurotransmitter release is a good example. These pathways converge on trans-activating factors that could deliver DNA and chromatin modifying enzymes to specific targets in the genome and modulate the DNA methylation pattern in a responsive organized manner.
It is becoming clear now that DNMTs are targeted to specific sequences in the genome and that the targeting factors are required not only for generating the patterns of methylation but also for maintaining the pattern of DNA methylation. For example, UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1), also known as NP95 in mouse and ICBP90 in human is required for targeting DNMT1 to newly replicating hemimethylated DNA.54 DNMTs are found in complexes with other proteins that include other chromatin modifying proteins such as HDAC1 and HDAC2.55,56 DNMT3A is allosterically activated by histone H3 tails lacking lysine 4 (K4) methylation.57 The discovery that DNMT1 and other DNMTs are targeted to specific sites by chromatin modifying enzymes or even by the state of chromatin suggests that DNA methylation is not exclusively automatic and provides a mechanism for a targeted change in DNA methylation in response to activation of signaling pathways. Similarly, demethylation can be targeted to specific sites in the genome in response to activation of signaling pathways. For example, it was suggested that the transcription factor nerve growth factor-induced protein A (NGFIA) is activated by a signaling pathway that is triggered by the serotonin receptor resulting in increase in cAMP. NGFIA recruits the histone acetyltransferase CBP, and the methylated DNA binding protein MBD2 58 to the GR promoter. Our hypothesis is that increased histone acetylation triggered by CREB binding protein (CBP) or by other recruited histone acetyltransferases (HATs) facilitates the demethylation of the gene by MBD2 or other DNA demethylases (Weaver IC, et al. unpublished data). Transcription factors that target DNA and chromatin modifying enzymes to genes might play an important role in experience-triggered DNA methylation changes.
Mechanisms that directly link neuronal activation signaling and gene-specific demethylation were proposed. Neuronal activation leads to CAMKII activation phosphorylation of MeCP2 and site specific demethylation of the BDNF promoter.59 This mechanism was suggested to explain how early life stress results in persistent life-long hypomethylation of the arginine vasopressin (AVP) gene. The AVP promoter is methylated and bound by the methyl CpG binding protein MECP2. Depolarization of hypothalamic neurons triggers phosphorylation of MeCp2 at Ser438 by calcium dependent CamKII (calmodulin kinase II).60 This facilitates demethylation of the AVP gene. The change in MeCp2 affinity to the methylated DNA by phosphorylation in response to neuronal activation was shown before to facilitate demethylation of the BDNF promoter.61 This signaling pathway delineates a direct link between neuronal activation and the phosphorylation state of a protein interacting with methylated genes in the brain. This could serve as a prototype for how external signal can modulate DNA methylation in a responsive organized manner.
Experience Driven Modulation of DNA Methylation and Behavior
Perhaps one of the most remarkable examples of how environments affect development after birth is the impact of the early life social environment on health trajectories later in life.12–14 If the social environment affects DNA methylation, its impact cannot result from stochastic inhibition of DNA methylation/demethylation enzymes; that could be an explanation for the impact of toxins or food ingredients but not social exposures. Social environments must evoke signaling pathways in the brain and the body that are associated with organized responses. There are several models that measure the impact of early life social environment on behavior and other health phenotypes later in life. Animal models could be used to test whether the impact of early life social environment on the phenotype is mediated by “genetic” or “epigenetic” mechanisms. Maternal behavior plays a cardinal role in the behavioral development of mammals. Models of maternal deprivation in primates and rodents and natural variation in maternal care in rodents were used to demonstrate the profound impact of maternal care and “nurture” on a panel of phenotypes in the offspring that last into adulthood.62,63
Hippocampal glucocorticoid receptor (GR) controls the negative feedback of the HPA axis by glucocorticoids. In the rat, the adult offspring of mothers that exhibit increased levels of pup licking/grooming (i.e., high LG mothers) over the first week of life show increased hippocampal (GR) expression, enhanced glucocorticoid feedback sensitivity, decreased hypothalamic corticotrophin releasing factor (CRF) expression and more modest HPA stress responses compared to animals reared by Low LG mothers.64,65 The GR/NR3C1 gene encoding the glucocorticoid receptor (GR exon 17 promoter) exhibits differences in DNA methylation and histone acetylation in the hippocampus of the offspring of high and low LG mothers. Differences in epigenetic programming in response to differences in maternal LG emerged early in life and remained stable into adulthood illustrating how epigenetic programming early in life could set up life-long behavioral trajectories.28
The basic concepts of this study were repeated more recently in several other models of early life social adversity. Exposure of infant rats to stressed caretakers that displayed abusive behaviour produced persisting changes in methylation of BDNF gene promoter in the adult prefrontal cortex.66 Early-life stress (ELS) in mice caused sustained DNA hypomethylation of an important regulatory region of the arginine vasopressin (AVP) gene.60
An extremely important question is whether the results in rodents could be translated to humans? The state of methylation of rRNA gene promoters and GR were examined a cohort of suicide victims in Quebec who were abused as children and their control group. Ribosomal RNA (rRNA) forms the skeleton of the ribosome, the protein synthesis machinery. Protein synthesis is essential for building new memories and creating new synapses in the brain. Our genome contains around 400 copies of the genes encoding rRNA. One possible way to control the protein synthesis capacity of a cell is through changing the fraction of active rRNA alleles in a cell.67 We have previously shown that the fraction of rRNA genes that is active and is associated with the RNA Pol1 transcription machinery is unmethylated while the fraction that is inactive is methylated.67 Our results showed that the suicide victims who experienced childhood abuse had higher overall methylation in their rRNA genes and expressed less rRNA. This difference in methylation was region specific: it was present in the hippocampus and was not observed in the cerebellum. Moreover, although significant methylation differences were observed between the controls and the suicide victims, no sequence differences were observed. The fact that the difference in methylation was brain-region specific and that no sequence differences were observed further strengthens the conclusion that this difference in methylation was driven by environmental rather than genetic variation.68 These data point to the possibility that the effects of early life adversity might not be limited to the usual suspects of highly brain specific genes but that ubiquitously expressed genes could be involved as well. Modulation of expression of ubiquitous genes might be important in modulating brain function.
Individuals with treatment-resistant forms of major depression show decreased GR expression and increased HPA activity. Site-specific differences in DNA methylation in the GR exon 1f promoter and its expression were detected between suicide completers who had reported social adversity early in life and suicide completers who did not experience social adversity early in life.69 Differences in DNA methylation of the GR promoter were observed also in peripheral blood cells; the GR promoter was more methylated in lymphocytes in newborns exposed prenatally to maternal depression than control newborns.70 This lends support to the hypothesis that DNA methylation differences in response to social adversity are system wide and are not limited to brain specific regions.
Epigenetic modulation of other candidate genes was implicated in suicide; the Gamma-aminobutyric acid A receptor alpha 1 subunit (GABRA1) promoter71 within the frontopolar cortex72 and Tropomyosin-related kinase B (TRKB) in the frontal cortex of suicide completers.73 It is unknown yet whether these changes in DNA are also associated with early life adversity.
Genome and System Wide Impact of Early Life Adversity
The first studies summarized above focused on a candidate gene approach. However, the large number of phenotypes that are associated with early life adversity both in animals and humans suggest that the impact of early adversity on the DNA methylation pattern will be broad. Moreover, it is clear that genes don't act independently but through functional gene circuitries. We therefore reasoned that adaptation of the DNA methylation pattern to early life adversity will be broad and that it will involve several systems in the body (Fig. 1). We tested this hypothesis in several studies.
First, we examined the state of DNA methylation, histone acetylation and gene expression in a 7 million base pair region of chromosome 18 containing the glucocorticoid receptor gene in the hippocampus of adult rats and showed that natural variations in maternal care in the rat are associated with coordinate changes in DNA methylation, chromatin and gene expression spanning over a hundred kilobase pairs. Interestingly, a chromosomal region containing a cluster of the PROTOCADHERIN α, -β and -γ (Pcdh) gene families implicated in synaptogenesis show the highest differential response to maternal care. The entire cluster reveals epigenetic and transcriptional changes in response to maternal care. These studies suggest that the DNA methylation response to early life maternal care is coordinated in clusters that cover broad areas in the genome and that the epigenetic response to early life maternal care involves not only single candidate gene promoters but includes transcriptional and intragenic sequences, as well as those residing distantly from transcription start sites and regions containing non-coding RNAs.74
Second, we showed that a similar pattern of response to childhood abuse is associated with DNA methylation differences throughout the genomic region spanning the six and a half million base-pair region centered at the NR3C1 gene in the hippocampus of adult humans. The DNA methylation differences associated with child abuse bear a striking resemblance to DNA methylation differences between adult offspring of high and low maternal care rats. This provides evidence for an analogous crossspecies epigenetic and transcriptional response to early life environment (Suderman et al. submitted 2011).
Third, we tested whether the response to early life adversity is system wide and includes T cells as well as the brain by examining in parallel the impact of differential maternal rearing in a rhesus model of maternal deprivation. We examined the impact of depriving maternal care on DNA methylation in the prefrontal cortex and T cells. Our results show that similar to the rat and human the changes associated with differences in rearing are widespread in the genome and that they are not limited to the brain and occur in T cells as well. Although the vast majority of DNA methylation changes that associate with rearing are different in T cells and prefrontal cortex, some similarities were detected. This data is consistent with the hypothesis that the response to early life adversity is genome wide and system wide, that multiple tissues respond to adversity early in life including immune cells found in circulating blood. Since changes in DNA methylation associated with maternal rearing were detected in T cells, it might be possible to perform population DNA methylation studies of behavior examining either whole blood or T cells. We have initiated a study of the impact of socioeconomic positioning on DNA methylation that examined blood DNA from the British birth cohort of 1958. This study detected a signature of DNA methylation that is associated with early life adversity (Borghol et. al. unpublished).
Two studies have recently demonstrated that epigenetic effects associated with behavioral adversity could be detected in blood cells. First, Pituitary adenylate cyclase-activating polypeptide (PACAP), a protein known to be involved in stress response in the pituitary was found to be differentially methylated in peripheral blood cells in humans with post traumatic stress syndrome.75 Second, telomere lengths differences were identified between orphans in the Bucharest Early Intervention Project who were placed under high quality foster care compared with those subjected to continued care in institutions.76 This study demonstrates that the effects of early life adversity could be molecularly detected in blood. These studies are encouraging since they point to the possibility that a DNA methylation response to early life adversity could be detected in peripheral blood cells.
The fact that changes in methylation of a gene relating to pituitary function is detected in peripheral blood cells is seemingly surprising. There is no question that DNA is differentially methylated in different tissues; therefore, responses to adversity that involve behavior would be expected to be limited to specific brain regions. Our preliminary data suggests that indeed this is the case; several differentially methylated regions associated with behavior are limited to specific brain regions. For example, changes in DNA methylation associated with early life child adversity in humans were found in the hippocampus but not in the cerebellum.68 However, this does not necessarily exclude the possibility that the response of the DNA methylation pattern to social adversity is system wide and affects several interacting physiological functions; specific genes will be differentially methylated in different tissues in a manner that is consistent with a system-wide response to adversity. For example, the relationship between stress and the immune system is well established. In this case, changes in DNA methylation that are detected in the periphery might be more than surrogate markers of changes in DNA methylation in the brain. The DNA methylation alterations might also teach us the intricate physiological adaptations and body-wide interrelationships involved in the response to early life social adversity. However, it is possible that in addition to changes in DNA methylation in response to adversity that are specific to distinct tissues, some genes will respond similarly in brain and peripheral tissues if they encode common functions in response to adversity in both the brain and periphery. As discussed above, a long line of data have established that the physiological response to early life socioeconomic adversity is not limited to the brain.13,77,78 There is no reason therefore to believe that DNA methylation changes in response to adversity should not occur in the periphery as well as the brain.
Summary
Although our understanding of the mechanisms linking external environmental signals and DNA methylation are rudimentary, few studies suggest a potential conduit between the external environment and a directed modulation of the DNA methylation pattern in neurons. Such a mechanism could be at the foundation of the manner by which experience shapes human development early in life and modulates it throughout life. These mechanisms extend the potential role of DNA methylation beyond the “classic” highly programmed and innate processes of embryonal development into a genome adaptation mechanism after birth and throughout life.
Acknowledgments
Work in M.S. laboratory reviewed here is supported by the Canadian Institutes of Health Research, The National Institute of Cancer Research Canada, the US National Institutes of Health (National Institute of Child Health and Human Development), the Sackler program for psychobiology and epigenetics at McGill University and the Canadian Institute for Advanced Research.
References
- 1.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 2.Razin A, Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984;782:331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- 3.Razin A, Webb C, Szyf M, Yisraeli J, Rosenthal A, Naveh-Many T, et al. Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc Natl Acad Sci USA. 1984;81:2275–2279. doi: 10.1073/pnas.81.8.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeltsch A. Reading and writing DNA methylation. Nat Struct Mol Biol. 2008;15:1003–1004. doi: 10.1038/nsmb1008-1003. [DOI] [PubMed] [Google Scholar]
- 7.Turnbull JF, Adams RL. DNA methylase: purification from ascites cells and the effect of various DNA substrates on its activity. Nucleic Acids Res. 1976;3:677–695. doi: 10.1093/nar/3.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruenbaum Y, Stein R, Cedar H, Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981;124:67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- 9.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 10.Szyf M, Rouleau J, Theberge J, Bozovic V. Induction of myogenic differentiation by an expression vector encoding the DNA methyltransferase cDNA sequence in the antisense orientation. J Biol Chem. 1992;267:12831–12836. [PubMed] [Google Scholar]
- 11.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 12.Hertzman C, Power C, Matthews S, Manor O. Using an interactive framework of society and lifecourse to explain self-rated health in early adulthood. Soc Sci Med. 2001;53:1575–1585. doi: 10.1016/s0277-9536(00)00437-8. [DOI] [PubMed] [Google Scholar]
- 13.Power C, Hertzman C, Matthews S, Manor O. Social differences in health: life-cycle effects between ages 23 and 33 in the 1958 British birth cohort. Am J Public Health. 1997;87:1499–1503. doi: 10.2105/ajph.87.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Power C, Li L, Hertzman C. Associations of early growth and adult adiposity with patterns of salivary cortisol in adulthood. J Clin Endocrinol Metab. 2006;91:4264–4270. doi: 10.1210/jc.2006-0625. [DOI] [PubMed] [Google Scholar]
- 15.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci USA. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases [letter] Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 18.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szyf M. A dynamic methylome; implications of non-CG methylation/demethylation. Cell Cycle. 2010;9:3846–3847. doi: 10.4161/cc.9.19.13563. [DOI] [PubMed] [Google Scholar]
- 20.Fuso A, Nicolia V, Cavallaro RA, Scarpa S. DNA methylase and demethylase activities are modulated by one-carbon metabolism in Alzheimer's disease models. J Nutr Biochem. 2011;22:242–251. doi: 10.1016/j.jnutbio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 22.Lucarelli M, Fuso A, Strom R, Scarpa S. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J Biol Chem. 2001;276:7500–7506. doi: 10.1074/jbc.M008234200. [DOI] [PubMed] [Google Scholar]
- 23.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 24.Wilks A, Seldran M, Jost JP. An estrogen-dependent demethylation at the 5′ end of the chicken vitellogenin gene is independent of DNA synthesis. Nucleic Acids Res. 1984;12:1163–1177. doi: 10.1093/nar/12.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szyf M, Theberge J, Bozovic V. Ras induces a general DNA demethylation activity in mouse embryonal P19 cells. J Biol Chem. 1995;270:12690–12696. doi: 10.1074/jbc.270.21.12690. [DOI] [PubMed] [Google Scholar]
- 26.Mastronardi FG, Noor A, Wood DD, Paton T, Moscarello MA. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J Neurosci Res. 2007;85:2006–2016. doi: 10.1002/jnr.21329. [DOI] [PubMed] [Google Scholar]
- 27.Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci USA. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 29.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 30.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 33.Jost JP. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci USA. 1993;90:4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Razin A, Szyf M, Kafri T, Roll M, Giloh H, Scarpa S, et al. Replacement of 5-methylcytosine by cytosine: a possible mechanism for transient DNA demethylation during differentiation. Proc Natl Acad Sci USA. 1986;83:2827–2831. doi: 10.1073/pnas.83.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 36.Shen JC, Rideout WMd, Jones PA. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992;71:1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 37.Zingg JM, Shen JC, Jones PA. Enzyme-mediated cytosine deamination by the bacterial methyltransferase M.MspI. Biochem J. 1998;332:223–230. doi: 10.1042/bj3320223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 39.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5 hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surani MA, Hajkova P. Epigenetic reprogramming of mouse germ cells toward totipotency. Cold Spring Harb Symp Quant Biol. 2010;75:211–218. doi: 10.1101/sqb.2010.75.010. [DOI] [PubMed] [Google Scholar]
- 46.Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, et al. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, et al. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujita H, Fujii R, Aratani S, Amano T, Fukamizu A, Nakajima T. Antithetic effects of MBD2a on gene regulation. Mol Cell Biol. 2003;23:2645–2657. doi: 10.1128/MCB.23.8.2645-2657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angrisano T, Lembo F, Pero R, Natale F, Fusco A, Avvedimento VE, et al. TACC3 mediates the association of MBD2 with histone acetyltransferases and relieves transcriptional repression of methylated promoters. Nucleic Acids Res. 2006;34:364–372. doi: 10.1093/nar/gkj400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex [see comments] Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 51.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation [see comments] Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 52.Detich N, Theberge J, Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem. 2002;277:35791–35794. doi: 10.1074/jbc.C200408200. [DOI] [PubMed] [Google Scholar]
- 53.Hamm S, Just G, Lacoste N, Moitessier N, Szyf M, Mamer O. On the mechanism of demethylation of 5-methylcytosine in DNA. Bioorg Med Chem Lett. 2008;18:1046–1049. doi: 10.1016/j.bmcl.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 54.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 55.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 56.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li BZ, Huang Z, Cui QY, Song XH, Du L, Jeltsch A, et al. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011;21:1172–1181. doi: 10.1038/cr.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weaver IC, D'Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, et al. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 61.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 62.Ruppenthal GC, Arling GL, Harlow HF, Sackett GP, Suomi SJ. A 10-year perspective of motherless-mother monkey behavior. J Abnorm Psychol. 1976;85:341–349. doi: 10.1037//0021-843x.85.4.341. [DOI] [PubMed] [Google Scholar]
- 63.Suomi SJ, Collins ML, Harlow HF, Ruppenthal GC. Effects of maternal and peer separations on young monkeys. J Child Psychol Psychiatry. 1976;17:101–112. doi: 10.1111/j.1469-7610.1976.tb00382.x. [DOI] [PubMed] [Google Scholar]
- 64.Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, et al. Maternal care, hippocampal glucocorticoid receptors and r hypothalamic-pituitaryadrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 65.Francis D, Diorio J, Liu D, Meaney MJ. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286:1155–1158. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- 66.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown SE, Szyf M. Epigenetic programming of the rRNA promoter by MBD3. Mol Cell Biol. 2007;27:4938–4952. doi: 10.1128/MCB.01880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, Ernst C, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 2008;3:2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 71.Linthorst AC, Flachskamm C, Muller-Preuss P, Holsboer F, Reul JM. Effect of bacterial endotoxin and interleukin-1 beta on hippocampal serotonergic neurotransmission, behavioral activity and free corticosterone levels: an in vivo microdialysis study. J Neurosci. 1995;15:2920–2934. doi: 10.1523/JNEUROSCI.15-04-02920.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poulter MO, Du L, Weaver IC, Palkovits M, Faludi G, Merali Z, et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry. 2008;64:645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 73.Ernst C, Deleva V, Deng X, Sequeira A, Pomarenski A, Klempan T, et al. Alternative splicing, methylation state and r expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch Gen Psychiatry. 2009;66:22–32. doi: 10.1001/archpsyc.66.1.22. [DOI] [PubMed] [Google Scholar]
- 74.McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6:14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Drury SS, Theall K, Gleason MM, Smyke AT, De Vivo I, Wong JY, et al. Telomere length and early severe social deprivation: linking early adversity and cellular aging. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.53. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cunha F, Heckman JJ. The economics and psychology of inequality and human development. J Eur Econ Assoc. 2009;7:320–364. doi: 10.1162/jeea.2009.7.2-3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Power C, Atherton K, Strachan DP, Shepherd P, Fuller E, Davis A, et al. Life-course influences on health in British adults: effects of socio-economic position in childhood and adulthood. Int J Epidemiol. 2007;36:532–539. doi: 10.1093/ije/dyl310. [DOI] [PubMed] [Google Scholar]