Abstract
Complex I is present in almost all aerobic species. Being the largest complex of the respiratory chain, it has a central role in energizing biological membranes and is essential for many organisms. Bacterial complex I is composed of 14 subunits that are sufficient to achieve the respiratory functions. Eukaryotic enzymes contain orthologs of the 14 bacterial subunits and around 30 additional subunits. This complexity suggests either that complex I requires more stabilizing subunits in mitochondria or that it fulfills additional functions. In many organisms recent work on complex I concentrated on the determination of its exact composition. This review summarizes the work done to elucidate complex I composition in the model plant Arabidopsis and proposes a model for the organization of its 44 confirmed subunits. The comparison of the different studies investigating the composition of complex I across species identifies sample preparation for the proteomic analysis as critical to differentiate between true subunits, assembly factors, or proteins associated with complex I. Coupling comparative proteomics with biochemical or genetic studies is thus required to define a subunit and its function within the complex.
Keywords: multiprotein complex, proteomics, plant mitochondria, complex I, respiratory chain
Introduction
The NADH-Ubiquinone oxidoreductase (EC 1.6.5.3) is part of the respiratory chain present in all aerobic organisms. It is also named NDH-1 in bacteria and complex I in eukaryotes because it is the first complex involved in the mitochondrial electron transfer chain. Its roles during respiration are to oxidize NADH, transfer the resulting electrons through a series of iron-sulfur (Fe/S) clusters to the quinone pool and export protons in the compartment located on the p-side of the membrane (reviewed in Brandt, 2006).
Complex I is known as the major entry point of electrons in the respiratory chain but surprisingly its mode of action is less understood than those of other respiratory complexes. It is composed of 14 subunits in bacteria and more than 40 subunits in higher eukaryotes. In E. coli the 14 subunits are named NuoA-N and are encoded by the nuo operon. They are sufficient for the oxidation of NADH, the transfer of the electrons onto quinone and the pumping of protons into the periplasm. The structure of the bacterial enzyme has been solved only recently (Efremov et al., 2010; Efremov and Sazanov, 2011b) and a functional mechanism, suggesting that complex I acts like a steam engine, has been proposed (Efremov and Sazanov, 2011a).
In eukaryotes, proteins homologous to these 14 “core” subunits are conserved and additional “accessory” subunits are found associated to the core subunits. Conservation of the core subunits suggests that the essential redox components and the basic mechanisms of electron and proton translocation described in bacteria are conserved across species. Structural work in Yarrowia lipolytica confirmed similarities between the bacterial complex and the core of eukaryotic complex I (Hunte et al., 2010) but provided no information on potential roles of the accessory subunits. These subunits can be divided into two groups. The first set contains subunits generally found in all eukaryotic complex I. In the second group are accessory subunits specific to plants, fungi, or animals. A comparative analysis of complex I composition in different organisms was undertaken to trace the evolution of the complex (Gabaldón et al., 2005). According to this analysis, bovine complex I contains 11 mammal-specific subunits while the Arabidopsis enzyme contains eight plant-specific subunits and Neurospora contains three fungi-specific subunits. The numbers of specific subunits have evolved with the release of new proteomic analyses [for example from 8 (Heazlewood et al., 2003) to 17 (Klodmann et al., 2010) in Arabidopsis]. The species-specific diversity in the composition of complex I suggests it has acquired additional functions in different organisms as mitochondria may have evolved differently in these organisms. However, a recent bioinformatic analysis questioned this variability in complex I composition and suggested a conserved subunit composition across species (Cardol, 2011). In the present review, an updated overview over proteomic approaches used to resolve the composition of Arabidopsis complex I is provided.
History of the Proteomic Analyses of Plant Complex I
Initial studies of higher plant complex I purified the enzyme from potato mitochondria using hydroxyapatite and gel filtration chromatography. Subsequent SDS-PAGE detected 32 protein bands, from which 10 N-terminal sequences were obtained (Herz et al., 1994). A similar number of bands was obtained when immunopurified wheat complex I was subjected to SDS-PAGE (Combettes and Grienenberger, 1999). The composition of complex I was later analyzed using Blue-Native gel electrophoresis (BN-PAGE). This technique separates mitochondrial complexes according to their size in their native form. The complexes are subsequently dissociated in order to separate the different subunits on a second dimension involving a SDS-PAGE. Using this approach, complex I from potato mitochondria was resolved into 30 distinct spots (Jänsch et al., 1996). The first proteomic analysis of Arabidopsis and rice complex I coupling BN-SDS-PAGE and mass spectrometry (MS) identified 30 and 24 proteins, respectively (Heazlewood et al., 2003). These proteins comprised several plant-specific subunits, defined as having no homologs among the subunits of complex I from bovine or Neurospora. The development of a three-dimensional BN-SDS-SDS-PAGE technique allowed the visualization of highly hydrophobic subunits. Using this technique, 37 distinct spots containing 44 different subunits were obtained for Arabidopsis complex I. This list included 14 plant-specific subunits (Meyer et al., 2008). The latest proteomic analysis of complex I subunits separated by BN-SDS-PAGE identified three new putative subunits (Klodmann et al., 2011).
In an alternative approach to determine its internal architecture, complex I was purified from Arabidopsis cell culture and fractionated using low concentrations of detergent. A series of subcomplexes was obtained, and a systematic proteomic analysis of the composition of all subcomplexes identified 40 subunits (Klodmann et al., 2010). This study not only provided data about the composition but it also gave key information on the internal organization of Arabidopsis complex I. Using Arabidopsis mutants disrupted in genes encoding complex I subunits, subcomplexes corresponding to assembly intermediates were identified, and similar data about the internal organization of complex I were obtained (Meyer et al., 2011). A consensus composition has been proposed, comprising 49 different subunits, 17 of which are specific to plants (Klodmann and Braun, 2011).
Arabidopsis Complex I Composition
In the original proteomic study, 30 proteins were identified by MS as part of complex I in Arabidopsis (Heazlewood et al., 2003). A search for orthologs of mammalian complex I subunits in Arabidopsis genome allowed the identification of 14 additional subunits (Heazlewood et al., 2003). Later proteomic studies confirmed some as being complex I subunits and identified other orthologs that had not been initially described. Still, a number of mammalian subunits have no confirmed orthologs in plants. Recently, using a PSI-Blast approach, the composition of complex I in different eukaryotes was investigated (Cardol, 2011). This analysis indicated that many subunits initially considered as plant-specific could be distant relatives of mammal-specific subunits. Combining proteomic and bioinformatic data, the subunit composition of complex I in Arabidopsis has been reevaluated (Table 1). This analysis shows that Arabidopsis complex I contains seven subunits absent for the bovine enzyme and bovine complex I contains eight mammal-specific subunits. Among the latter, six have orthologs in Arabidopsis, however these orthologs were never found experimentally to be associated with complex I and may thus not be part of the complex in plants. An example is the Arabidopsis homolog of the bovine 42 kDa subunit, encoded by the gene At1g72040. This protein, which is annotated as a member of the nucleoside triphosphate hydrolases superfamily, is predicted to be mitochondrial but has only been identified in the nuclear proteome in plants (Bae et al., 2003). The protein orthologous to the bovine B9 subunit, encoded by At2g46540, was identified in the mitochondrial proteome (Brugière et al., 2004; Heazlewood et al., 2004). The closest homolog of the B14.5a subunit, encoded by At5g08050, has been experimentally found in plastids (Ferro et al., 2010) but not in mitochondria. The protein homologous to the B22 subunit can drive a GFP fusion protein into mitochondria in vivo (Han et al., 2010) and has been found associated with a subcomplex of complex I (Sunderhaus et al., 2006). A mutant in the gene encoding this protein showed a respiration defect but the presence of complex I in this mutant was not investigated (Han et al., 2010). The closest homolog of the MLQR subunit, encoded by At3g29970, has not been identified in any proteomic study. Finally, the Acyl Carrier Protein (ACP, subunit SDAP in bovine) was found to be a soluble protein of the mitochondrial matrix in Arabidopsis (Meyer et al., 2007). In contrast, 43 proteins were identified in at least three proteomic approaches directed at the composition of Arabidopsis complex I. With the addition of the small hydrophobic mitochondrial-encoded ND4L, 44 Arabidopsis proteins are considered as bona fide subunits. A model of the organization of these subunits in the complex is proposed in Figure 1. The function of the accessory subunits has not been investigated in plants. Indeed, complex I mutants were used to analyze the consequences of the absence of complex I on respiration and plant metabolism (Dutilleul et al., 2003; Meyer et al., 2009) and to determine the assembly of complex I (Meyer et al., 2011).
Table 1.
Comparative analysis of the composition of complex I in bovine and Arabidopsis.
| Arabidopsis identifier | Bovine | Comments | Heazlewood et al. (2003) | Sunderhaus et al. (2006) | Meyer et al. (2008) | Klodmann et al. (2010) | Klodmann and Braun (2011) | Klodmann et al. (2011) | Concensus (44 subunits) |
|---|---|---|---|---|---|---|---|---|---|
| AtMg00990 | ND3 | E. coli NUOA | X | X | X | X | |||
| At5g11770 | 20 kDa (PSST) subunit | E. coli NUOB | X | X | X | X | X | X | |
| AtMg00070 | ND9/30 kDa subunit | E. coli NUOC | X | X | X | X | X | X | |
| AtMg00510 | ND7/49 kDa subunit | E. coli NUOD | X | X | X | X | X | X | X |
| At4g02580 | 24 kDa subunit | E. coli NUOE | X | X | X | X | X | X | |
| At5g08530 | 51 kDa subunit | E. coli NUOF | X | X | X | X | X | X | |
| At5g37510 | 75 kDa subunit | E. coli NUOG | X | X | X | X | X | X | X |
| AtMg00516, AtMg01120, AtMg01275 | ND1 | E. coli NUOH | X | X | X | X | X | X | X |
| At1g79010, At1g16700 | 23 kDa (TYKY) subunit | E. coli NUOI | X | X | X | X | X | X | |
| AtMg00270 | ND6 | E. coli NUOJ | X | X | X | X | |||
| AtMg00650 | ND4L | E. coli NUOK | X* | ||||||
| AtMg00060, AtMg00513, AtMg00665 | ND5 | E. coli NUOL | X | X | X | X | X | X | |
| AtMg00580 | ND4 | E. coli NUOM | X | X | X | X | X | ||
| AtMg00285, AtMg01320 | ND2 | E. coli NUON | X | X | X | X | X | ||
| At3g03070 | 13 kDa subunit | X | X | X | X | ||||
| At2g47690, At3g62790 | 15 kDa (PFFD) subunit | X | X | X | X | X | X | X | |
| At5g67590 | 18 kDa subunit | X | X | X | X | X | |||
| At2g20360 | 39 kDa subunit | X | X | X | X | X | X | X | |
| At1g76200 | AGGG subunit | X | X | X | X | X | |||
| At5g47570 | ASHI subunit | X | X | X | X | X | |||
| At1g14450, At2g02510 | B12 subunit | X | X | X | X | X | X | ||
| At5g52840 | B13 subunit | X | X | X | X | X | X | X | |
| At3g12260 | B14 subunit | X | X | X | X | X | |||
| At4g20150 | B14.5b subunit | X | X | X | X | X | X | ||
| At2g42210 | B14.7 subunit | X | X | X | X | ||||
| At2g31490 | B15 subunit | X | X | X | X | X | X | X | |
| At2g33220, At1g04630 | B16.6 subunit | X | X | X | X | X | X | X | |
| At3g03100 | B17.2 subunit (DAP13) | X | X | X | X | X | |||
| At2g02050 | B18 subunit | X | X | X | X | X | X | X | |
| At5g47890 | B8 subunit | X | X | X | X | X | |||
| At2g42310, At3g57785 | ESSS subunit | X | X | X | X | X | X | X | |
| At4g00585 | KFYI subunit | X | X | X | X | X | X | ||
| At4g16450 | MNLL subunit | X | X | X | X | X | X | X | |
| At3g08610 | MWFE subunit | X | X | X | X | X | X | ||
| At3g18410, At1g49140 | PDSW subunit | X | X | X | X | X | X | X | |
| At3g06310, At5g18800 | PGIV subunit | X | X | X | X | X | |||
| At1g67785 | SGDH subunit | X | X | X | X | X | |||
| ARABIDOPSIS PROTEINS NOT FOUND IN BOVINE COMPLEX I | |||||||||
| At1g19580 | CA1: carbonic anhydrase 1 | X | X | X | X | X | X | ||
| At1g47260 | CA2: carbonic anhydrase 2 | X | X | X | X | X | X | X | |
| At5g66510 | CA3:carbonic anhydrase 3 | X | X | X | X | X | X | X | |
| At3g48680, At5g63510 | CAL: carbonic anhydrase-like | X | X | X | X | X | X | X | |
| At1g67350 | P1 | X | X | X | X | X | X | ||
| At2g27730 | P2 | X | X | X | X | X | X | X | |
| At5g14105 | P3 | X | X | X | X | ||||
| At1g68680 | X | ||||||||
| At1g72170 | X | ||||||||
| At3g10110, At1g18320 | TIM22 | X | |||||||
| At3g47930 | GLDH | X$ | |||||||
| BOVINE SUBUNITS NOT FOUND IN MATURE ARABIDOPSIS COMPLEX I | |||||||||
| At1g72040 | 42 kDa subunit | ||||||||
| At2g46540 | B9 subunit | ||||||||
| At5g08050 | B14.5a subunit | ||||||||
| At4g34700 | B22 subunit | X$ | |||||||
| At3g29970 | MLQR subunit | ||||||||
| At1g65290, At2g44620, At5g47630 | SDAP subunit | ||||||||
| NON-CONSERVED BOVINE COMPLEX I | |||||||||
| 10 kDa subunit | |||||||||
| B17 subunit | |||||||||
From six proteomic analyses of Arabidopsis complex I, 49 different proteins were identified (the identification is indicated with ‘X’). From this list, 44 proteins are part of a consensus composition as they were identified in at least three studies, with the exception of ND4L (*) that was never identified by MS, most probably due to its small size and extreme hydrophobicity. $These proteins were identified in subcomplexes but not in mature complex I.
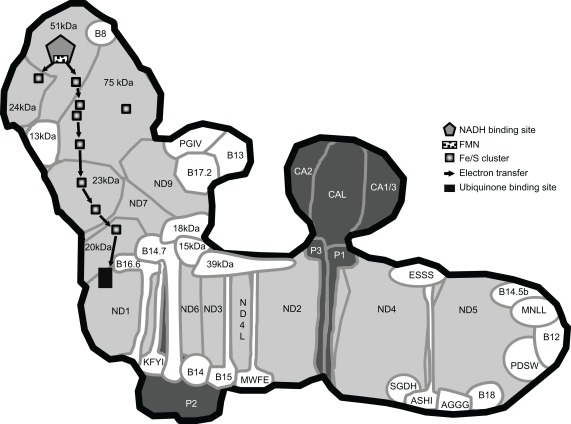
Figure 1.
Model of Arabidopsis complex I. This model includes the 44 consensus subunits defined in Table 1. It was built using the shape of the complex obtained by single particle analysis (Dudkina et al., 2005), the presence of CAs in the extra matrix-facing domain (Sunderhaus et al., 2006), the structure of the bacterial enzyme for the localization of the 14 core subunits and the Fe/S clusters (Efremov and Sazanov, 2011b), the co-localization of subunits within the same subcomplex (Klodmann et al., 2010; Meyer et al., 2011) as well as bioinformatic prediction of transmembrane domains and topology using the TMHMM prediction service (Krogh et al., 2001). The nomenclature used is the one determined for bovine and plant-specific subunits are names as in Table 1. The core subunits are shown in light gray. Accessory subunits present in bovine and Arabidopsis complex I are shown in white and accessory subunits present in Arabidopsis but absent from bovine complex I are shown in dark gray.
Carbonic anhydrases
The main difference between plant and animal complex I is the presence of gamma-carbonic anhydrases (CAs) in the plant enzyme. The CAs were proposed to be involved in CO2 metabolism in plant mitochondria (Braun and Zabaleta, 2007). To date, no direct proof of this function has been obtained (Klodmann and Braun, 2011). Among the Arabidopsis CAs, two carbonic anhydrase-like (CAL) proteins have been identified and were considered to represent two distinct complex I subunits. Other higher plants, such as rice, maize, or poplar, possess only one gene encoding CAL. This suggests that the presence of two CALs in Arabidopsis is the result of a recent gene duplication event, and that the two CALs are isoforms of the same subunit. A similar analysis of the genes encoding CAs revealed that plants possess two (maize, sorghum) or three (Arabidopsis, rice, poplar) genes encoding CAs. Additional biochemical data and particularly the characterization of mutants are required to determine the exact composition of Arabidopsis complex I regarding CAs and CALs. To date, only CA2 and CAL2 have been studied. The ca2 mutant showed highly reduced levels of complex I (Perales et al., 2005) and a trimmed CA domain (Sunderhaus et al., 2006), suggesting that CA2 is not easily replaced by other CAs. The cal2 mutant contained wild-type levels of complex I (Meyer et al., 2011), in agreement with both CALs being isoforms. The CA domain of complex I may thus include CA2, CAL, and either CA1 or CA3.
Other putative plant-specific subunits
Four other putative proteins have been described as complex I subunits in proteomic studies. The l-galactono-1,4-lactone dehydrogenase (GLDH, At3g47930) was found in a smaller version of complex I but not in the holocomplex (Heazlewood et al., 2003). GLDH was later identified as a complex I assembly factor as a mutant lacking this protein did not contain a fully assembled complex I (Pineau et al., 2008). Because GLDH is absent from the mature complex (Meyer et al., 2008; Klodmann et al., 2010), it cannot be considered as a complex I subunit. A protein encoded by the gene At1g68680 has only been detected in one proteomic study of complex I (Meyer et al., 2008). It was recently identified in the proteome of the mitochondrial outer membrane (Duncan et al., 2011) and may therefore not be a complex I subunit. TIM22 (encoded by At1g18320 and At3g10110) has been identified in the vertical containing complex I subunits on a BN-SDS-PAGE (Klodmann et al., 2011). As this protein acts as an import channel during the import of cytoplasmically translated carriers into the inner mitochondrial membrane (Murcha et al., 2007), a role of TIM22 in the assembly of complex I is plausible. Another protein, encoded by At1g72170, has also been identified by Klodmann et al. (2011) as a putative new complex I subunit. This protein had previously been found in the Arabidopsis mitochondrial proteome (Heazlewood et al., 2004) and in the mitochondrial membrane proteome (Brugière et al., 2004) but no data are available regarding its function. More biochemical evidence is required to validate these proteins as complex I subunits.
How to Define a Subunit?
Distinguishing subunits from assembly factors
Defining the subunit composition of a large complex is a challenge, not only because of technological limitations. It is generally assumed that a subunit is essential for either the presence or the function (or both) of the complex. However, not all proteins that correspond to this definition are subunits. For example, all the assembly factors identified in mammals (for review see McKenzie and Ryan, 2010) or in plants (Bych et al., 2008; Pineau et al., 2008) are essential for the presence of the mature complex but are not present in the mature complex. On the other hand, assembly factors can be accessory subunits. For example, the MWFE subunit is required for the insertion of some mitochondrial-encoded subunits within the membrane (Yadava et al., 2004; Meyer et al., 2011). Importantly, while a combination of proteomic and genetic tools are suitable to define if a protein is present in and essential for the biogenesis of complex I, additional biochemical or proteomic studies are required to identify “true” subunits and distinguish them from assembly factors or interactants. For example, the characterization of a mutant will elucidate if a protein is important for the assembly/stability of the complex, defining interaction partners will determine if the protein is involved in other metabolic pathways and in vitro activity assay will highlight new functions of the complex.
Importance of the techniques used for complex I preparation
The principal characteristic of a subunit is its presence in the fully assembled complex. Proteomic analyses of the composition of a multiprotein complex critically depend on the availability of a highly purified complex. Remarkably, proteomic approaches using different techniques to isolate complex I identified sets of accessory subunits that only partially overlap. Several factors can influence the outcome of the purification strategy. While detergents, salts and buffers used for the solubilization of mitochondrial membranes and extraction of the complex are critical, the fractionation technique used to isolate the complex (immunoprecipitation, chromatography, or BN-PAGE) is also important. Notably, in all studies that identified ACP as part of complex I, the sample for the proteomic analysis was prepared by non-gel based techniques (Carroll et al., 2003; Murray et al., 2003; Abdrakhmanova et al., 2004; Bridges et al., 2010). In contrast, studies using BN-PAGE have not found ACP associated with complex I (Heazlewood et al., 2003; Cardol et al., 2004; Klodmann et al., 2010; Meyer et al., 2011). Interestingly, ACP has been found in abundance in the soluble mitochondrial matrix fraction in mammals (Cronan et al., 2005) and in plants (Meyer et al., 2007). ACP therefore seems to be a soluble matrix protein that is loosely associated with complex I and whose interaction with complex I is stable during chromatography purification or immunopurification of the complex but not during the separation of complexes on BN-PAGE. Similarly, subunits found in bovine complex I (purified by chromatography) and conserved in Arabidopsis mitochondria but never found in Arabidopsis complex I preparations (always analyzed on BN-PAGE) could be soluble mitochondrial proteins that are loosely associated with the complex and detached during the electrophoresis.
Alternative proteomic approaches
Several MS techniques have been recently developed to discriminate between true subunits and proteins transiently interacting with a complex. Such approaches are based on differential labeling of the two samples to compare (i.e., comparing complex with interactors to complex alone) and the ratio labeled/non-labeled protein defines stable and dynamic components of complexes (Kito et al., 2008; Pflieger et al., 2008). Alternatively, the determination of the stoichiometry of the different subunits of a complex could help defining true subunits from interactors. In a total extract, an interactor will not be associated with all the complexes and thus have a lower stoichiometry than true subunits. Several approaches have been developed to determine the stoichiometry of proteins (Gerber et al., 2003; Hochleitner et al., 2004; Beynon et al., 2005; Nanavati et al., 2008). So far, none of these proteomic approaches have been tested to investigate the composition of respiratory complexes. The transient association between complex I and other mitochondrial proteins, such as ACP involved in lipid biosynthesis, implies possible interactions of the respiratory chain with other metabolic pathways.
Conclusions
More than 15 years after the first efforts to determine the subunit composition of complex I in plants, 44 confirmed complex I subunits are known in the model organism Arabidopsis thaliana. Knowledge of the composition of complex I is a prerequisite for defining strategies to characterize the functions that complex I fulfills in plant mitochondria.
Analyses of the composition of a multiprotein complex require proteomic approaches. The comparison of proteomic investigations of complex I composition in different organisms indicates that sample preparation is critical. In addition, thorough proteomic investigations are indispensable for determining the subunit composition of the complex and can provide indications toward which metabolic pathways are associated with the respiratory chain. Importantly, coupling proteomics with biochemical and genetic analyses will be essential for i) functional studies of proteins found in preparations of the complex, and for ii) defining if these proteins are true subunits, assembly factors or proteins that interact with the complex.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Kristina Kühn is thanked for critical reading of the manuscript. Etienne H. Meyer is supported by the Centre National de la Recherche Scientifique and a Marie Curie International Reintegration Grant (PIRG256398).
References
- Abdrakhmanova A., Zickermann V., Bostina M., Radermacher M., Schägger H., Kerscher S., Brandt U. (2004). Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim. Biophys. Acta 1658, 148–156 10.1016/j.bbabio.2004.04.019 [DOI] [PubMed] [Google Scholar]
- Bae M. S., Cho E. J., Choi E.-Y., Park O. K. (2003). Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 36, 652–663 10.1046/j.1365-313X.2003.01907.x [DOI] [PubMed] [Google Scholar]
- Beynon R. J., Doherty M. K., Pratt J. M., Gaskell S. J. (2005). Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2, 587–589 10.1038/nmeth774 [DOI] [PubMed] [Google Scholar]
- Brandt U. (2006). Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 10.1146/annurev.biochem.75.103004.142539 [DOI] [PubMed] [Google Scholar]
- Braun H.-P., Zabaleta E. (2007). Carbonic anhydrase subunits of the mitochondrial NADH dehydrogenase complex (complex I) in plants. Physiol. Plant 129, 114–122 10.1111/j.1399-3054.2006.00773.x [DOI] [Google Scholar]
- Bridges H. R., Fearnley I. M., Hirst J. (2010). The subunit composition of mitochondrial NADH:ubiquinone oxidoreductase (complex I) from Pichia pastoris. Mol. Cell Proteomics 9, 2318–2326 10.1074/mcp.M110.001255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière S., Kowalski S., Ferro M., Seigneurin-Berny D., Miras S., Salvi D., Ravanel S., d’ Hérin P., Garin J., Bourguignon J., Joyard J., Rolland N. (2004). The hydrophobic proteome of mitochondrial membranes from Arabidopsis cell suspensions. Phytochemistry 65, 1693–1707 10.1016/j.phytochem.2004.03.028 [DOI] [PubMed] [Google Scholar]
- Bych K., Kerscher S., Netz D. J. A., Pierik A. J., Zwicker K., Huynen M. A., Lill R., Brandt U., Balk J. (2008). The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 27, 1736–1746 10.1038/emboj.2008.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P. (2011). Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes: a highly conserved subunit composition highlighted by mining of protein databases. Biochim. Biophys. Acta 1807, 1390–1397 10.1016/j.bbabio.2011.06.015 [DOI] [PubMed] [Google Scholar]
- Cardol P., Vanrobaeys F., Devreese B., Van Beeumen J., Matagne R. F., Remacle C. (2004). Higher plant-like subunit composition of mitochondrial complex I from Chlamydomonas reinhardtii: 31 conserved components among eukaryotes. Biochim. Biophys. Acta 1658, 212–224 10.1016/j.bbabio.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Carroll J., Fearnley I. M., Shannon R. J., Hirst J., Walker J. E. (2003). Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell Proteomics 2, 117–126 10.1074/mcp.M300014-MCP200 [DOI] [PubMed] [Google Scholar]
- Combettes B., Grienenberger J. M. (1999). Analysis of wheat mitochondrial complex I purified by a one-step immunoaffinity chromatography. Biochimie 81, 645–653 10.1016/S0300-9084(99)80121-7 [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Fearnley I. M., Walker J. E. (2005). Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett. 579, 4892–4896 10.1016/j.febslet.2005.07.077 [DOI] [PubMed] [Google Scholar]
- Dudkina N. V., Eubel H., Keegstra W., Boekema E. J., Braun H.-P. (2005). Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. U.S.A. 102, 3225–3229 10.1073/pnas.0408870102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan O., Taylor N. L., Carrie C., Eubel H., Kubiszewski-Jakubiak S., Zhang B., Narsai R., Millar A. H., Whelan J. (2011). Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol. 157, 1093–1113 10.1104/pp.111.183160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C., Garmier M., Noctor G., Mathieu C., Chétrit P., Foyer C. H., de Paepe R. (2003). Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15, 1212–1226 10.1105/tpc.009464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov R. G., Baradaran R., Sazanov L. A. (2010). The architecture of respiratory complex I. Nature 465, 441–445 10.1038/nature09066 [DOI] [PubMed] [Google Scholar]
- Efremov R. G., Sazanov L. A. (2011a). Respiratory complex I: “steam engine” of the cell? Curr. Opin. Struct. Biol. 21, 532–540 10.1016/j.sbi.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Efremov R. G., Sazanov L. A. (2011b). Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 10.1038/nature10330 [DOI] [PubMed] [Google Scholar]
- Ferro M., Brugière S., Salvi D., Seigneurin-Berny D., Court M., Moyet L., Ramus C., Miras S., Mellal M., Le Gall S., Kieffer-Jaquinod S., Bruley C., Garin J., Joyard J., Masselon C., Rolland N. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell Proteomics 9, 1063–1084 10.1074/mcp.M900325-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T., Rainey D., Huynen M. A. (2005). Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 348, 857–870 10.1016/j.jmb.2005.02.067 [DOI] [PubMed] [Google Scholar]
- Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003). Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 10.1073/pnas.1432927100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Qin G., Kang D., Chen Z., Gu H., Qu L.-J. (2010). A nuclear-encoded mitochondrial gene AtCIB22 is essential for plant development in Arabidopsis. J. Genet. Genomics 37, 667–683 10.1016/S1673-8527(09)60085-0 [DOI] [PubMed] [Google Scholar]
- Heazlewood J. L., Howell K. A., Millar A. H. (2003). Mitochondrial complex I from Arabidopsis and rice: orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim. Biophys. Acta 1604, 159–169 10.1016/S0005-2728(03)00045-8 [DOI] [PubMed] [Google Scholar]
- Heazlewood J. L., Tonti-Filippini J. S., Gout A. M., Day D. A., Whelan J., Millar A. H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16, 241–256 10.1105/tpc.016055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz U., Schröder W., Liddell A., Leaver C. J., Brennicke A., Grohmann L. (1994). Purification of the NADH:ubiquinone oxidoreductase (complex I) of the respiratory chain from the inner mitochondrial membrane of Solanum tuberosum. J. Biol. Chem. 269, 2263–2269 [PubMed] [Google Scholar]
- Hochleitner E. O., Sondermann P., Lottspeich F. (2004). Determination of the stoichiometry of protein complexes using liquid chromatography with fluorescence and mass spectrometric detection of fluorescently labeled proteolytic peptides. Proteomics 4, 669–676 10.1002/pmic.200300668 [DOI] [PubMed] [Google Scholar]
- Hunte C., Zickermann V., Brandt U. (2010). Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 10.1126/science.1191046 [DOI] [PubMed] [Google Scholar]
- Jänsch L., Kruft V., Schmitz U. K., Braun H. P. (1996). New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 9, 357–368 10.1046/j.1365-313X.1996.09030357.x [DOI] [PubMed] [Google Scholar]
- Kito K., Kawaguchi N., Okada S., Ito T. (2008). Discrimination between stable and dynamic components of protein complexes by means of quantitative proteomics. Proteomics 8, 2366–2370 10.1002/pmic.200800182 [DOI] [PubMed] [Google Scholar]
- Klodmann J., Braun H.-P. (2011). Proteomic approach to characterize mitochondrial complex I from plants. Phytochemistry 72, 1071–1080 10.1016/j.phytochem.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Klodmann J., Senkler M., Rode C., Braun H.-P. (2011). Defining the protein complex proteome of plant mitochondria. Plant Physiol. 157, 587–598 10.1104/pp.111.182352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J., Sunderhaus S., Nimtz M., Jänsch L., Braun H.-P. (2010). Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22, 797–810 10.1105/tpc.109.073726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- McKenzie M., Ryan M. T. (2010). Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life 62, 497–502 10.1002/iub.335 [DOI] [PubMed] [Google Scholar]
- Meyer E. H., Heazlewood J. L., Millar A. H. (2007). Mitochondrial acyl carrier proteins in Arabidopsis thaliana are predominantly soluble matrix proteins and none can be confirmed as subunits of respiratory complex I. Plant Mol. Biol. 64, 319–327 10.1007/s11103-007-9156-9 [DOI] [PubMed] [Google Scholar]
- Meyer E. H., Solheim C., Tanz S. K., Bonnard G., Millar A. H. (2011). Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 286, 26081–26092 10.1074/jbc.M110.209601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E. H., Taylor N. L., Millar A. H. (2008). Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J. Proteome Res. 7, 786–794 10.1021/pr700595p [DOI] [PubMed] [Google Scholar]
- Meyer E. H., Tomaz T., Carroll A. J., Estavillo G., Delannoy E., Tanz S. K., Small I. D., Pogson B. J., Millar A. H. (2009). Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 151, 603–619 10.1104/pp.109.141770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha M. W., Elhafez D., Lister R., Tonti-Filippini J., Baumgartner M., Philippar K., Carrie C., Mokranjac D., Soll J., Whelan J. (2007). Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol. 143, 199–212 10.1104/pp.106.090688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J., Zhang B., Taylor S. W., Oglesbee D., Fahy E., Marusich M. F., Ghosh S. S., Capaldi R. A. (2003). The subunit composition of the human NADH dehydrogenase obtained by rapid one-step immunopurification. J. Biol. Chem. 278, 13619–13622 10.1074/jbc.M305694200 [DOI] [PubMed] [Google Scholar]
- Nanavati D., Gucek M., Milne J. L. S., Subramaniam S., Markey S. P. (2008). Stoichiometry and absolute quantification of proteins with mass spectrometry using fluorescent and isotope-labeled concatenated peptide standards. Mol. Cell Proteomics 7, 442–447 [DOI] [PubMed] [Google Scholar]
- Perales M., Eubel H., Heinemeyer J., Colaneri A., Zabaleta E., Braun H.-P. (2005). Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 350, 263–277 10.1016/j.jmb.2005.04.062 [DOI] [PubMed] [Google Scholar]
- Pflieger D., Jünger M. A., Müller M., Rinner O., Lee H., Gehrig P. M., Gstaiger M., Aebersold R. (2008). Quantitative proteomic analysis of protein complexes: concurrent identification of interactors and their state of phosphorylation. Mol. Cell Proteomics 7, 326–346 [DOI] [PubMed] [Google Scholar]
- Pineau B., Layoune O., Danon A., De Paepe R. (2008). L-galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 283, 32500–32505 10.1074/jbc.M805320200 [DOI] [PubMed] [Google Scholar]
- Sunderhaus S., Dudkina N. V., Jänsch L., Klodmann J., Heinemeyer J., Perales M., Zabaleta E., Boekema E. J., Braun H.-P. (2006). Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J. Biol. Chem. 281, 6482–6488 10.1074/jbc.M511542200 [DOI] [PubMed] [Google Scholar]
- Yadava N., Houchens T., Potluri P., Scheffler I. E. (2004). Development and characterization of a conditional mitochondrial complex I assembly system. J. Biol. Chem. 279, 12406–12413 10.1074/jbc.M313588200 [DOI] [PubMed] [Google Scholar]