Abstract
Supplementation of branched chain amino acids, especially leucine, is critical to improve malnutrition by regulating protein synthesis and degradation. Emerging evidence has linked leucine deprivation induced protein breakdown to autophagy. In this study, we aimed to establish a cell-free assay recapitulating leucine-mediated autophagy in vitro and dissect its biochemical requirement. We found that in a cell-free assay, membrane association of Barkor/Atg14(L), a specific autophagosome-binding protein, is suppressed by cytosol from nutrient-rich medium and such suppression is released by nutrient deprivation. We also showed that rapamycin could efficiently reverse the suppression of nutrient rich cytosol, suggesting an essential role of mTORC1 in autophagy inhibition in this cell-free system. Furthermore, we demonstrated that leucine supplementation in the cultured cells blocks Barkor puncta formation and autophagy activity. Hence, we establish a novel cell-free assay recapitulating leucine-mediated autophagy inhibition in an mTORC1-dependent manner; this assay will help us to dissect the regulation of amino acids in autophagy and related human metabolic diseases.
Key words: autophagy, autophagosome, PtdIns3K/Vps34, Barkor/Atg14(L), rubicon, LC3, p62, S6K, rapamycin
Introduction
Amino acids are required for activation of the mammalian target of rapamycin (mTOR) kinase, which is an atypical serine/threonine kinase that plays an indispensable role in the control of protein metabolism, cell growth, and autophagy.1–3 When complexed with the interacting proteins raptor (for regulatory associated protein of mTOR) and GβL (also termed mLST8) in the mammalian target of rapamycin complex 1 (mTORC1), mTOR serves as an integrator of cellular signals to control the balance between cellular anabolism and cellular catabolism.3 mTORC1 serves as an integration hub for a variety of upstream growth factor, nutrient, and stress signals and modulates a variety of anabolic (e.g., protein biosynthesis) and catabolic (e.g., autophagy) processes to adapt to the cellular environment.4,5 Biochemical approaches in mammalian cells showed that GTP-loaded Ras homolog enriched in brain (Rheb) stimulates mTORC1 activity both in vitro and in vivo, while GAP activity of the TSC1/TSC2 complex results in GTP hydrolysis and Rheb inhibition.6–9 The mTORC1 pathway is a major contributor to the anabolic response following essential amino acids (EAA) or leucine ingestion, although multiple pathways are involved.10
Cells adapt to changes in their environment by adjusting anabolic and catabolic pathways.11 In times of nutrient shortage, for example, macromolecules are degraded to produce substrates for energy production.1 Central among the responses to nutrient deprivation is autophagy, which is characterized by formation of double-membrane vesicles (autophagosomes) that capture intracellular cargoes and deliver them to lysosomes for degradation. Autophagy plays an important role in a variety of human diseases and intracellular nutrient recycling.12–14 One of the most important upstream regulators of autophagy is the protein mTOR.2,4,15 When extracellular amino acids are limiting, autophagy recycles intracellular constituents as a way to provide an alternative source of amino acids.16 Several recent studies demonstrated that mTORC1 suppresses autophagy through regulation of the ULK1-Atg13-FIP200 complex.17–19
The signaling pathway responsible for leucine-mediated autophagic response is intensively studied. It has been suggested that the class III phosphatidylinositol 3-kinase (PtdIns3K) might be a major downstream effect in amino acid deprivation-induced autophagy.1 PtdIns3K plays a critical role in autophagy and endocytosis.20–25 Its specific regulation in autophagy relies on an autophagic-specific adaptor protein Barkor, also known as Atg14 or Atg14L. Barkor serves as a targeting factor for PtdIns3K recruitment to autophagosomes. Barkor is essential for autophagosome formation and its recruitment to early autophagosome structures,20–23 which is considered as a necessary step in catastrophic assembly of protein complexes on autophagosome for its nucleation, elongation and completion. Barkor directly binds to autophagic membranes through its C-terminal BATS domain and senses membrane curvature in a phosphatidylinositol 3-phosphate (PtdIns(3)P)-dependent manner.26
Given the specific role of Barkor in autophagy activation, its association with autophagic membrane could serve as readout to dissect the signaling pathway regulated by leucine. To test this possibility, we established an in vitro cell-free assay for Barkor membrane association. Importantly, Barkor membrane association is negatively regulated by an inhibitory activity present in the nutrient-rich cell extracts, and this inhibition could be reserved by the mTORC1 inhibitor rapamycin. We further dissect the requirement of mTOR subunits in leucine-mediated autophagy inhibition. This study therefore describes the first cell-free assay recapitulating the leucine effect on autophagy activation.
Results
Reconstitution of leucine effect on Barkor membrane association in a cell-free system.
Upon autophagy initiation, PtdIns3K is recruited by Barkor to the phagophore, the early autophagosome structure.20–23 This relocation ensures the nucleation of the initial membrane and promotes membrane extension by attracting downstream protein complexes to autophagosomes. We aim to design an in vitro assay utilizing the Barkor autophagic membrane association as readout to study the leucine effect on autophagy in mammalian cells.
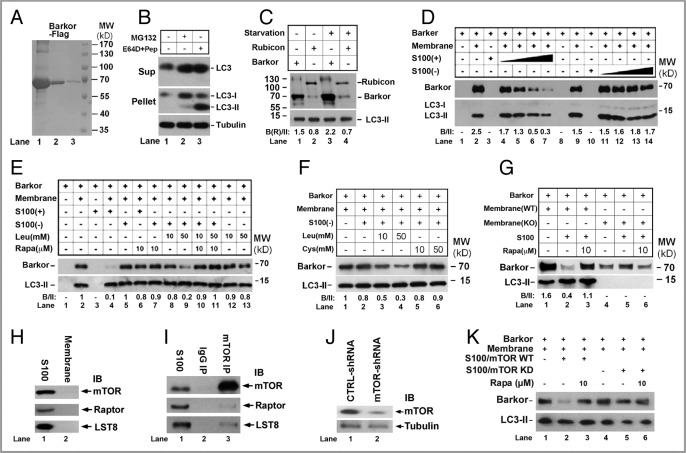
For this purpose, the recombinant full-length Flag-tagged Barkor was purified from baculovirus-infected insect cells (Fig. 1A). Rubicon, another PtdIns3K subunit that predominantly localizes to endosomal structures,22–25 was also purified and used as a control in this assay. To prepare the autophagic membrane structures, we first tested a biochemical fractionation assay to isolate the autophagosome-enriched membrane. U2OS cells were treated with either lysosomal protease inhibitors E64D and Pepstatin A to block lysosomal degradation, or MG132 to prevent proteasomal degradation. Cells were lysed by hypotonic buffer and homogenization, and separated into cytosolic (supernatant) and nuclear/organelles (pellet) fractions by centrifugation. As shown in Figure 1B, the free form of LC3 was detected in the cytosol (Fig. 1B, lane 1), and LC3-PE conjugated form was only detected in the pellet fraction of cells treated with lysosome inhibitor (Fig. 1B, lane 3). MG132 treatment led to a minor accumulation of LC3-II since proteasome inhibition could lead to moderate activation of autophagy (Fig. 1B, lane 2). This result confirms that autophagosome membranes are enriched in the pellet fraction. Hence, the pellet fraction could serves as a source for autophagosome enriched membrane in the in vitro assay.
Figure 1.
In vitro reconstitution of a cell-free system for leucine-mediated Barkor membrane association. (A) Purification of Barkor protein from Sf9 cells. Different amounts (10 µg, 3 µg and 1 µg) of purified recombinant Barkor-Flag protein are analyzed by SDS/PAGE and Coomassie Blue staining. (B) Fractionation of the cytosolic and membrane fractions. U2OS cells were treated with MG132 (10 µM), or E64D (10 µM) and Pepstatin A (10 µg/ml) for 6 h; lysed by hypertonic buffer; and fractionated by centrifugation. LC3 and tubulin were detected by individual antibodies respectively. (C) Recombinant Barkor association with the autophagosome-enriched membrane is induced by starvation. Cells were treated with nutrient-rich medium or EBSS medium (nutrient-poor) for one hour and the membrane pellets were isolated, and incubated with recombinant Barkor or Rubicon (1 µM) at 4°C for one hour. The pellets were recollected and subjected to SDS-PAGE followed by western blot analysis. Barkor (B) or Rubicon (R) vs. LC3-II are quantified and shown as B(R)/II. (D) The inhibitory activity in S-100 blocks Barkor membrane association. S100 was fractionated from nutrient-rich [S100(+)] or nutrient-poor [S100(−)] HeLa cells, and incubated with recombinant Barkor (1µM) and membrane fractions from starved HeLa cells at RT for one hour. After centrifugation, the Barkor associated membrane fractions were analyzed by western blotting. Endogenous LC3 was detected as a loading marker for autophagosomes. Barkor (B) vs. LC3-II are quantified and shown as B/II. (E) Leucine inhibition on Barkor membrane association is dependent on mTOR activity. Different doses of rapamycin and leucine as indicated were added into the reactions. Barkor and LC3 were detected by western blotting. (F) Leucine specific effect on Barkor membrane association. Different concentrations of leucine and cysteine are included in the reactions as indicated. (G). Autophagic or nonautophagic membrane binding of Barkor. The membrane fractions were collected from Atg5+/+ (WT) or Atg5−/− (KO) MEFs and incubated with recombinant Barkor (1 µM), HeLa S100 and rapamycin (10 µM) as indicated. (H) Detection of mTOR, Raptor and LST8 in the cytosolic and membrane fractions of HeLa cells by western blotting. (I) Detection of mTOR, Raptor and LST8 in the immunoprecipitated mTOR complex from the cytosolic fraction of HeLa cells. (J) HeLa cells were transfected by control siRNA or siRNA against mTOR, the resulting S100 was probed for mTOR and tubulin. (K) S100 was collected from HeLa cells transfected by control siRNA (WT) or mTOR siRNA (KD), and added in the cell-free assay for Barkor membrane association. The membrane associated Barkor and LC3 were detected by western blotting.
We then examined the in vitro membrane association of Barkor and Rubicon in a co-sedimentation assay. We incubated purified recombinant Barkor and Rubicon with membrane pellets isolated from unstressed or starved HeLa cells. The association of Barkor to membrane from starved cells is stronger than that from unstressed cells (Fig. 1C, lane 3 compared with lane 1), suggesting that Barkor membrane association is starvation induced. In contrast, endosome-localized Rubicon membrane association is not altered (Fig. 1C, lanes 2 and 4). This result suggests that Barkor membrane association in vitro correlates with its stress-inducible autophagosome targeting in vivo. In this experiment, endogenous LC3 lipidation form (LC3-II) was used a loading control for equal amounts of autophagosomes.
We further tested whether cytosolic factors can regulate Barkor membrane association in vitro. In the reactions with recombinant Barkor and the membrane fractions from starved HeLa cells, S-100 (membrane-free cytosolic fraction after 100,000 g ultra-centrifugation) collected from nutrient-rich [S100(+)] or nutrient-deprived [S100(−)] HeLa cells was added. Nutrient-rich S-100 could efficiently block the binding of Barkor to autophagosomes in a dose-dependent manner (Fig. 1D, lanes 4–7). The inhibitory activity was significantly reduced when S100 was prepared from starved HeLa cells (Fig. 1D, lanes 11–14). This result indicates that the inhibitory activity of S100 is robust in the cell extract from nutrient-rich HeLa cells but much weaker in the cell extract from nutrient-deprived HeLa cells.
Since inhibition of mTORC1 promotes autophagosome formation in vivo, we speculated the inhibitory activity in S-100 might arise from the mTORC1 signaling pathway. In the in vitro cell-free assay, we incubated the reaction mixture with the small-molecule mTORC1 inhibitor rapamycin. Rapamycin efficiently reversed the inhibitory effect of S-100 from unstressed HeLa cells on Barkor membrane association (Fig. 1E, lane 6 compared with lane 4). In contrast, rapamycin had little effect on S-100 from starved HeLa cells (Fig. 1E, lane 7 compared with lane 5). Hence mTORC1 is probably the major stress-adjustable inhibitor of Barkor membrane association in the unstressed S-100.
To mimic the leucine inhibition on autophagy, we supplemented starved S-100 with different concentration of leucine in the cell-free assay. In the presence of S-100 from starved HeLa cells, Barkor membrane association activity was robust, which could be efficiently blocked by leucine addition (Fig. 1E, lanes 8 and 9). The inhibitory effect is also mediated through mTORC1, since rapamycin could reverse the leucine inhibition (Fig. 1E, lanes 10 and 11). Without S-100, leucine supplementation alone failed to inhibit Barkor membrane association (Fig. 1E, lanes 12 and 13). Not all amino acids have shown a strong inhibition on mTOR signaling pathway and autophagy. Leucine is the most robust one, whereas cysteine barely has any effect. We further investigated if cysteine could inhibit Barkor membrane association as efficient as leucine in vitro. At the same concentration that leucine inhibits Barkor membrane association (Fig. 1F, lanes 3 and 4), cysteine had no inhibitory effect (Fig. 1F, lanes 5 and 6), indicating that the inhibition is leucine-specific. These results therefore recapitulate the in vivo requirement of leucine inhibition in a cell-free system, and delineate a cellular pathway from mTORC1 to Barkor for signal transduction.
Since the membrane fraction might also contain membranes other than autophagic membranes, we examined the nonautophagic membrane binding of Barkor. In Atg5 knockout (KO) MEFs, no autophagosome is formed.27,28 Barkor association with the membrane fraction from Atg5 KO cells is much less than that from Atg5 wild-type (WT) cells (Fig. 1G, lane 4 compared with lane 1). Also the nonautophagic Barkor membrane binding is not regulated by S100 or rapamycin (Fig. 1G, lanes 5 and 6). These data indicate that the majority of Barkor associates with autophagic membranes in the mTOR-dependent manner in the cell-free assay.
To ensure the mTORC1 is intact and functional in the cytosolic fraction (S100), we detected the distribution of mTORC1 in different fractions. As expected, the majority of mTORC1 subunits including mTOR, Raptor and LST8 cofractionates in the cytosolic fraction (S100) (Fig. 1H), and exists in the complex form demonstrated by the immunoprecipitation assay (Fig. 1I). To consolidate mTOR as the major inhibitor of Barkor membrane binding assay, we depleted mTOR from HeLa cells by RNA interference (Fig. 1J). Barkor membrane binding could be efficiently blocked by the mTOR proficient S100 (Fig. 1K, lane 2), but cannot be inhibited by the mTOR depleted S100 (Fig. 1K, lane 5). These data further confirm that mTOR is the major inhibitor present in cytosolic extract for blocking Barkor membrane association.
Leucine deprivation induces autophagy in HEK 293T cells.
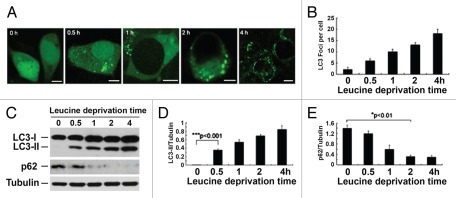
To determine whether leucine deprivation induces autophagy, we generated a HEK 293T cell line stably expressing EGFP-LC3. LC3 serves as an immunohistological marker for the autophagosome.27,28 EGFP-LC3 expressing cells were cultured in the medium without leucine for different time periods. Initially, EGFP-LC3 displayed mostly diffused cytoplasmic staining indicating a very low level of basic autophagy (Fig. 2A). Upon leucine deprivation, the average number of LC3 puncta per cell representing the magnitude of in vivo autophagy activity was significantly augmented in a time-dependent manner, peaking at 4 h upon leucine deprivation (Fig. 2A and B). Consistently, the conversion of LC3-I to LC3-II was also increased (Fig. 2C and D). In addition, the cellular level of p62, an autophagy substrate,27,28 was reduced upon leucine deprivation (Fig. 2C and E). In summary, leucine deprivation elicits substantial autophagy activation in HEK 293T cells.
Figure 2.
Leucine deprivation activates autophagy. (A) Fluorescent EGFP-LC3 detected in transfected HEK 293T cells upon incubation in the leucine-free medium for 0 h, 0.5 h, 1 h, 2 h and 4 h, respectively. (B) Quantification of EGFP-LC3 dots per cell as described in (A). (C) Western blot of LC3, p62 and tubulin in HEK 293T cells treated by the leucine-free medium as indicated. (D and E) Quantification of LC3-II/tubulin and p62/tubulin as described in (C). Data are means ± SEM for at least three different experiments.
Leucine supplementation suppresses autophagy induced by leucine deprivation.
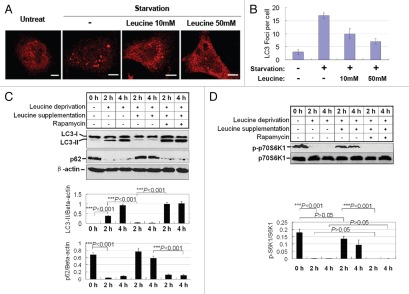
We further tested if leucine supplementation inhibits autophagy. We used a human osteosarcoma U2OS cell line stably expressing Myc-LC3 to perform a leucine-deprivation and re-supplementation assay. Consistent with the previous observations in HEK293T cells, leucine deprivation also dramatically increased the number of autophagic vacuoles decorated by LC3 in U2OS cells (Fig. 3A and B). Re-supplementation with different concentration of leucine repressed autophagy activity in a dose-dependent manner (Fig. 3A and B).
Figure 3.
Leucine supplementation inhibits autophagy inhibition via the mTORC1 pathway. (A) Leucine supplementation blocks autophagosome formation. U2OS cells were treated with leucine-free medium alone or different doses of leucine (10 mM and 50 mM). Myc-LC3 was detected by anti-Myc antibody. (B) Autophagosomes marked by Myc-LC3 described in (A) were counted under a fluorescence microscope (Data are means ± SEM of 20 cells). (C) Leucine supplementation inhibits autophagy. Leucine was added to the leucine-free medium with or without rapamycin (2 µM) for 2 h and 4 h in HEK 293T cells. LC3, p62 and β-actin (used as loading control) protein levels were assessed in HEK 293T cells by western blot (top). Quantification of LC3-II/tubulin and p62/tubulin is shown as the relative expression levels (bottom). Data are means ± SEM for at least three different experiments. (D). Leucine supplementation inhibits mTOR activity. Cells with the same treatments as described in (C) are probed with phosphorylated p70S6K (p-p70S6K), p70S6K, and β-actin (top), and the quantification of p-p70S6K/p70S6K is plotted (bottom).
To validate the autophagy inhibition by leucine supplementation, we examined the LC3-II and p62 level in U2OS cells deprived and re-supplemented with leucine. As shown in Figure 3C, leucine deprivation significantly increased the level of LC3-II, which was accompanied with p62 reduction (Fig. 3C). Consistently, 10 mM leucine supplementation restored the autophagy activity to basal levels (Fig. 3C). As expected, rapamycin potently reversed the leucine supplementation effect, proving that rapamycin-sensitive mTORC1 activation is required for leucine-mediated autophagy inhibition (Fig. 3C).
To ensure that mTOR activity is essential for leucine inhibition, we examine the phosphorylation of S6K, one of the mTOR physiological substrates.29 As expected, S6K phosphorylation was dramatically suppressed by leucine deprivation and restored by leucine replenishment (Fig. 3D).
Leucine supplementation can suppress Barkor autophagosome targeting in vivo.
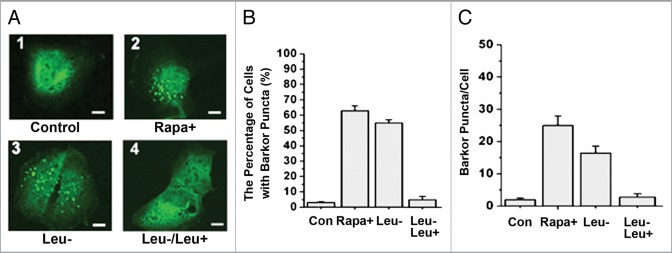
Since Barkor autophagosome targeting is an essential step in autophagosome formation,26 we investigated if leucine deprivation and supplementation affect this step. We examined the subcellular localization of Barkor in the absence or presence of leucine. The percentage of U2OS cells with Barkor puncta and puncta per cell were dramatically increased after 6 h incubation with leucine-free medium (Fig. 4). As a positive control, the mTORC1 inhibitor rapamycin also stimulated the Barkor puncta formation. Leucine supplementation (3 mM) dramatically decreased Barkor puncta (Fig. 4). These data indicate that Barkor autophagosome targeting is induced by leucine deprivation and suppressed by leucine supplementation in vivo.
Figure 4.
Leucine regulates Barkor puncta formation in vivo. (A) For these treatments, U2OS cells were transfected with pCDNA5-GFP-Barkor. Twenty-four hour later, cells were subjected to nutrient-rich medium for 6 h [Control, (1)], nutrient-rich + 250 nM rapamycin for 6 h (2), leucine-free medium for 6 h (3), leucine-free medium + 3 mM leucine for 6 h (4), and then were fixed using 3.5% paraformaldehyde and observed by confocal microscope. (B) Quantification of the percentage of cells with Barkor puncta (%) in cells described in (A). (C) Quantification of Barkor puncta per cell in cells described in (A).
Requirement of mTOR adaptors in leucine-mediated autophagy inhibition.
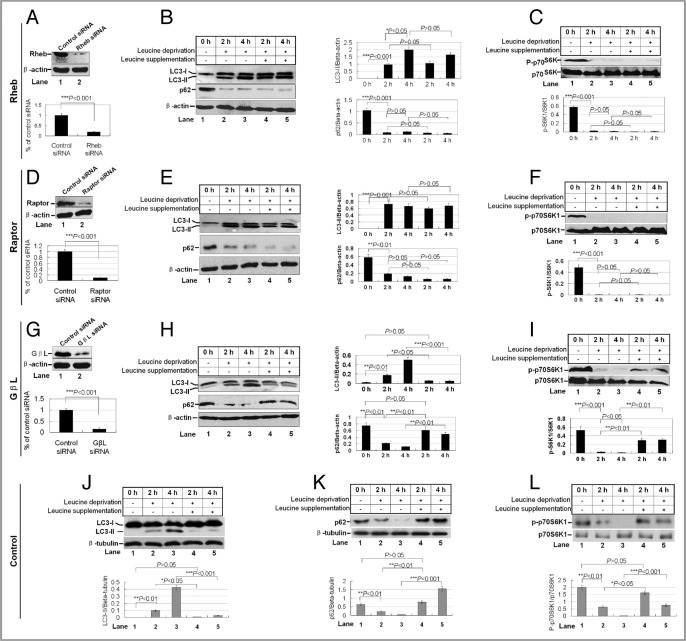
The rapamycin-sensitive mTOR activity is regulated by several adaptors, including Rheb, Raptor and GβL, through distinct mechanisms. To determine the roles of these adaptors in leucine regulation, we depleted the individual adaptors in HEK 293T cells by RNA interference (Fig. 5). mTOR activity was compromised but still detectable in the unstressed cells depleted with Rheb, Raptor and GβL(Fig. 5 and data not show). The small GTPase Rheb is the key downstream effector of TCS1/2 in controlling mTORC1 activity, and Raptor is an essential scaffold for the mTOR kinase activity in vivo.29 In Rheb (Fig. 5A) or Raptor depleted cells (Fig. 5D), autophagy activity, presented as LC3-II conversion and p62 degradation, were strongly stimulated by leucine deprivation, but leucine supplementation failed to inhibit autophagy activity (Fig. 5B, C, E and F). Interestingly, depletion of another positive regulator of the mTOR signaling complex, GβL,30 still respond to leucine deprivation and supplementation (Fig. 5G–I). In the cells treated with a control siRNA, autophagy and mTOR activity is reversibly controlled by leucine deprivation and supplementation (Fig. 5J–L).
Figure 5.
The contribution of mTORC1 adaptors in leucine-mediated inhibition of autophagy and mTOR activity. HEK 293T cells were transduced with pGCsilencer™ RNAi vectors expressing shRNA against Rheb. (A, D and G) Western blotting for Rheb, Raptor and GβL expression (top) and quantification of these proteins compared with β-actin (bottom). (B, E and H) Leucine deprivation and supplementation on autophagy. HEK 293T cells were incubated in leucine-free medium in the absence or presence of 30 mM leucine for 2 h and 4 h. Cell lysates were analyzed by western blotting with the indicated antibodies (left), and the quantification of LC3-II/β-actin and p62/β-actin as the relative expression levels (right). Data are means ± SEM for at least three different experiments. (C, F and I) Leucine deprivation and supplementation on mTOR activity. The same cell lysates as described in (B, E and H) were probed with p-p70S6K and p70S6K antibodies (top) and quantified (bottom). (J–L). A control siRNA was used in the cells with leucine deprivation and supplementation. The protein levels of LC3-II, p62 and p-p70S6K were detected (top) and quantified (bottom).
Discussion
In this study, we demonstrate that leucine regulates Barkor membrane association, an essential event in autophgaosome biogenesis. Recently, Barkor/Atg14(L) was identified as an autophagy-specific subunit of the class III PI3-kinase complex in mammalian cells by four research groups including us.20–23 In this study, we demonstrate that Barkor membrane association is regulated by an inhibitory activity in the unstressed cytosol but not in the starved cytosol. Since this activity could be reversed by rapamycin, mTOR is very likely the responsible inhibitor. In the starved cells with low mTOR activity, leucine supplementation could stimulate mTOR activity and inhibit Barkor membrane association in the cell-free system. This data therefore provide the direct evidence that leucine controls autophagy through a mTOR-Barkor pathway. Reconstitution of leucine effect on autophagy in the cell-free system provides an effective system to further dissect the signaling pathway regulated by leucine.
Although the mTOR activity is an essential part of the leucine-mediated signaling pathway, interestingly, we have observed different response to nutrients in the cells depleted with different mTOR adaptors. Leucine deprivation in the Rheb or Raptor deficient cells strongly induces autophagy and mTOR inactivation that cannot be reversed by leucine supplementation, whereas in GβL depleted cell both autophagy and mTOR activity is sensitive to leucine deprivation and supplementation. It is possible that the residual GβL might still sustain some function, despite that GβL protein level is significantly reduced in siRNA treated cells. Nevertheless, this observation is coincident with the nutrient-sensitive association of Raptor-mTOR and nutrient-insensitive interaction of GβL-mTOR.30 It is also likely that GβL has Rheb and Raptor-independent functions. Knockout of GβL/mLST8 is accompanied by a selective loss of mTORC2 function, whereas mTORC1 function appears to be preserved.31 In additional to these mTORC1 adaptors, it will be interesting to explore the contribution of Ulk1 and Atg13 in our cell-free assays, considering the substantial functional and physical interaction among these molecules. The detailed mechanism will be further explored in the future study. It is probable that unknown mechanism(s) is also involved in this regulation.
In summary, our data show that leucine inhibits autophagy via mTOR-Barkor signaling pathway. Investigating how autophagosome formation is initiated or inhibited, and proceeded remains difficult. The findings presented here provide an insight into the role of leucine in regulating mammalian autophagic activity and will serve as a resource for further mechanistic analysis of intracellular amino acids recycling and metabolism so critical for protein homeostasis.
Materials and Methods
Reagents and Antibodies.
Rapamycin (R8781) and leucine (L8912) were purchased from Sigma Aldrich. Rabbit anti-Phospho-S6K1 (Thr389) (9234), rabbit anti-S6K1 (2708), rabbit anti-Rheb (4935), rabbit anti-Raptor (2280), rabbit anti-GβL (3274), rabbit anti-Flag (2044), rabbit anti-p62 (5114) antibodies, rabbit anti-β-tubulin (5346), mouse monoclonal anti-β-actin antibodies (3700) and mTOR (2983) were obtained from Cell Signaling Technology. Rabbit anti-LC3 (L7543) was obtained from Sigma Aldrich.
Cell culture, cell transfection and cell lysate preparation.
HEK 293T and U2OS cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma Aldrich, D9443) containing 10% fetal bovine serum (Hyclone, 30396) and 1% penicillin-streptomycin (Invitrogen, 15070) in 5% CO2 at 37°C. For leucine-deprivation assay, HEK 293T and U2OS cells were cultured in DMEM-low glucose without L-leucine medium (MP Biomedicals, 1642149) supplemented with 10% dialysis FBS (Sigma, 12105C). Cell transfection was performed with Lipofectin 2000 (Invitrogen, 11668) according to the protocol provided by the manufacturer. Whole-cell lysates (WCLs) used for immunoblotting of different cell lines were prepared in tandem affinity purification buffer [20 mM Tris·HCl (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40, 1 mM NaF, 1 mM Na3VO4, 1 mM EDTA, protease inhibitor mixture (Roche, 04693124001)].21
Immunofluorescence staining.
Cells were transfected with different plasmids. Twelve hours after transfection, cells were trypsinized and transferred to 6-well dishes with coverslips. Twenty-four hours later, cells grown on coverslips were fixed with 4% paraformaldehyde solution in PBS at room temperature for 20 min. After permeabilization with PBS buffer containing 0.1% Triton X-100 at room temperature for 20 min, cells were incubated with primary antibodies (anti-Myc or anti-GFP) at 37°C for 2 h. After washing with PBS buffer containing 0.1% Triton X-100, cells were incubated with rhodamine red-conjugated secondary antibodies at 37°C for 2 h. Slides were examined by using a laser scanning confocal microscope (Zeiss LSM 510 META UV/Vis).21
Vector-based small interference RNA expression.
Construction of the vector PGCsilencer H1/Neo/GFP/RNAi (control-shRNA, Rheb-shRNA, Raptor-shRNA and GβL-shRNA) was fulfilled by Shanghai GeneChem Co., Ltd. The shRNA sequences were designed with “siRNA Target Finder” provided by Ambion. The shRNA coding sequence for human Rheb knockdown is 5′-UCAGUGUAGUUUGUUGUUUAA-3′, for raptor is 5′-GGACAACGGCCACAAGUACTT-3′, and for GβL is 5′-AGCACATCCGCATGTATGATCTC-3′.
Western blotting and treatments.
Rapamycin and leucine were added in fresh media as indicated in text or figure legends. Treatments were terminated by rapid removal of medium with cells on ice, followed by cell lysate preparation. Protein extracts were separated in 7.5%, 12% or 15% SDS-polyacrylamide gels and transferred to PDVF membranes. Blocking was at room temperature for 1 h in 5% fat-free milk, and membranes were incubated overnight at 4°C with primary antibody, and then for 1 h at room temperature with a secondary antibody, and developed with enhanced chemiluminescence method (ECL) and visualized by Kodak Image Station 2000MM. Images were employed for densitometric analysis.
Preparation of S-100 Fraction from human HeLa cells.
We set up HeLa cells at 5 × 105 cells per 100 mm dish in DMEM medium supplemented with 10% FBS and 1% penicillin-streptomycin. After incubation for 48 h at 37°C in a 5% CO2 incubator, the cells were harvested, collected by centrifugation (1000 g for 10 min at 4°C). The cell pellets of HeLa cells were washed once with ice-cold PBS and resuspended in 5 volume of ice-cold buffer Q [20 mM Hepes-KOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, 1 mM dithiothreitol (DTT), 10% glycerol, and 0.1 mM PMSF] supplemented with protease inhibitors (5 µg/ml pepstatin A, 10 µg/ml leupeptin, and 2 µg/ml aprotinin). After sitting on ice for 15 min, the cells were broken by passing 15 times through a G22 needle. After centrifugation in a microcentrifuge for 5 min at 4°C, the supernatants were further centrifuged at 105 × g for 30 min in a benchtop ultracentrifuge (Beckman). The resulting supernatant (S-100) was stored at −80°C and used for the in vitro cell-free assay.
Purification of autophagosome-enriched membrane from human HeLa cells.
Human HeLa cells were cultured with DMEM (10% FBS, and 1% penicillin-streptomycin). The media were changed into EBSS for starvation when cell confluence was up to 80∼90%. After incubation for 3 h at 37°C in a 5% CO2 incubator, the cells were harvested, and collected by centrifugation (1000 g for 10 min at 4°C). The cell pellets of HeLa cells were washed three times with ice-cold PBS and resuspended in 5 volume of ice-cold buffer Q supplemented with protease inhibitors. After sitting on ice for 30 min, the cells were broken by passing 15 times through a G25 ½ needle. After centrifugation (500 g) in a microcentrifuge for 5 min at 4°C, the supernatants were collected and centrifuged (21,000 rpm) for 30 min at 4°C. After removing the supernatant, the pellets were washed three times using ice-cold PBS and stored at −80°C for the in vitro cell-free assay.
Recombinant Barkor protein expression and purification.
The recombinant ZZ-Barkor-His6-Flag protein was overexpressed using suspension cultures of Sf9. One liter sf9 cells were infected with recombinant baculovirus at a multiplicity of infection (MOI) of 5 and allowed to incubate at 27°C for ∼48 h. Cells were collected by centrifugation, washed three times with PBS and resuspended in 20 ml of hypotonic buffer [20 mM TRIS-HCl (pH 7.6), 5 mM KCl; 2 mM MgCl2, 0.5% NP-40] supplemented with protease inhibitors cocktail (Roche, 04693124001). The resuspended cells were lysed by treating (20 times) with Dounce cell homogenizer and centrifuged at 12000 g for 15 min. Both supernatant and the pellet were collected. The pelleted fraction was treated with 16 ml of high salt buffer [20 mM TRIS-HCl (pH 7.6), 450 mM NaCl, 0.1% NP-40, 25% glycerol, 100 U DNase I/ml, protease inhibitor cocktail] and vortexed vigorously and then centrifuged at 12000 g for 20 min. The supernatant was combined with the first supernatant (collected earlier) to generate the final lysate. The recombinant His6-Flagtagged Barkor protein was purified from the soluble supernatant fraction by absorption to IgG-Sepharose resin (GE Healthcare, 17096901) according to the manufacturer's instructions. Briefly, 1 ml of IgG-Sepharose resin pre-equilibrated with 20 ml TEV protease cleavage buffer [10 mM TRIS-HCl (pH 8.0), 150 mM NaCl, 0.1% NP-40, 1 mM DTT], was added to the cell lysate and rotate at 4°C for 1 to 2 h at 30 rpm. Beads were then pelleted by centrifugation for 10 min at 1000 g, washed three times with TEV protease cleavage buffer to remove unbound proteins and then the recombinant protein was eluted from the resin by incubating at 4°C overnight with homemade TEV protease (20 U/ml) to cleave the ZZ domain from the recombinant Barkor. IgG-Sepharose eluent was collected after centrifugation at 500 g for 10 min at 4°C, and then incubated with 0.5 ml Ni-NTA Sepharose (Qiagen, 30210), and incubated at 4°C for 2 h rotating at 30 rpm. The beads were pelleted by centrifugation at 500 g for 5 min at 4°C, and then the beads were washed with 20 ml buffer containing 20 mM imidazole. Barkor-His6-Flag was eluted with 3 ml buffer containing 250 mM imidazole. The final eluent was dialyzed against 500 ml storage buffer [25 mM TRIS-HCl (pH 7.5), 125 mM NaCl, 1 mM DTT, 10% glycerol].
Cell-free assay for Barkor membrane association in vitro.
The autophagosome-enriched membranes were prepared from human HeLa cells. The membrane fractions were incubated with Barkor protein (1 µM) and S-100 in a final volume of 100 µl of buffer Q for 30 min at 37°C with gentle shaking (300 rpm). The reaction was stopped by transferring the reaction tubes on ice. The reaction mixture was subsequently subjected to centrifugation (21,000 rpm) in order to separate the autophagosomal pellet from the soluble proteins and washed twice with 100 µl buffer Q. The binding of Barkor to autophagosome-enriched membrane was assessed by western blotting using rabbit anti-Flag antibody.
Statistical analysis.
Experiments were performed at least three times, with each condition run in duplicate. Statistical significance was determined by paired Student's t-test and/or with analysis of variance (ANOVA) test. Differences were considered significant with a p-value < 0.05.
Acknowledgments
We thank all of the Zhong laboratory members for helpful discussion and technical assistance. This work was supported by University of California Cancer Research Coordinating Committee funds, a New Investigator Award for Aging from the Ellison Medical Foundation, Hellman Family Fund, American Cancer Society grant (RSG-11-274-01-CCG) and NIH RO1 (CA133228) (to Q.Z.) and by the grants from the National Natural Science Foundation of China (No.31172290, No.31072036 and No.30700580 to X.Y.) and Huazhong Agricultural University Scientific and Technological Self-innovation Foundation (No. 2010PY011 to X.Y.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Meijer AJ. Amino acid regulation of autophagosome formation. Methods Mol Biol. 2008;445:89–109. doi: 10.1007/978-1-59745-157-4_5. [DOI] [PubMed] [Google Scholar]
- 2.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 5.Guertin DA, Sabatini DM. Encylopedia of Life Sciences. John Wiley & Sons, Ltd: Chichester; 2006. Cell Size Control; pp. 1–10. [Google Scholar]
- 6.Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin-and farnesylation-dependent manner. J Biol Chem. 2003;278:32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- 7.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/S1097-2765(03)00220-X. [DOI] [PubMed] [Google Scholar]
- 8.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/S0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 10.Glynn EL, Fry CS, Drummond MJ, Timmerman KL, Dhanani S, Volpi E, et al. Excess leucine intake enhances muscle anabolic signaling but not net protein anabolism in young men and women. J Nutr. 2010;140:1970–1976. doi: 10.3945/jn.110.127647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guertin DA, Sabatini DM. An expanding role for mTOR in cancer. Trends Mol Med. 2005;11:353–361. doi: 10.1016/j.molmed.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Ganley IG. Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidy-linositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 23.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci USA. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286:185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci USA. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 30.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/S1097-2765(03)00114-X. [DOI] [PubMed] [Google Scholar]
- 31.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]