Abstract
Objective:
To identify a new genetic cause of distal hereditary motor neuropathy (dHMN), which is also known as a variant of Charcot-Marie-Tooth disease (CMT), in a Chinese family.
Methods:
We investigated a Chinese family with dHMN clinically, electrophysiologically, and genetically. We screened for the mutations of 28 CMT or related pathogenic genes using an originally designed microarray resequencing DNA chip.
Results:
Investigation of the family history revealed an autosomal dominant transmission pattern. The clinical features of the family included mild weakness and wasting of the distal muscles of the lower limb and foot deformity, without clinical sensory involvement. Electrophysiologic studies revealed motor neuropathy. MRI of the lower limbs showed accentuated fatty infiltration of the gastrocnemius and vastus lateralis muscles. All 4 affected family members had a heterozygous missense mutation c.2677G>A (p.D893N) of alanyl-tRNA synthetase (AARS), which was not found in the 4 unaffected members and control subjects.
Conclusion:
An AARS mutation caused dHMN in a Chinese family. AARS mutations result in not only a CMT phenotype but also a dHMN phenotype.
Distal hereditary motor neuropathy (dHMN) is also known as distal spinal muscular atrophy or a variant of Charcot-Marie-Tooth disease (CMT). dHMN is genetically and clinically heterogeneous. It has been classified into 7 subtypes according to age at onset, mode of inheritance, and the presence of additional features.1 To date, at least 11 genes have been shown to be involved in dHMN: heat shock 27 kDa protein 1 (HSPB1), heat shock 22 kDa protein 8 (HSPB8), heat shock 27 kDa protein 3 (HSPB3), dynactin 1 (DCTN1), glycyl-tRNA synthetase (GARS), pleckstrin homology domain containing, family G (with RhoGef domain) member 5 (PLEKHG5), Berardinelli-Seip congenital lipodystrophy 2 (BCSL2), senataxin (SETX), immunoglobulin mu binding protein 2 (IGHMBP2), ATPase and Cu2+ transporting, alpha polypeptide (ATP7A), and transient receptor potential cation channel, subfamily V, member 4 (TRPV4).2 Interestingly, 5 of these genes (HSPB8, HSPB1, GARS, TRPV4, and BCSL2) have been described in CMT2–6; in some patients, dHMN and CMT phenotypes have been found to coexist.7
Four aminoacyl-tRNA synthetases (AARSs) have been implicated in CMT/dHMN:) glycyl (GARS; MIM 601472) in CMT2D and dHMN5A; 2) tyrosyl (YARS; MIM 608323) in dominant intermediate CMT type C; 3) alanyl (AARS; MIM 613287) in CMT2N; and 4) lysyl (KARS; MIM 601421) in CMT-recessive intermediate B and hereditary neuropathy with liability to pressure palsies.5,8–10 Although mutations in AARS cause axonal CMT, no published reports linking AARS mutations to the dHMN phenotype exist.
We report clinical and electrophysiologic findings in 3 patients with dHMN from a Chinese family carrying a novel missense mutation (D893N) in AARS.
METHODS
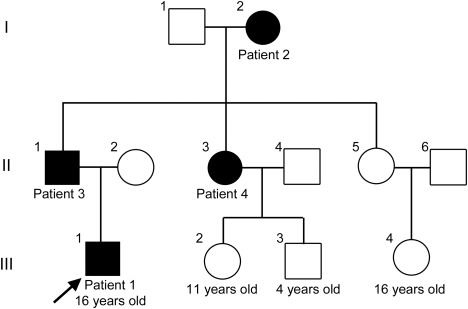
We studied 3 generations of a Chinese family that included 4 affected and 8 unaffected members ascertained by neurologic examination (figure 1).
Figure 1. Pedigree of the distal hereditary motor neuropathy family.
The arrow indicates the proband. Affected individuals are represented by solid black symbols; open symbols represent healthy individuals.
Patients.
Patient 1.
Patient 1 (III-1), now a 16-year-old boy, was referred to our neuromuscular disease department at the age of 11 years. He reported frequent falling and difficulty in rising from the squatting position since the age of 2 years; however, his condition had not deteriorated. Neurologic examination was initially performed at the first referral. His gait was almost normal, with no ataxia, but standing on his heels was difficult, and his heels could not touch the ground when squatting. Mild atrophy and weakness in the distal muscles of the lower limbs were observed, with a muscle strength score of 4 of 5 (Medical Research Council [MRC] scale) for the extensor digitorum brevis muscles, whereas the muscle strength scores of the iliopsoas, quadriceps femoris, biceps femoris, anterior tibial, and gastrocnemius muscles were 5 of 5 (MRC scale). However, the muscle strength of the quadriceps femoris and gastrocnemius muscles was relatively weaker than that of the iliopsoas or anterior tibial muscles. Pes cavus and toe clawing were noted (figure 2A). Sensory examination, including pain sensation, light touch sensation, position sensation, and vibration sensation of the 4 limbs was unremarkable. Deep tendon reflexes were decreased in the knees and absent in the ankles. Examination of the upper limbs was normal. There was no evidence of tremor or pyramidal tract signs.
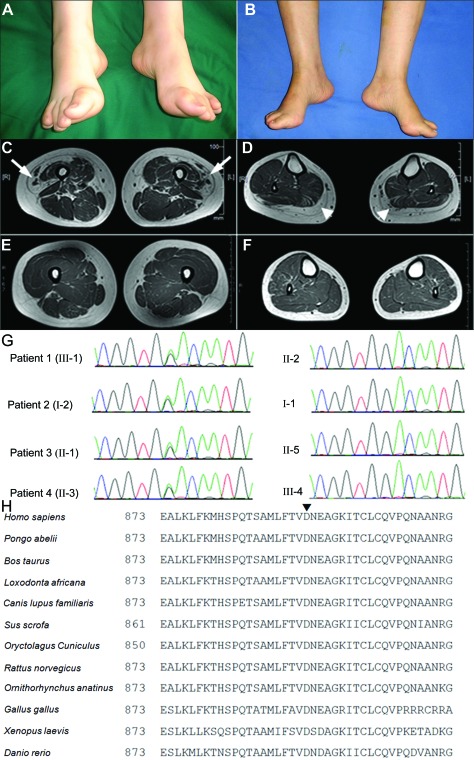
Figure 2. Clinical, radiologic, and genetic findings of the distal hereditary motor neuropathy family.
(A) A picture of patient 1 shows moderate pes cavus and toe clawing. (B) A picture of patient 2 shows pes cavus. (C and D) Axial T1-weighted images of the lower limbs in patient 1. (C) Axial image of the thighs, illustrating marked fatty replacement of the vastus lateralis muscle (arrows). (D) Axial image of the legs demonstrating complete fatty replacement of the gastrocnemius muscle (arrowheads). (E and F) Axial T1-weighted images of the lower limbs of a healthy control subject. (G) Chromatogram of the heterozygous c.2677G>A (p.D893N) mutation in exon 19 of AARS: left, 4 affected members; right, 4 unaffected relatives. (H) Comparison of AARS from different species. Arrowhead (▾) on top of the alignment indicates 893 amino acids.
Patient 2.
Patient 2 (I-2), the 67-year-old grandmother of the proband, first showed mild motor disability of the lower limbs at 55 years of age. Physical examination revealed distal motor weakness, wasting in the lower limbs, and pes cavus (figure 2B). Results of sensory examination including pain sensation, light touch sensation, position sensation, and vibration sensation of the 4 limbs were unremarkable. Deep tendon reflexes were absent in the lower limbs. Examination of the upper limbs was normal. There was no evidence of ataxia, tremor, or pyramidal tract signs.
Patient 3 and patient 4.
Neither patient 3 (II-1), the 44-year-old father, nor patient 4 (II-3), the 38-year-old aunt, of the proband experienced symptoms; however, neurologic examination revealed foot deformity, mild atrophy, and weakness of the lower limbs. The results of the sensory examination of patients 3 and 4 were similar to those of patients 1 and 2. Deep tendon reflexes were decreased in the knees and absent in the ankles.
Standard protocol approvals, registrations, and patient consent.
The patients and family members included in this study gave written informed consent, and the study was approved by the Third Hospital of Hebei Medical University and the Institutional Review Board of Kagoshima University.
Electrophysiologic study.
Needle EMG and nerve conduction velocity studies were performed in patients 1, 2, and 3.
MRI study.
Skeletal muscle MRI of the lower limbs was performed in patient 1.
Mutation screening.
Genomic DNA of 8 family members (I-1, I-2, II-1, II-2, II-3, II-5, III-1, and III-2) was extracted from the peripheral blood obtained using standard methods. The purpose-built GeneChip CustomSeq Resequencing Array (Affymetrix, Inc., Santa Clara, CA) was designed to screen for CMT and related diseases such as ataxia with oculomotor apraxia types 1 and 2, spinocerebellar ataxia with axonal neuropathy, and dHMN. We designed 363 primer sets to cover the entire coding regions and flanking sequences of the following 28 pathogenic genes: early growth response 2 (EGR2), peripheral myelin protein 22 (PMP22), myelin protein zero (MPZ), gap junction protein beta 1 (GJB1), periaxin (PRX), lipopolysaccharide-induced TNFα factor (LITAF), neurofilament light chain polypeptide (NEFL), ganglioside-induced differentiation-associated protein 1 (GDAP1), myotubularin-related protein 2 (MTMR2), SH3 domain and tetratricopeptide repeats 2 (SH3TC2), SET-binding factor 2 (SBF2), N-myc downstream regulated 1 (NDRG1), mitofusin 2 (MFN2), Ras-related GTPase 7 (RAB7), GARS, HSPB1, HSPB8, lamin A/C (LMNA), dynamin 2 (DNM2), YARS, AARS, KARS, aprataxin (APTX), senataxin (SETX), tyrosyl-DNA phosphodiesterase 1 (TDP1), desert hedgehog (DHH), gigaxonin 1 (GAN1), and K-Cl cotransporter 3 (KCC3) and 9 other candidate genes. The 363 PCR amplicons were amplified in 32 multiplex PCR reactions using the Qiagen Multiplex PCR system (Qiagen, Venlo, The Netherlands). Each reaction required 120 ng of genomic DNA, 10 pmol of primer sets, dNTP, and Qiagen Multiplex PCR Master Mix (Qiagen). The following conditions were used for multiplex PCR: 15 minutes at 95°C; 42 cycles of amplification (94°C for 30 s, 60°C for 3 minutes, and 72°C for 90 s); and 15 minutes at 68°C. Pooling, DNA fragmentation, labeling, and chip hybridization were performed according to the CustomSeq Resequencing protocol (Affymetrix, Inc.). Chips were washed using a Fluidics Station 450 (Affymetrix, Inc.) using CustomSeq Resequencing wash protocols. Analysis of microarray data was performed using GeneChip Sequence Analysis Software, version 4.0 (Affymetrix, Inc.).11
To confirm the mutation revealed by our DNA chip, the proband and 7 members of the family underwent genetic analysis by direct sequencing. In brief, 50 ng of genomic DNA from the patients was amplified using the hot-start PCR method. Using a presequencing kit (USB Corp., Cleveland, OH), PCR products were purified and sequenced by dye terminator chemistry using an ABI Prism 377 DNA Sequencer (Applied Biosystems, Foster City, CA). The resulting sequences were then aligned, and mutations were evaluated using Sequencher version 4.8 sequence alignment software (Gene Codes, Ann Arbor, MI).
RESULTS
Electrophysiologic study.
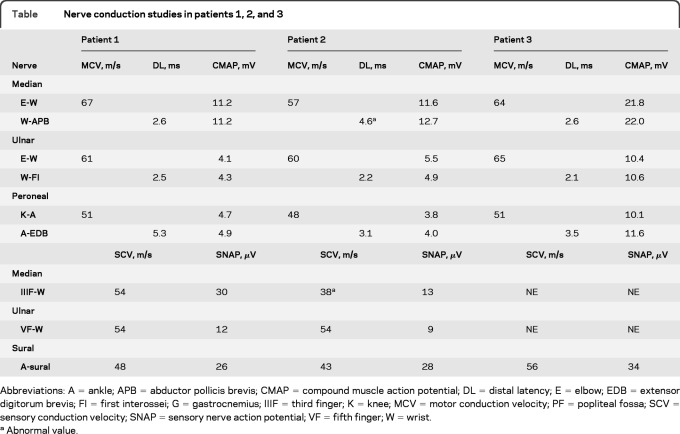
Needle EMG revealed a neurogenic pattern, with a high frequency of large motor unit potentials recorded from the lower limbs of all 3 patients tested. Patient 1 showed normal sensory nerve conduction velocities (SCVs) and sensory nerve action potential of the median, ulnar, and sural nerves. Furthermore, he showed normal motor nerve conduction velocities and amplitudes of the compound muscle action potentials of the median, ulnar, and peroneal nerves. Patient 2 showed normal SCV, with a slight increase in distal latency in the median nerve and a mild decrease in SCV, suggestive of bilateral carpal tunnel syndrome. Patient 3 showed results similar to those of patient 1 (table). These findings indicate a chronic neurogenic pattern, suggesting that this family had inherited motor neuropathy.
Table.
Nerve conduction studies in patients 1, 2, and 3
Abbreviations: A = ankle; APB = abductor pollicis brevis; CMAP = compound muscle action potential; DL = distal latency; E = elbow; EDB = extensor digitorum brevis; FI = first interossei; G = gastrocnemius; IIIF = third finger; K = knee; MCV = motor conduction velocity; PF = popliteal fossa; SCV = sensory conduction velocity; SNAP = sensory nerve action potential; VF = fifth finger; W = wrist.
Abnormal value.
MRI study.
An axial T1-weighted MRI showed accentuated fatty infiltration of the gastrocnemius and vastus lateralis muscles (figure 2, C and D).
Resequencing analysis and control study.
We identified one missense mutation, c.2677G>A (designated p.D893N), in exon 19 of AARS. All 4 family members considered to be clinically affected proved to have the heterozygous AARS p.D893N mutation, whereas none of the 4 unaffected relatives harbored this mutation (figure 2G). In addition, the mutation was not found in 220 East Asian (120 Chinese and 100 Japanese) control chromosomes or the chromosomes of patients with 850 inherited neuropathy nor did we find the D893N mutation in the 1000 Genomes Web site, which catalogs human genetic variations using 1,197 samples including 300 East Asian (200 Chinese) samples (http://browser.1000genomes.org).
DISCUSSION
We report that an AARS mutation caused dHMN. A detailed investigation of the history of a family revealed 9 members over 3 generations, 4 of whom were affected individuals, consistent with a pattern of autosomal dominant transmission (figure 1). Clinical examination revealed benign wasting and weakness of the lower limbs. The diagnosis of dHMN was based on the history of autosomal dominant inheritance and electrophysiologic studies.
AARSs are a ubiquitously expressed, essential family of enzymes responsible for attaching amino acids to their cognate tRNAs in all cells and tissues. Mutations in 4 genes, GARS, YARS, AARS, and KARS, that encode AARSs have been implicated in CMT/dHMN.5,8–10 Mutations in GARS, YARS, and AARS cause autosomal dominant CMT/dHMN. Two KARS mutations were detected in the compound heterozygous state in a patient with autosomal recessive CMT, developmental delay, self-abusive behavior, dysmorphic features, and vestibular schwannoma.10 To elucidate the reason that mutations in ubiquitously expressed AARSs result in peripheral neuropathy, the effects of GARS, YARS, AARS, and KARS mutations in CMT/dHMN were investigated. A common pathologic mechanism for genetic disorders is a loss of gene function through altered mRNA or protein levels, although this is unusual for neurodegenerative diseases inherited in an autosomal dominant manner. Studies on G240R GARS heterozygous mutated lymphoblastoid cells did not reveal severely altered transcription, translation, or protein stability.12 Pathogenic mechanisms such as defective aminoacylation, abnormal distribution in axons, or a combination of both are postulated to underlie CMT/dHMN, based on functional and protein localization studies of heterozygous GARS and AARS mutants.9,12 Functional analyses of compound heterozygous mutations in KARS revealed severely affected enzyme activity.10
AARS catalyzes the attachment of alanine to its cognate tRNAs during protein synthesis. Investigation of the structure of AARS by X-ray crystallography revealed that it contains, from the N to the C terminus, an aminoacylation domain, a middle helical domain, an editing domain, and a so-called C-Ala domain.13 The reported variant (R329H) is located in the middle helical domain, which together with the aminoacylation domain, is responsible for the complete and specific aminoacylation of AARS. The D893N variation is located in the C-Ala domain, which facilitates efficient editing by bringing together the aminoacylation and editing domains. A sequence homology search was performed to align protein sequences from multiple species, using a constraint-based multiple alignment tool (COBALT) (http://www.ncbi.nlm.nih.gov/tools/cobalt/). Aspartic acid 893 was conserved among all species analyzed (figure 2H). Thus, the D893N mutation identified in the Chinese dominant dHMN family is located in a remarkably well-conserved sequence of amino acids, suggesting that it may have a potential functional impact on AARS. Furthermore, we were able to computationally predict the effect of the D893N mutation on protein function using the MuPro (http://www.ics.uci.edu/∼baldig/mutation.html) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) algorithms, which gave scores of −0.334 and 0.802, respectively. MUPro scores of less than 0 indicate a decrease in protein stability, and PolyPhen-2 scores of approximately 1 indicate a prediction of pathogenicity. The D893N mutation is probably a pathogenic mutation, based on the degree of conservation of the affected residues.
Five genes (HSPB8, HSPB1, GARS, TRPV4, and BCSL2) have been described in both dHMN and CMT.2–7 We add AARS on the basis of the present report. GARS and AARS are involved in common processes during protein synthesis, and the mutations reported to date were all missense mutations. Pathogenic missense mutations for autosomal dominant disease usually have a gain-of-function or dominant-negative effect. These pathogenic mutations of tRNA synthetases may directly disrupt protein synthesis.
Our data confirm that a mutation in the AARS gene (designated p.D893N) is associated with dominant dHMN in a Chinese family. This observation adds to a growing body of evidence that implicates specific genes/proteins in peripheral nerve function and delineates the pathologic consequences of their dysfunction.
ACKNOWLEDGMENT
The authors thank the families described in this report for their cooperation and A. Yoshimura of Kagoshima University for her excellent technical assistance.
GLOSSARY
- AARS
alanyl-tRNA synthetase
- CMT
Charcot-Marie-Tooth
- dHMN
distal hereditary motor neuropathy
- MRC
Medical Research Council
- SCV
sensory nerve conduction velocity
AUTHOR CONTRIBUTIONS
Z. Zhao and Dr. Hashiguchi contributed equally to this work. Dr. Hu and Dr. Takashima designed the research. Z. Zhao, Dr. Hashiguchi, Dr. Tokunaga, Dr. Sakiyama, and Dr. Okamoto performed genetic studies. L. Zhu, H. Shen, and Dr. Hu performed clinical research and provided patient information. Z. Zhao, Dr. Hu, and Dr. Takashima wrote the article.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Harding AE. Inherited neuronal atrophy and degeneration predominantly of lower motor neurons. In: Dyck PJ, Thomas PK, eds. Peripheral Neuropathy. Philadelphia: WB Saunders; 1993 [Google Scholar]
- 2.Drew AP, Blair IP, Nicholson GA. Molecular genetics and mechanisms of disease in distal hereditary motor neuropathies: insights directing future genetic studies. Curr Mol Med 2011;11:650–665 [DOI] [PubMed] [Google Scholar]
- 3.Irobi J, Van Impe K, Seeman P, et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet 2004;36:597–601 [DOI] [PubMed] [Google Scholar]
- 4.Houlden H, Laura M, Wavrant-De Vrièze F, et al. Mutations in the HSP27 (HSPB1) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2. Neurology 2008;71:1660–1668 [DOI] [PubMed] [Google Scholar]
- 5.Antonellis A, Ellsworth RE, Sambuughin N, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 2003;72:1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Windpassinger C, Auer-Grumbach M, Irobi J, et al. Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat Genet 2004;36:271–276 [DOI] [PubMed] [Google Scholar]
- 7.Sambuughin N, Sivakumar K, Selenge B, et al. Autosomal dominant distal spinal muscular atrophy type V (dSMA-V) and Charcot-Marie-Tooth disease type 2D (CMT2D) segregate within a single large kindred and map to a refined region on chromosome 7p15. J Neurol Sci 1998;161:23–28 [DOI] [PubMed] [Google Scholar]
- 8.Jordanova A, Irobi J, Thomas FP, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet 2006;38:197–202 [DOI] [PubMed] [Google Scholar]
- 9.Latour P, Thauvin-Robinet C, Baudelet-Méry C, et al. A major determinant for binding and aminoacylation of tRNAAla in cytoplasmic alanyl-tRNA synthetase is mutated in dominant axonal Charcot- Marie-Tooth disease. Am J Hum Genet 2010;86:77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLaughlin HM, Sakaguchi R, Liu C, et al. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet 2010;87:560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutler DJ, Zwick ME, Carrasquillo MM, et al. High-throughput variation detection and genotyping using microarrays. Genome Res 2001;11:1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonellis A, Lee-Lin SQ, Wasterlain A, et al. Functional analyses of glycyl-tRNA synthetase mutations suggest a key role for tRNA-charging enzymes in peripheral axons. J Neurosci 2006;26:10397–10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swairjo MA, Otero FJ, Yang XL, et al. Alanyl-tRNA synthetase crystal structure and design for acceptor-stem recognition. Mol Cell 2004;13:829–841 [DOI] [PubMed] [Google Scholar]