Abstract
Summary
Background
UMOD mutations cause familial juvenile hyperuricemic nephropathy (FJHN) and medullary cystic kidney disease (MCKD), although these phenotypes are nonspecific.
Design, setting, participants, & measurements
We reviewed cases of UMOD mutations diagnosed in the genetic laboratories of Necker Hospital (Paris, France) and of Université Catholique de Louvain (Brussels, Belgium). We also analyzed patients with MCKD/FJHN but no UMOD mutation. To determine thresholds for hyperuricemia and uric-acid excretion fraction (UAEF) according to GFR, these parameters were analyzed in 1097 patients with various renal diseases and renal function levels.
Results
Thirty-seven distinct UMOD mutations were found in 109 patients from 45 families, all in exon 4 or 5 except for three novel mutations in exon 8. Median renal survival was 54 years. The type of mutation had a modest effect on renal survival, and intrafamilial variability was high. Detailed data available in 70 patients showed renal cysts in 24 (34.3%) of nonspecific localization in most patients. Uricemia was >75th percentile in 31 (71.4%) of 42 patients not under dialysis or allopurinol therapy. UAEF (n = 27) was <75th percentile in 70.4%. Among 136 probands with MCKD/FJHN phenotype, UMOD mutation was found in 24 (17.8%). Phenotype was not accurately predictive of UMOD mutation. Six probands had HNF1B mutations.
Conclusions
Hyperuricemia disproportionate to renal function represents the hallmark of renal disease caused by UMOD mutation. Renal survival is highly variable in patients with UMOD mutation. Our data also add novel insights into the interpretation of uricemia and UAEF in patients with chronic kidney diseases.
Introduction
For many years, genes involved in adult hereditary tubulointerstitial nephritis (TIN) remained unknown. Clinicians identified several phenotypes that appeared to be distinct and labeled these as familial juvenile hyperuricemic nephropathy (FJHN), medullary cyst kidney disease (MCKD), or adult nephronophthisis, all of autosomal dominant inheritance (1).
Two different loci for MCKD were subsequently mapped: MCKD1 on chromosome 1q21 (2) and MCKD2 on chromosome 16p12 (3). To date, the gene involved in MCKD1 remains unknown. Then it was demonstrated that FJHN and MCKD2 represented two facets of a single disease secondary to mutations in the UMOD gene encoding uromodulin (1,4–6). Additionally, in one patient with glomerulocystic disease, a UMOD mutation was identified (7). Taken together, these observations showed a large variability of diseases associated to the UMOD gene, reminiscent of situations known with other genes (e.g., FBN1) (8). Thus, the conditions caused by mutations in UMOD gene are nowadays preferentially called uromodulin-associated diseases (9).
In a few families with FJHN/MCKD, linkage to the MCKD1 and MCKD2 loci could be excluded, suggesting further genetic heterogeneity (10–12). Recently, mutations in preprorenin signal peptide (1q32) were reported as a new cause of autosomal dominant TIN (13). Mutations in HNF1B gene were also shown to cause a wide variety of renal and extrarenal manifestations including renal cysts and, rarely, FJHN (14,15). Although the last decade has witnessed important advances into the understanding of the genetic substratum of hereditary TIN, other genes involved remain to be discovered. Hereditary TIN may be underrecognized, because hallmarks of these diseases are common in patients with chronic kidney disease (CKD) and because family history may be absent or unknown. We aimed to report our experience in diagnosis of UMOD mutations, along with detailed descriptions of phenotype.
Materials and Methods
Study Population
First, we reviewed all cases of patients in whom the UMOD mutation was found between January 2003 and September 2009 in the genetic laboratories of the Necker Hospital (Paris, France) and of the Université Catholique de Louvain (Brussels, Belgium). For the majority of patients, full medical charts were reviewed to collect clinical data. In other cases, data were obtained from a specific questionnaire that was systematically asked of clinicians who sent a DNA sample. GFR was estimated using the Modification of Diet in Renal Disease equation (16) or the Schwartz formula for children (17). Data on renal function evolution were collected until last follow-up available or estimated GFR <10 ml/min per 1.73 m2.
Second, we retrospectively reviewed all cases of patients screened for the UMOD mutation at Necker Hospital. Given that the Necker Hospital laboratory is the only one to perform UMOD gene studies in France, the selection of patients investigated reflected diagnostic procedures for hereditary TIN in France. The criteria for UMOD sequencing remained the same during all of the period studied. Patients included were those with full clinical data available (including renal ultrasound) and phenotype suggesting FJHN or MCKD on the basis of the following inclusion criteria: TIN with either: (1) gout or hyperuricemia before 40 years; (2) renal cysts; and (3) family history of kidney disease in at least one first-degree relative. Patients with proteinuria >1 g/d, hematuria, or diabetes and those with well-defined hereditary nephropathies, especially polycystic kidney diseases, were excluded.
Uric Acid Levels in CKD Population
Variation of uricemia and uric acid excretion fraction (UAEF) according to GFR were studied in 1097 consecutive adult patients (613 men and 484 women) who underwent GFR measurement in the physiology department of Bichat Hospital between 2006 and 2009. These parameters were routinely measured in all patients. Patients included in this analysis had CKD of various etiology (excluding FJHN) with GFR >10 ml/min per 1.73 m2 and were not treated by allopurinol. GFR was assessed by 51Cr-EDTA renal clearance as described previously (18). Determination of UAEF was based on fasting urine samples. We used the following formula for calculation: UAEF = urinary uric acid/serum uric acid multiplied by urinary creatinine/serum creatinine. The data on uricemia were analyzed separately in men and women, because the values were higher in men.
DNA Mutational Analysis
Informed consent was obtained from the affected patients. Genomic DNA was isolated from peripheral blood leukocytes by standard procedure. All of the exons covering UMOD (10 coding exons) and HNF1B coding regions were amplified by PCR for direct sequencing. Primers used for UMOD gene have been previously reported (4,19). Bidirectional sequencing was carried out using a Big Dye Terminator kit (Applied Biosystem, Foster City). Additionally, screening for large rearrangements in HNF1B gene was performed using quantitative multiplex PCR amplification (15,20). The nomenclature for the description of sequence variants was as recommended in www.hgvs.org (last modified May 06, 2009). Nucleotide numbering reflects cDNA numbering with + 1 corresponding to the A of the ATG translation initiation codon in the reference sequence (NM_003361).
Statistical Analyses
Quantitative parameters are presented as median and interquartile range (25th to 75th percentiles), and qualitative parameters are presented as number and percentage. Categorical variables were compared using the chi-squared test or Fisher exact test when appropriate. Continuous variables were compared using the Mann–Whitney U test (two groups); P < 0.05 was considered statistically significant. A log-rank test was used for comparison of survival curves.
Results
Spectrum of Germline Mutations in the UMOD Gene
Overall, 37 distinct UMOD mutations, 19 of which are novel, were found in 109 patients belonging to 45 families (Table 1). Thirty-one (83.8%) changes were located within exon 4 of UMOD gene, whereas the remaining mutations occurred in exon 5 (n = 3, 8.1%) and exon 8 (n = 3, 8.1%) (Figure 1A). Mutation types were highly stereotyped including mainly missense changes (35 of 37, 95.6%) with in-frame deletions (2 of 37, 5.4%). Each missense mutation affected a highly conserved nucleotide, was absent in more than 150 control DNA (300 chromosomes), and fully segregated in the family (familial cases). To assess putative correlations between phenotype and genotype, mutations were further classified in five groups: (1) substitutions affecting cysteine residues (gain or loss, n = 17), (2) substitution affecting a polar residue (n = 12), (3) substitutions affecting an amino acid other than cysteine or polar charged residues (n = 3), (4) in-frame deletion (n = 2), and (5) mutations occurring within exon 8 (n = 3) (Table 1). In two patients with known UMOD mutation, neomutation was demonstrated by genetic testing of parents, which revealed that none of them was carrying a mutation.
Table 1.
Mutations in UMOD gene were found in 109 patients belonging to 45 families
| Family | No. of Cases | Exon | Nucleotide Change | Effect on Coding Sequence | Reference | Type of Mutation |
|---|---|---|---|---|---|---|
| V2 | 1 | 4 | c0.95G>A | p.C32Y | Vylet'al et al. (9) | 1 |
| F2 | 3 | 4 | c0.100G>A | p.E34K | Novel | 2 |
| D2 | 2 | 4 | c0.176A>C | p.D59A | Dahan et al. (4) | 2 |
| S1 | 1 | 4 | c0.205T>C | p.C69R | Novel | 1 |
| D1 | 3 | 4 | c0.334T>C | p.C112R | Dahan et al. (4) | 1 |
| B1 | 3 | 4 | c0.376T>C | p.C126R | Dahan et al. (4), Vylet'al et al. (9), Turner et al. (8) | 1 |
| F4 | 1 | 4 | c0.403T>A | p.C135Y | Kudo et al. (6) | 1 |
| L3 | 6 | 4 | c0.442T>C | p.C148R | Novel | 1 |
| F3 | 2 | 4 | c0.449G>C | p.C150S | Rampoldi et al. (7) | 1 |
| G6 | 1 | 4 | c0.449G>C | p.C150S | Rampoldi et al. (7) | 1 |
| C3 | 3 | 4 | c0.459C>T | (r.spl?)a | Novel | 4 |
| L5 | 3 | 4 | c0.509G>A | p.C170Y | Dahan et al. (4) | 1 |
| W1 | 3 | 4 | c0.509G>A | p.C170Y | Dahan et al. (4) | 1 |
| H1 | 1 | 4 | c0.514G>C | p.D172H | Novel | 2 |
| B2 | 3 | 4 | c0.552G>C | p.W184C | Novel | 1 |
| P1 | 1 | 4 | c0.553C>T | p.R185C | Novel | 1 |
| L1 | 5 | 4 | c0.554G>A | p.R185H | Novel | 3 |
| J1 | 8 | 4 | c0.553C>A | p.R185Sa | Dahan et al. (4) | 1 |
| G3 | 3 | 4 | c0.563_661del | p.E188_L221del | Dahan et al. (4) | 4 |
| G1 | 2 | 4 | c0.585_586CG>TA | p.D196N | Williams et al. (21) | 2 |
| R2 | 3 | 4 | c0.586G>T | p.D196Y | Lhotta et al. (37) | 2 |
| P2 | 1 | 4 | c0.610G>C | p.R204P | Novel | 2 |
| A1 | 2 | 4 | c0.610C>G | p.R204G | Dahan et al. (4) | 2 |
| D4 | 2 | 4 | c0.628G>A | p.G210S | Novel | 2 |
| P3 | 1 | 4 | c0.628G>A | p.G210S | Novel | 2 |
| B3 | 2 | 4 | c0.649T>G | p.C217G | Novel | 1 |
| P4 | 9 | 4 | c0.665G>C | p.R222P | Dahan et al. (4) | 2 |
| G4 | 1 | 4 | c0.674C>T | p.T225M | Dahan et al. (4) | 2 |
| L2 | 8 | 4 | c0.674C>T | p.T225M | Dahan et al. (4) | 2 |
| L6 | 1 | 4 | c0.674C>T | p.T225M | Dahan et al. (4) | 2 |
| L4 | 2 | 4 | c0.707C>T | p.P236L | Kudo et al. (6) | 3 |
| B4 | 1 | 4 | c0.707C>T | p.P236L | Kudo et al. (6) | 3 |
| D3 | 1 | 4 | c0.710C>G | p.S237C | Novel | 1 |
| R1 | 2 | 4 | c0.749A>T | p.H250L | Novel | 2 |
| V1 | 2 | 4 | c0.817G>A | p.V273L | Novel | 3 |
| G2 | 2 | 4 | c0.844T>C | p.C282R | Dahan et al. (4) | 1 |
| P5 | 2 | 4 | c0.855C>A | p.A285E | Novel | 2 |
| S2 | 1 | 5 | c0.891T>G | p.C297W | Schaffer et al. (38) | 1 |
| G5 | 1 | 5 | c0.893G>A | p.C297Y | Novel | 1 |
| F1 | 3 | 5 | c0.944G>A | p.C315Y | Novel | 1 |
| L7 | 1 | 5 | c0.944G>A | p.C315Y | Novel | 1 |
| L8 | 2 | 5 | c0.944G>A | p.C315Y | Novel | 1 |
| C2 | 2 | 8 | c0.1382C>A | p.A461E | Novel | 5 |
| M1 | 1 | 8 | c0.1406C>T | p.T469M | Novel | 5 |
| C1 | 1 | 8 | c0.1462G>A | p.G488R | Williams et al. (21) | 5 |
The exon-mutated nucleotide changes in genomic DNA and their consequences on protein sequence for the mutated allele are provided for each family. Thirteen families were previously reported by Dahan et al. (4). The references are indicated for the 23 mutations previously described by other groups. Types of mutations were classified as follows: 1, cysteine substitution in exon 4 or 5 (n = 17); 2, polar residue substitution in exon 4 or 5 (n = 12); 3, other residue substitution in exon 4 or 5 (n = 3); 4, in-frame deletion (n = 2); and 5, mutation in exon 8 (n = 3).
Using the HSF website (http://www.umd.be/HSF/), we observed the creation of a new donor site at position c0.458 (new splice site: CGGgttcct) responsible, if the cryptic site is used, for a strong variation of the UMOD exon 4 length with the loss of 408 nucleotides. The silent change c0.459C>T may then cause aberrant splicing with the appearance of an in-frame UMOD transcript lacking 135 amino acids (r.spl?).
Figure 1.
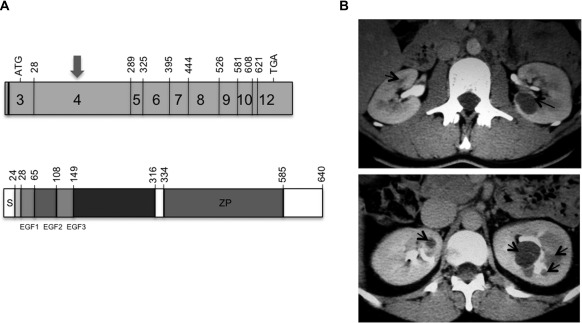
Representation of UMOD gene and uromodulin protein and radiological features associated to UMOD or HNF1B mutation. (A) Schematic representation of UMOD gene and uromodulin protein. The shaded boxes represent the exons of UMOD gene encoding uromodulin (4,5). The number of the first codon of each exon is indicated. The codon that initiates traduction is in exon 3 (nucleotide 106). More than 80% of mutations are in exon 4 (arrow) (upper panel). Uromodulin protein is a 640-amino acid glycoprotein. The N-terminal region (mainly exon 4) contains three calcium-binding EGF domains followed by a highly conserved cysteine-rich sequence of 166 residues. The C-terminal region contains the zona pellucida (ZP) domain and a phosphatidylinositol anchor (lower panel) (4,5). (B) Injected computed tomography (CT) in two patients with UMOD and HNF1B mutation, respectively. CT in one patient with UMOD mutation showed bilateral corticomedullary cysts (5 mm on the right kidney and 20 mm on the left kidney) (arrows), and kidneys were of normal size (left panel). CT in one patient with HNF1B mutation showed normal kidney size with multiple bilateral medullary cysts (arrows), predominantly on the left kidney (right panel).
Clinical Features of Individuals Carrying a UMOD Mutation
Detailed data on clinical features and renal imaging at diagnosis of UMOD mutation were available in 70 patients belonging to 38 families (Table 2). Sixty-two (88.6%) patients had a family history of gout or renal disease compatible with autosomal dominant inheritance.
Table 2.
Clinical features at diagnosis in 70 patients with UMOD mutation
| Characteristic | Statistic |
|---|---|
| Number of patients | 70 |
| Number of families | 38 |
| Family ethnicity (n (%)) | |
| Caucasian | 36 (94.7) |
| north African | 2 (5.3) |
| Age (years) | 31 (21 to 44) |
| Systolic blood pressure (mmHg)a | 144 (130 to 150) |
| Diastolic blood pressure (mmHg)a | 90 (79 to 100) |
| Male gender | 44 (62.9) |
| Familial history of gout and/or renal disease | 62 (88.6) |
| Patients on dialysis | 4 (5.7) |
| Estimated GFR (ml/min per 1.73 m2) | 42 (30 to 60) |
| Uric acid level and renal excretion | |
| under allopurinol therapy | 27 (38.6) |
| uricemia in men (μmol/L)b | 550 (493 to 634) |
| uricemia in women (μmol/L)b | 507 (462 to 535) |
| uricemia >90th percentileb | 21/42 (50) |
| uricemia >75th percentileb | 30/42 (71.4) |
| UAEF (n = 27) (%) | 4.6 (4 to 6) |
| UAEF <10th percentile (n = 27) | 13 (48.1) |
| UAEF <25th percentile (n = 27) | 19 (70.4) |
| Gout | |
| history of gout in men | 33/44 (75) |
| history of gout in women | 13/26 (50) |
| either gout or preemptive allopurinol therapy | 57 (81.4) |
| age at first gout episode (years) | 21 (16 to 31) |
| Renal cysts | |
| presence | 24 (34.3) |
| bilateral | 12 (17.1) |
| unilateral | 12 (17.1) |
| Renal cyst localization | |
| medullary only | 2 (2.8) |
| cortical only | 12 (17.1) |
| corticomedullary | 6 (8.6) |
| undetermined | 4 (5.7) |
| Kidney size <9 cm | 19 (27.1) |
Quantitative parameters are presented as medians (25th to 75th percentiles), and qualitative parameters are presented as numbers (%). UAEF, uric acid excretion fraction; IQR, interquartile ratio.
In 48 patients without antihypertensive therapy.
In 42 patients not under dialysis or allopurinol therapy.
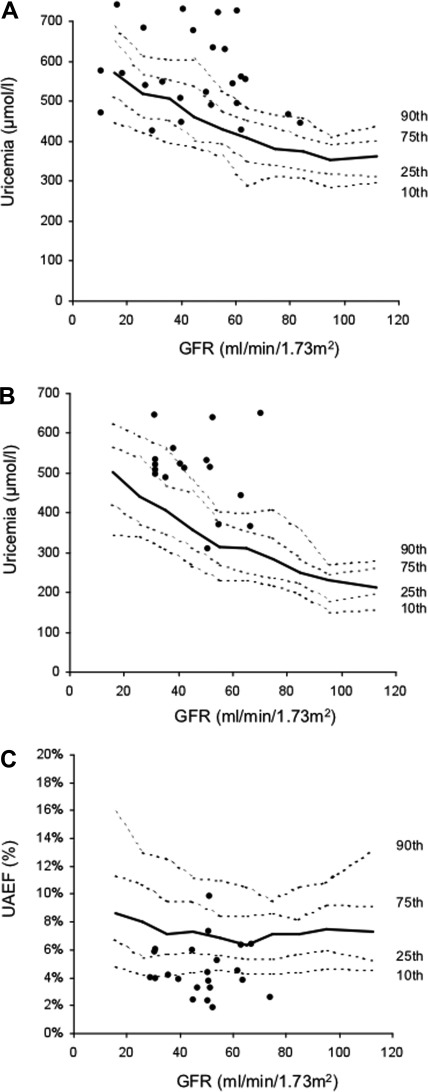
Uricemia and UAEF values were studied in 1097 patients with various forms of CKD to determine usual values and percentiles for a given GFR (Figure 2). Values in patients with UMOD mutation were compared with those obtained from this cohort. In men (n = 25 patients not under allopurinol or dialysis), uricemia was >90th percentile in 48% and >75th percentile in 60%. In women (n = 17), uricemia was >90th percentile value in 53% and >75th percentile value in 88% (Figure 2, A and B). Median UAEF (n = 27) was 4.6% (interquartile ratio, 4 to 6). UAEF was <10th percentile in 48.1% and <75th percentile in 70.4%.
Figure 2.
Serum uric acid levels and uric acid excretion fraction (UAEF) in chronic kidney disease population and in patients with UMOD mutation. Median and 10th, 25th, 75th, and 90th percentile values for uricemia in men (A), uricemia in women (B), and UAEF (C) according to GFR are provided. Circles indicate values observed in patients with UMOD mutation.
A history of gout was present in 75% of men and 50% of women. Age at first episode ranged between 3 and 51 years (median, 21 years; interquartile ratio, 16 to 31). Eleven (15.4%) patients received pre-emptive allopurinol therapy and never developed gout. Renal cysts were detected in 24 (34.3%) patients. Fifteen (62.5%) of them had estimated GFR ≥30ml/min per 1.73 m2. Renal cysts were bilateral in only 12 (17.1%) patients, and localization was cortical or corticomedullary (Figure 1B). Multiple medullary cysts evoking MCKD were observed in only two (2.8%) patients (D59A mutation). Kidney size was <9 cm in 19 patients (27.1%), most of them with significantly decreased renal function. No correlation was observed between the presence of cysts and reduced kidney size. One patient had unilateral renal agenesis, but no urinary tract anomalies were detected in others.
Decline in Renal Function in Patients with UMOD Mutation
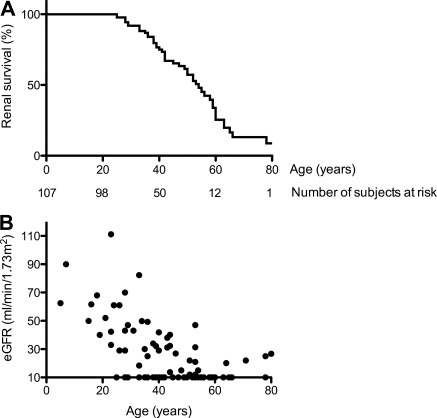
Data on renal survival and evolution of GFR were available in 107 and 93 patients from 44 families, respectively. Forty-six (43%) patients reached ESRD. Renal survival ranged between 25 and more than 70 years (median 54 years) (Figure 3A). The evolution of renal function was highly variable (Figure 3B).
Figure 3.
Renal survival and renal function at last follow-up. (A) Kaplan–Meier curve of renal survival in 107 patients with UMOD mutation. (B) Estimated GFR (eGFR) estimated by MDRD formula was available in 93 patients with UMOD mutation.
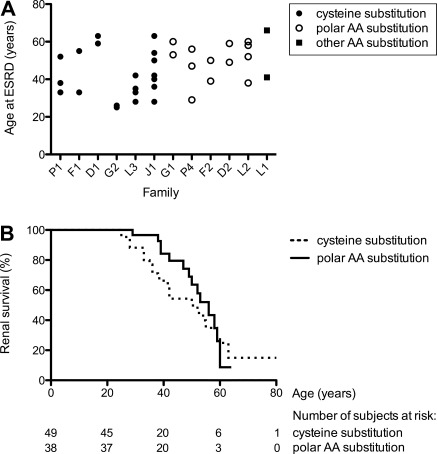
Intrafamilial variability in renal survival was investigated in families with at least two members reaching ESRD. Thirty-five patients belonging to 12 families were included. Important differences in age at onset of ESRD were observed between members belonging to the same family (Figure 4A).
Figure 4.
Intrafamilial variability and renal survival according to the type of mutation. (A) Age at onset of ESRD was analyzed in 12 families with at least two members reaching ESRD. Type of mutation as stated in Table 1 is provided. (B) Renal survival in probands was analyzed according to the type of mutation in UMOD gene. AA, amino acid.
Renal survival was also analyzed according to the mutation types defined above. Only probands were included in this comparison to exclude potential bias related to variability in familial screening that may have lead to diagnosis of UMOD mutation in pauci-symptomatic individuals. A slight trend toward higher renal survival was observed in probands with polar residue substitution (n = 14) compared with those with cysteine substitution (n = 21) (P = 0.41) (Figure 4B). The evolution was similar in four probands with other residue substitution. The disease appeared more severe in two probands with deletion, ESRD occurring at the age of 38 and 45 years, respectively. ESRD occurred in one proband with exon 8 mutation at 63 years of age. We observed no other correlation between phenotype and genotype.
UMOD and HNF1B Gene Screening in Patients with FJHN/MCKD Phenotype
We aimed to provide a picture of “everyday practice” of investigating patients with phenotype suggestive of UMOD mutation. For this purpose, we retrospectively reviewed patients presenting FJHN or MCKD phenotype screened for UMOD mutation at Necker Hospital.
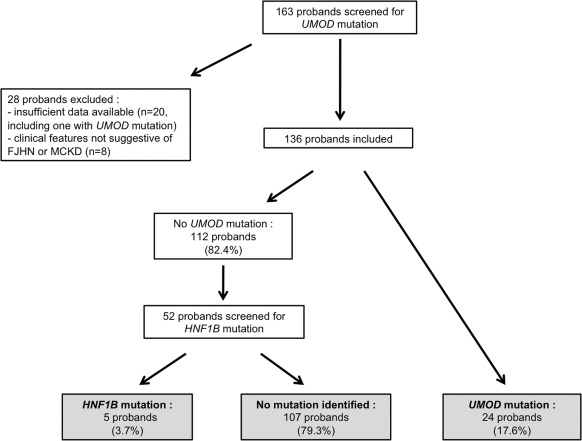
Over the period studied, 164 probands were screened for UMOD mutation. Twenty-eight were excluded from the analysis, because of insufficient clinical data available (n = 20) or clinical features not suggestive of FJHN or MCKD (n = 8), as stated in the Materials and Methods section. A UMOD mutation was detected in 17.6% of the 136 probands included. Fifty-two probands without UMOD mutation (preferentially those with renal cysts) were screened for HNF1B mutation, which was detected in five (Figure 5).
Figure 5.
Screening for UMOD and HNF1B mutation at Necker Hospital in probands with FJHN or MCKD phenotype.
We compared the phenotype of 24 probands with UMOD mutation to one of 107 probands with no mutation identified (Table 3). Median age at screening was 28.5 and 42 years in probands with and without mutation, respectively (P = 0.002). A family history of gout or renal disease compatible with autosomal dominant inheritance was reported by the majority of probands with or without UMOD mutation (P = 0.21).
Table 3.
Comparison of phenotype at diagnosis in probands with UMOD mutation and in those without mutation identified
| Characteristics of Probands | UMOD Mutation | No Mutation | P |
|---|---|---|---|
| Number of probands | 24 | 107 | |
| Age (years) | 28.5 (18.5 to 39.2) | 42 (30 to 53.5) | 0.01 |
| Caucasian ethnicity | 24 (100) | 95 (88.7) | 0.12 |
| Male gender | 20 (83.3) | 65 (60.7) | 0.06 |
| Familial history | |||
| familial history of gout and/or renal disease | 20 (83.3) | 74 (69.1) | 0.21 |
| at least two relatives affected | 14 (58.3) | 41 (38.3) | 0.11 |
| Body mass index (kg/m2)a | 23.9 (21.4 to 24.7) | 23.8 (22.1 to 25.9) | 0.38 |
| Estimated GFR (ml/min per 1.73 m2) | 43 (30 to 54) | 41.7 (27.7 to 53.8) | 0.57 |
| Systolic blood pressure (mmHg)a | 147 (130 to 150) | 140 (126 to 150) | 0.84 |
| Diastolic blood pressure (mmHg)a | 86 (80 to 92) | 85 (80 to 94) | 0.55 |
| Uric acid level and renal excretion | |||
| under allopurinol therapy | 4 (16.7) | 29 (27.1) | 0.43 |
| uricemia in men (μmol/L)b | 570 (504 to 666) | 482 (390 to 595) | 0.02 |
| uricemia in women (μmol/L)b | 520 (491 to 562) | 422 (360 to 488) | 0.24 |
| uricemia >90th percentile | 11/19 (57.8) | 17/74 (23) | 0.01 |
| uricemia >75th percentile | 14/19 (73.6) | 25/74 (33.7) | 0.01 |
| UAEF (%)b | 4.5 (3.3 to 5.8) | 5 (4 to 6.3) | 0.18 |
| UAEF <10th percentile | 7/15 (47.7) | 12/41 (29.3) | 0.34 |
| UAEF <25th percentile | 10/15 (66.7) | 30/41 (73.2) | 0.74 |
| Gout | |||
| history of gout | 16 (66.7) | 55 (51.4) | 0.26 |
| either gout or preemptive allopurinol therapy | 20 (83.3) | 58 (54.2) | 0.01 |
| age at first gout episode (years) | 22 (17.2 to 34) | 28 (20 to 40) | 0.14 |
| Renal cysts | |||
| presence | 7 (29.1) | 59 (54.2) | 0.02 |
| bilateral | 3 (12.5) | 44 (41.1) | 0.01 |
| unilateral | 4 (16.7) | 14 (13.1) | 0.74 |
| Renal cyst localization | |||
| medullary only | 0 | 8 (7.5) | 0.35 |
| cortical only | 6 (25) | 19 (17.8) | 0.4 |
| corticomedullary | 0 | 23 (21.5) | 0.01 |
| undetermined | 1 (4.2) | 8 (7.5) | 1.00 |
| Kidney size <9 cm | 8 (33.3) | 26 (24.3) | 0.44 |
Quantitative parameters are presented as medians (25th to 75th percentiles), and qualitative parameters are presented as numbers (%).
In patients without antihypertensive therapy: n = 22 in the UMOD mutation group, and n = 95 in the no-mutation group.
n = 15 in the UMOD mutation group, and n = 41 in the no-mutation group.
In patients not under allopurinol or on dialysis, n = 19 (14 men and 5 women) in the UMOD mutation group, and n = 74 (37 men and 37 women) in the no-mutation group.
Probands with UMOD mutation were more prone to have uricemia >75th percentile (as defined above) (P = 0.003) or >90th percentile (P = 0.005). Surprisingly, UAEF did not allow discriminating between probands with or without UMOD mutation. The frequency of a history of gout and the age at first episode were similar in both groups. However, 20 (83.3%) probands with UMOD mutation met the combined criteria “either gout or preemptive treatment of hyperuricemia by allopurinol,” that was significantly more frequent than in others (n = 58, 54.2%) (P = 0.01).
The prevalence of bilateral cysts was higher in probands without mutation (41.1% versus 13%, P = 0.009). In most patients, the topography of cysts was rather nonspecific, cortical, or corticomedullary, and kidney size was normal. Nonetheless, there was a large overlap in phenotype between probands with or without UMOD mutation. Phenotype was also analyzed in five probands found with HNF1B mutation. They displayed clinical features close to those associated with UMOD mutations (Table 4) except a high frequency of cysts (4 of 5, 80%) (Figure 1B). Except for the mother of patient F5, who developed diabetes at age 46 years, family history of diabetes was reported in none of these families.
Table 4.
FJHN/MCKD phenotype in five probands with mutations in HNF1B gene
| Family | F5 | D5 | L9 | M2 | L10 |
|---|---|---|---|---|---|
| Age at diagnosis | 20 | 52 | 27 | 31 | 38 |
| Estimated GFR at diagnosis (ml/min per 1.73 m2) | 33 | 40 | 50 | 31 | 38 |
| Diabetes/age | No | No | No | Yes/38 | No |
| Renal imaging | Unilateral, multiple cysts (7 to 15 mm)a | Bilateral, multiple, medullary cystsa | Bilateral, multiple, medullary (<10 mm)a | No cysts, pelvicaliceal dilationa | Bilateral, small kidneys (80 mm) |
| Gout/age | Yes/14 | No | No | Yes/11 | No |
| Uricemia (μmol/L) | 481 | 363 | 312 | 420b | 388 |
| Other involvement | No | No | No | No | No |
| Age at last follow-up | 30 | 54 | 30 | 58 | 41 |
| Estimated GFR at last follow-up (ml/min per 1.73 m2) | 22 | 33 | 29 | 15 | 35 |
| Mutation | G349_M402del | R295C | K71fs108X | G349_M402dupc | A317fs326X |
FJHN, familial juvenile hyperuricemic nephropathy; MCKD, medullary cystic kidney disease.
Renal imaging showed normal kidney size.
Under allopurinol therapy.
Reported by Carette et al. (15).
Discussion
Our study represents the largest cohort of patients with UMOD mutation reported to date. Thirty-seven distinct mutations in UMOD gene were identified in 45 families, of which 19 were new. In accordance with previous reports (4,7,9,19,21), about 80% of mutations were in exon 4. Mutations are likely to cause misfolding and structural destabilization, leading to impaired trafficking of the mutant protein, which probably plays a key role in the development of TIN (7,21,22). Exons 4 and 5 encode a highly conserved cysteine rich sequence and three calcium-binding EGF domains (23). Interestingly, three mutations were identified in exon 8, which encodes a zona pellucida domain (23) (Figure 1A). Mutations in exons encoding the zona pellucida domain were reported in two families: one in exon 6 (24) and one in exon 7 (21). Considering these data, screening only exons 4 to 8 (which represent 70% of coding cDNA) appears to be a reasonable option for everyday practice.
Autosomal dominant inheritance, renal dysfunction secondary to TIN, hyperuricemia, and renal cysts characterize the renal disease caused by UMOD mutation. Incomplete penetrance and neomutation probably explain the absence of family history observed in about 10% of our patients. Data on uricemia and renal function were not available in all parents without known renal disease, and only a few of them could be tested for UMOD mutation. This prevented us from determining the rate of neomutations.
Hyperuricemia was reported in about 80% of patients (4,25). However, it is often difficult to determine whether uric acid level is really out of proportion with GFR. To overcome the issue of uricemia increase secondary to renal dysfunction, we modeled the threshold relationship between GFR and uric acid level. A curve of uric acid level according to GFR was built in a large cohort of patients with various CKD. Serum uric acid level higher than the 75th and 90th percentiles values corresponding to GFR were observed, respectively, in 71.4% and 50% of patients with UMOD mutation and no allopurinol treatment.
Hyperuricemia is typically linked to low UAEF in patients with UMOD mutation and was hypothesized to be related to increased proximal tubule reabsorption secondary to urine salt wasting (1). However, the diagnostic value of UAEF is unclear. Values obtained in our CKD cohort showed that UAEF was little influenced by GFR. The 25th percentile was around 5.5%, and UAEF only slightly increased for GFR values below 25 ml/min per 1.73 m2. Therefore, 5.5% may be considered as “usual lower value” for most of patients with CKD. Our data are discordant with those published several decades ago, showing an important increase of UAEF with progressive GFR reduction below 40 ml/min per 1.73 m2 (26,27). The large number of patients analyzed and the accurate estimate of renal function by 51Cr-EDTA clearance (18) make our results more reliable. As expected, UAEF was abnormally low in the majority of patients with UMOD mutation. However, UAEF was >25th percentile value in 30% of them. This emphasizes that normal UAEF should not exclude diagnosis. In addition, the proportion of patients with abnormally low UAEF was similar in patients with FJHN but no UMOD mutation. Previous studies highlighted that low UAEF was common in patients with gout (28) and may also be observed in healthy subjects (29).
Medullary cysts were thought to be a hallmark of UMOD-related nephropathy, also called type 2 medullary cystic kidney disease. Renal cysts were reported in 30% to 70% of patients with UMOD mutation (4,9,19). However, an accurate description of the cysts was rarely provided, and several reports suggested that typical MCKD was seen in only a minority of patients with cysts (7,9,19). Cysts were detected in 34% of our patients, although radiologic pattern was nonspecific in most of them. Moreover, among probands screened for UMOD mutation, cysts were more frequent in patients without mutation. These results emphasize that cysts are not a hallmark of the disease, although renal imaging was limited to ultrasound in the majority of patients, and a more systematic use of tomodensitometry or magnetic resonance imaging may have provided different results. Considering our data, we propose to screen for UMOD mutation in all patients with TIN associated with uricemia >75th value corresponding to GFR and/or with a family history of gout or renal disease.
The course of TIN secondary to UMOD mutation is highly variable. ESRD often occurs during the fourth or fifth decade (4,9,25,30), although exceptional cases of patients reaching ESRD before 20 years (30) or later than 80 years have been reported (4,31). Because renal survival curves have not been published before, our study adds important insights into the prognosis. Median renal survival was 54 years. We observed a high intrafamilial variability of renal survival and only a small effect of mutation type. This suggests that important environmental factors or modifier genes modulate phenotype.
An UMOD mutation was detected in 17.6% of the 136 probands with the MCKD/FJHN phenotype screened. This is consistent with previous reports, which included about 20 to 25 families and where UMOD mutations were detected in 12% to 31% (9,19,21). Many of the probands with no UMOD mutation were clinically indistinguishable from those carrying UMOD mutations, although our data in the general CKD population showed that uricemia was less frequently out of proportion with GFR in probands without mutation.
A mutation in HNF1B gene was found in five probands, which displayed clinical features similar to those of patients with UMOD mutation, similarly to previously reported cases (14). Only one of them developed diabetes in the years after diagnosis of renal disease. Although renal manifestations secondary to mutations HNF1B encompass a wide clinical spectrum, the absence of cysts observed in one proband is very rare (32,33).
The cause of renal disease in probands with no mutation detected in UMOD or HNF1B genes remains unknown. Clinical presentation (including symptoms in related individuals, renal imaging, and uric acid levels) did not suggest all autosomal dominant polycystic kidney disease in almost all patients. It is possible that some patients had MCKD1 disease, which is responsible for TIN resembling those associated to UMOD mutations (34–36). Mutations in preprorenin signal peptide were recently reported in three families as a new cause of TIN of autosomal dominant inheritance (13). Further work will be required to determine what proportion of unexplained hereditary TIN is related to mutations in these genes. At any rate, it is clear that new genes remain to be discovered.
In summary, we provide a detailed description of phenotype in a large cohort of patients with UMOD mutations. Hyperuricemia represents the hallmark of renal disease. Our data also add novel insights into the interpretation of uricemia and UAEF in patients with renal diseases.
Disclosures
None.
Acknowledgments
We are very grateful for excellent assistance provided from all physicians who sent us clinical data and blood samples of patients, especially Dr. Laura Labriola (Université Catholique de Louvain, Brussels, Belgium), Prof. Christian Jacquot (Hôpital Européen Georges Pompidou University Hospital, Paris, France), Prof. Didier Lacombe (Bordeau University Hospital, Bordeaux, France), Prof. Marie-Noëlle Peraldi (Saint-Louis University Hospital, Paris, France), Dr. Renato Demontis (Creil Hospital, Creil, France), Dr. Florence Vende (Bichat University Hospital, Paris, France), Dr. Jean-Pierre Charmes (Limoges University Hospital, Limoges, France), Dr. Philippe Vanhille (Valenciennes Hospital, Valenciennes, France), Dr. Dominique Chauveau (Toulouse University Hospital, Toulouse, France), Dr. Catherine Bessin (Dieppe Hospital, Dieppe, France), and Dr. Claire Maynard (Chamberry Hospital, Chamberry, France).
This study was supported by funds from the Health Ministry (Programme Hospitalier de Recherche Clinique PO81258), the Association Demain; Fondation Groupama pour la Santé and the Association Demain; Fondation Groupama pour la, the Association pour l'Information et la Recherche sur les Maladies Rénales Génétiques. K.D., Y.P., and O.D. are supported by the Belgian agencies Fonds National de la Recherche Scientifique and Fonds de la Recherche Scientifique Médicale, the Fondation Alphonse & Jean Forton, Concerted Research Action 05/10-328, Interuniversity Attraction Pole IUAP P6/05, and EUNEFRON Program Grant FP7, GA#201590 of the European Community.
G.B. and K.D. contributed equally to this work and should be considered as co-first authors.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1. Scolari F, Caridi G, Rampoldi L, Tardanico R, Izzi C, Pirulli D, Amoroso A, Casari G, Ghiggeri GM: Uromodulin storage diseases: Clinical aspects and mechanisms. Am J Kidney Dis 44: 987–999, 2004 [DOI] [PubMed] [Google Scholar]
- 2. Christodoulou K, Tsingis M, Stavrou C, Eleftheriou A, Papapavlou P, Patsalis PC, Ioannou P, Pierides A, Constantinou Deltas C: Chromosome 1 localization of a gene for autosomal dominant medullary cystic kidney disease. Hum Mol Genet 7: 905–911, 1998 [DOI] [PubMed] [Google Scholar]
- 3. Scolari F, Puzzer D, Amoroso A, Caridi G, Ghiggeri GM, Maiorca R, Aridon P, De Fusco M, Ballabio A, Casari G: Identification of a new locus for medullary cystic disease, on chromosome 16p12. Am J Hum Genet 64: 1655–1660, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dahan K, Devuyst O, Smaers M, Vertommen D, Loute G, Poux JM, Viron B, Jacquot C, Gagnadoux MF, Chauveau D, Buchler M, Cochat P, Cosyns JP, Mougenot B, Rider MH, Antignac C, Verellen-Dumoulin C, Pirson Y: A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol 14: 2883–2893, 2003 [DOI] [PubMed] [Google Scholar]
- 5. Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, Shirts B, Xu L, Zhu H, Barmada MM, Bleyer AJ: Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kudo E, Kamatani N, Tezuka O, Taniguchi A, Yamanaka H, Yabe S, Osabe D, Shinohara S, Nomura K, Segawa M, Miyamoto T, Moritani M, Kunika K, Itakura M: Familial juvenile hyperuricemic nephropathy: Detection of mutations in the uromodulin gene in five Japanese families. Kidney Int 65: 1589–1597, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Rampoldi L, Caridi G, Santon D, Boaretto F, Bernascone I, Lamorte G, Tardanico R, Dagnino M, Colussi G, Scolari F, Ghiggeri GM, Amoroso A, Casari G: Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet 12: 3369–3384, 2003 [DOI] [PubMed] [Google Scholar]
- 8. Turner CL, Emery H, Collins AL, Howarth RJ, Yearwood CM, Cross E, Duncan PJ, Bunyan DJ, Harvey JF, Foulds NC: Detection of 53 FBN1 mutations (41 novel and 12 recurrent) and genotype-phenotype correlations in 113 unrelated probands referred with Marfan syndrome, or a related fibrillinopathy. Am J Med Genet A 149A:161–170, 2009 [DOI] [PubMed] [Google Scholar]
- 9. Vylet'al P, Kublova M, Kalbacova M, Hodanova K, Baresova V, Stiburkova B, Sikora J, Hulkova H, Zivny J, Majewski J, Simmonds A, Fryns JP, Venkat-Raman G, Elleder M, Kmoch S: Alterations of uromodulin biology: A common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int 70: 1155–1169, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Auranen M, Ala-Mello S, Turunen JA, Jarvela I: Further evidence for linkage of autosomal-dominant medullary cystic kidney disease on chromosome 1q21. Kidney Int 60: 1225–1232, 2001 [DOI] [PubMed] [Google Scholar]
- 11. Hodanova K, Majewski J, Kublova M, Vyletal P, Kalbacova M, Stiburkova B, Hulkova H, Chagnon YC, Lanouette CM, Marinaki A, Fryns JP, Venkat-Raman G, Kmoch S: Mapping of a new candidate locus for uromodulin-associated kidney disease (UAKD) to chromosome 1q41. Kidney Int 68: 1472–1482, 2005 [DOI] [PubMed] [Google Scholar]
- 12. Kroiss S, Huck K, Berthold S, Ruschendorf F, Scolari F, Caridi G, Ghiggeri GM, Hildebrandt F, Fuchshuber A: Evidence of further genetic heterogeneity in autosomal dominant medullary cystic kidney disease. Nephrol Dial Transplant 15: 818–821, 2000 [DOI] [PubMed] [Google Scholar]
- 13. Zivna M, Hulkova H, Matignon M, Hodanova K, Vylet'al P, Kalbacova M, Baresova V, Sikora J, Blazkova H, Zivny J, Ivanek R, Stranecky V, Sovova J, Claes K, Lerut E, Fryns JP, Hart PS, Hart TC, Adams JN, Pawtowski A, Clemessy M, Gasc JM, Gubler MC, Antignac C, Elleder M, Kapp K, Grimbert P, Bleyer AJ, Kmoch S: Dominant renin gene mutations associated with early-onset hyperuricemia, anemia, and chronic kidney failure. Am J Hum Genet 85: 204–213, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bingham C, Ellard S, van't Hoff WG, Simmonds HA, Marinaki AM, Badman MK, Winocour PH, Stride A, Lockwood CR, Nicholls AJ, Owen KR, Spyer G, Pearson ER, Hattersley AT: Atypical familial juvenile hyperuricemic nephropathy associated with a hepatocyte nuclear factor-1beta gene mutation. Kidney Int 63: 1645–1651, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Carette C, Vaury C, Barthelemy A, Clauin S, Grunfeld JP, Timsit J, Bellanne-Chantelot C: Exonic duplication of the hepatocyte nuclear factor-1beta gene (transcription factor 2, hepatic) as a cause of maturity onset diabetes of the young type 5. J Clin Endocrinol Metab 92: 2844–2847, 2007 [DOI] [PubMed] [Google Scholar]
- 16. Brosius FC, 3rd, Hostetter TH, Kelepouris E, Mitsnefes MM, Moe SM, Moore MA, Pennathur S, Smith GL, Wilson PW: Detection of chronic kidney disease in patients with or at increased risk of cardiovascular disease: A science advisory from the American Heart Association Kidney and Cardiovascular Disease Council; the Councils on High Blood Pressure Research, Cardiovascular Disease in the Young, and Epidemiology and Prevention; and the Quality of Care and Outcomes Research Interdisciplinary Working Group: developed in collaboration with the National Kidney Foundation. Circulation 114: 1083–1087, 2006 [DOI] [PubMed] [Google Scholar]
- 17. Schwartz GJ, Haycock GB, Edelmann CM, Jr, Spitzer A: A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics 58: 259–263, 1976 [PubMed] [Google Scholar]
- 18. Froissart M, Rossert J, Jacquot C, Paillard M, Houillier P: Predictive performance of the modification of diet in renal disease and Cockcroft-Gault equations for estimating renal function. J Am Soc Nephrol 16: 763–773, 2005 [DOI] [PubMed] [Google Scholar]
- 19. Wolf MT, Mucha BE, Attanasio M, Zalewski I, Karle SM, Neumann HP, Rahman N, Bader B, Baldamus CA, Otto E, Witzgall R, Fuchshuber A, Hildebrandt F: Mutations of the uromodulin gene in MCKD type 2 patients cluster in exon 4, which encodes three EGF-like domains. Kidney Int 64: 1580–1587, 2003 [DOI] [PubMed] [Google Scholar]
- 20. Heidet L, Decramer S, Pawtowski A, Moriniere V, Bandin F, Knebelmann B, Lebre AS, Faguer S, Guigonis V, Antignac C, Salomon R: Spectrum of HNF1B mutations in a large cohort of patients who harbor renal diseases. Clin J Am Soc Nephrol 5: 1079–1090, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams SE, Reed AA, Galvanovskis J, Antignac C, Goodship T, Karet FE, Kotanko P, Lhotta K, Moriniere V, Williams P, Wong W, Rorsman P, Thakker RV: Uromodulin mutations causing familial juvenile hyperuricaemic nephropathy lead to protein maturation defects and retention in the endoplasmic reticulum. Hum Mol Genet 18: 2963–2974, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jennings P, Aydin S, Kotanko P, Lechner J, Lhotta K, Williams S, Thakker RV, Pfaller W: Membrane targeting and secretion of mutant uromodulin in familial juvenile hyperuricemic nephropathy. J Am Soc Nephrol 18: 264–273, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Serafini-Cessi F, Malagolini N, Cavallone D: Tamm-Horsfall glycoprotein: Biology and clinical relevance. Am J Kidney Dis 42: 658–676, 2003 [DOI] [PubMed] [Google Scholar]
- 24. Tinschert S, Ruf N, Bernascone I, Sacherer K, Lamorte G, Neumayer HH, Nurnberg P, Luft FC, Rampoldi L: Functional consequences of a novel uromodulin mutation in a family with familial juvenile hyperuricaemic nephropathy. Nephrol Dial Transplant 19: 3150–3154, 2004 [DOI] [PubMed] [Google Scholar]
- 25. Bleyer AJ, Woodard AS, Shihabi Z, Sandhu J, Zhu H, Satko SG, Weller N, Deterding E, McBride D, Gorry MC, Xu L, Ganier D, Hart TC: Clinical characterization of a family with a mutation in the uromodulin (Tamm-Horsfall glycoprotein) gene. Kidney Int 64: 36–42, 2003 [DOI] [PubMed] [Google Scholar]
- 26. Calabrese G, Simmonds HA, Cameron JS, Davies PM: Precocious familial gout with reduced fractional urate clearance and normal purine enzymes. Q J Med 75: 441–450, 1990 [PubMed] [Google Scholar]
- 27. Danovitch GM, Weinberger J, Berlyne GM: Uric acid in advanced renal failure. Clin Sci 43: 331–341, 1972 [DOI] [PubMed] [Google Scholar]
- 28. Perez-Ruiz F, Calabozo M, Erauskin GG, Ruibal A, Herrero-Beites AM: Renal underexcretion of uric acid is present in patients with apparent high urinary uric acid output. Arthritis Rheum 47: 610–613, 2002 [DOI] [PubMed] [Google Scholar]
- 29. Stiburkova B, Pospisilova E, Kmoch S, Sebesta I: Analysis of excretion fraction of uric acid. Nucleosides Nucleotides Nucleic Acids 25: 1301–1304, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Wolf MT, Beck BB, Zaucke F, Kunze A, Misselwitz J, Ruley J, Ronda T, Fischer A, Eifinger F, Licht C, Otto E, Hoppe B, Hildebrandt F: The uromodulin C744G mutation causes MCKD2 and FJHN in children and adults and may be due to a possible founder effect. Kidney Int 71: 574–581, 2007 [DOI] [PubMed] [Google Scholar]
- 31. Yokota N, Yamanaka H, Yamamoto Y, Fujimoto S, Eto T, Tanaka K: Autosomal dominant transmission of gouty arthritis with renal disease in a large Japanese family. Ann Rheum Dis 50: 108–111, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bellanne-Chantelot C, Chauveau D, Gautier JF, Dubois-Laforgue D, Clauin S, Beaufils S, Wilhelm JM, Boitard C, Noel LH, Velho G, Timsit J: Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann Intern Med 140: 510–517, 2004 [DOI] [PubMed] [Google Scholar]
- 33. Ulinski T, Lescure S, Beaufils S, Guigonis V, Decramer S, Morin D, Clauin S, Deschenes G, Bouissou F, Bensman A, Bellanne-Chantelot C: Renal phenotypes related to hepatocyte nuclear factor-1beta (TCF2) mutations in a pediatric cohort. J Am Soc Nephrol 17: 497–503, 2006 [DOI] [PubMed] [Google Scholar]
- 34. Cohn DH, Shohat T, Yahav M, Ilan T, Rechavi G, King L, Shohat M: A locus for an autosomal dominant form of progressive renal failure and hypertension at chromosome 1q21. Am J Hum Genet 67: 647–651, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kiser RL, Wolf MT, Martin JL, Zalewski I, Attanasio M, Hildebrandt F, Klemmer P: Medullary cystic kidney disease type 1 in a large Native-American kindred. Am J Kidney Dis 44: 611–617, 2004 [PubMed] [Google Scholar]
- 36. Stavrou C, Koptides M, Tombazos C, Psara E, Patsias C, Zouvani I, Kyriacou K, Hildebrandt F, Christofides T, Pierides A, Deltas CC: Autosomal-dominant medullary cystic kidney disease type 1: Clinical and molecular findings in six large Cypriot families. Kidney Int 62: 1385–1394, 2002 [DOI] [PubMed] [Google Scholar]
- 37. Lhotta K, Gehringer A, Jennings P, Kronenberg F, Brezinka C, Andersone I, Strazdins V: Familial juvenile hyperuricemic nephropathy: Report on a new mutation and a pregnancy. Clin Nephrol 71: 80–83, 2009 [DOI] [PubMed] [Google Scholar]
- 38. Schäffer P, Gombos E, Meichelbeck K, Kiss A, Hart PS, Bleyer AJ: Childhood course of renal insufficiency in a family with a uromodulin gene mutation. Pediatr Nephrol 25: 1355–1360, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]