Abstract
Thyroid hormones regulate brain development and function through the control of gene expression, mediated by binding of T3 to nuclear receptors. Brain T3 concentration is tightly controlled by homeostatic mechanisms regulating transport and metabolism of T4 and T3. We have examined the role of the inactivating enzyme type 3 deiodinase (D3) in the regulation of 43 thyroid hormone-dependent genes in the cerebral cortex of 30-d-old mice. D3 inactivation increased slightly the expression of two of 22 positively regulated genes and significantly decreased the expression of seven of 21 negatively regulated genes. Administration of high doses of T3 led to significant changes in the expression of 12 positive genes and three negative genes in wild-type mice. The response to T3 treatment was enhanced in D3-deficient mice, both in the number of genes and in the amplitude of the response, demonstrating the role of D3 in modulating T3 action. Comparison of the effects on gene expression observed in D3 deficiency with those in hypothyroidism, hyperthyroidism, and type 2 deiodinase (D2) deficiency revealed that the negative genes are more sensitive to D2 and D3 deficiencies than the positive genes. This observation indicates that, in normal physiological conditions, D2 and D3 play critical roles in maintaining local T3 concentrations within a very narrow range. It also suggests that negatively and positively regulated genes do not have the same physiological significance or that their regulation by thyroid hormone obeys different paradigms at the molecular or cellular levels.
Thyroid hormones are important for brain development and function (1–3). Most of their effects in the brain and other tissues are mediated by regulating gene expression at the transcriptional and post transcriptional levels (4). Thyroid hormone regulation of gene expression in the brain is extremely complex and depends on age and the particular brain region and cell type (5). The active thyroid hormone is T3, which binds to specific nuclear receptors to either repress or activate gene transcription (6). T3 is formed in the thyroid gland but also in peripheral tissues from the precursor T4 by the action of the 5′-deiodinase type 1 and 5′-deiodinase type 2 (D2) (7). In the brain and other tissues, such as brown adipose tissue and pituitary, local generation of T3 by D2 is an important step in thyroid hormone-dependent biological effects (8–11). D2 is encoded by the Dio2 gene, and its activity is regulated by T4 through a nongenomic effect on protein degradation (12). This posttranslational effect is the main control of D2 activity and does not imply changes in the mRNA concentration. However, pretranslational effects involving changes in the concentration of Dio2 mRNA also occur in response to hypothyroidism and T3 treatment (13). The Dio2 gene is expressed in astrocytes and in the specialized glial cells called tanycytes lining the inferior walls of the third ventricle (14, 15). Cellular targets of T3 also include neurons and oligodendrocytes, which express T3 receptors and, in normal physiological conditions, lack D2 activity (16).
The T3 reaching the target cells in the central nervous system has two origins. One is the circulation, from which T4 and T3 cross the blood-brain barrier (BBB) through specific transporters (17). In the astrocytes, T3 is also generated from T4 by the action of D2. It is thought that the Slco1c1 transporter (18), expressed in the membrane of endothelial capillary cells, facilitates the passage of T4 through the BBB (19). Slco1c1 is also expressed in the astrocytic end feet, which are in close contact with the surface of the capillary endothelium (19). Therefore, it is likely that T4 entering the brain from the circulation through this route finds its way directly to the astrocytes, in which T3 is formed. Thus, it is presumed that no significant diffusion of T4 to the interstitial fluid does occur. On the other hand, T4 and T3 also cross the BBB through a very important transporter, the monocarboxylate 8 (Mct8, Slc16a2) transporter (20). Mct8 is specific for T4 and T3 and is not present in the astrocytic end feet (19). Therefore, it is likely that T4 and T3 crossing the BBB through Mct8 are delivered to the interstitial fluid from which they can reach the target cells (21).
Another determinant of brain thyroid hormone action is type 3 deiodinase (D3), which is encoded by the imprinted gene Dio3 (7, 22). D3 catalyzes the formation of the inactive metabolites rT3 from T4 and T2 from T3 (7, 22). It is believed that D3 has an important role in limiting the amount of T3 reaching target tissues and preventing excessive exposure to the hormone (9). According to this view, the high D3 activities present in the uterus during implantation and in the placenta (23) would limit the amounts of thyroid hormone reaching the conceptus. In the brain Dio3 is expressed in neurons (24, 25), and during perinatal development, neuronal D3 would tightly control the amount of T3 accumulating in the brain. We have recently proposed that one role of D3 could be to restrict the availability of T3 from the circulation so that most T3 acting in the brain during the fetal, and perhaps early postnatal periods, would be derived from T4 in the astrocytes (21). Consistent with the proposed role of D3, Dio3 knockout (KO) mouse neonates manifest elevated T3 concentration in the brain and increased expression of the T3 target genes Hr and Nrgn (26).
Previous findings using a mouse model of D2 deficiency suggest that T3-regulated genes expressed in the cerebral cortex exhibit different sensitivity in their response to T3, depending on whether they are positively or negatively regulated genes (27). In the present studies, we have analyzed the expression of a large set of those genes in response to hyperthyroidism and D3 deficiency. Our results further support the notion of differential T3 sensitivity between those groups of genes and reveal an unsuspected complexity in the mechanisms by which thyroid hormone metabolism at the local level finely regulates gene expression in the central nervous system.
Materials and Methods
In vivo studies
Protocols for animal handling were approved by the local institutional Animal Care Committee, following the rules of the European Union and the National Institutes of Health. Animals were housed in temperature- (22 ± 2 C) and light (12-h light, 12-h dark cycle; lights on at 0700 h)-controlled conditions and had free access to food and water. Chemical hypothyroidism was induced in wild-type (WT) mice of the C57/BL/6J strain by administering 0.02% 1-methyl-2-mercapto-imidazol (Sigma Chemical Co., St. Louis, MO) plus 1% KClO4 in the drinking water ad libitum. These antithyroid drugs were given to pregnant and lactating dams from gestational d 17, until postnatal (P) d 21. Dio3KO mice were in a mixed C57/BL/6J and 129/Sv background as described (26). Experiments involving comparisons between WT and Dio3KO mice, as well as T3 treatments, were performed using WT and Dio3KO littermates obtained from heterozygous parents. Hyperthyroidism was induced by administration of 0.5 μg/ml of T3 and 0.1 μg/ml of T4 in the drinking water containing 0.1% BSA from P21 to P30. The corresponding doses were 1.0 μg T3 and 0.2 μg T4 per mice per day. Although T3 was in excess, the T4 dose was physiological. The rationale of this treatment was to maintain physiological circulating concentrations of T4, as the D2 substrate, in the face of T3-induced hyperthyroidism. Handling of the Dio2KO mice was as previously described (27).
The pups were killed by decapitation on P21, to analyze the effects of hypothyroidism and Dio2 deletion, or on P30 to analyze the effects of hyperthyroidism and Dio3 inactivation. The whole neocortex was rapidly dissected out from underlying structures, cut in two halves through the sagittal plane, frozen on dry ice, and kept at −80 C. RNA was isolated from individual hemicortices. Procedures for RNA isolation and quantitative PCR (qPCR) on TaqMan arrays (Applied Biosystems, Foster City, CA), were as previously described (27). Data were expressed relative to the values obtained for the control WT, which was given a value of 1.0 after correction for 18S RNA and Ppia mRNA. Thyroid hormone concentration in serum was determined as previously described (26).
Cell culture and transfection
Mouse neuroblastoma (N2a CA3 clone) cell lines expressing the thyroid hormone receptor-α1 isoform (TRα1) were cultured as described (28). The cells were seeded in each of six-well plates with DMEM containing 10% thyroid hormone-deprived fetal calf serum (29) up to 80% confluence. The cells were transfected with 0.5 μg of a Dio3-expression clone in pcDNA (Invitrogen, Carlsbad, CA) using Lipofectamine 2000 as described by the manufacturer (Invitrogen). Control cells were transfected with empty pcDNA plasmid. Transfection efficiency was 18%. Twenty-four hours after transfection, T3 was added at final concentrations of 0.1 and 0.5 nm in triplicate, and the cells were incubated for a further 24 h before harvesting and RNA isolation. The response to T3 was analyzed by qPCR using as target the Hr gene.
D2 and D3 activities
D2 and D3 enzymatic activities were determined as previously described (30, 31). In brief, tissues were homogenized in a 10 mm Tris-HCl, 0.25 sucrose (pH 7.4) buffer. A suitable amount of tissue homogenate was used in the enzymatic reaction to ensure that deiodination did not exceed 20% and was proportional to the amount of protein content. Tissue homogenates were incubated at 37 C for an hour with the appropriate [125I]-labeled substrate (PerkinElmer, Waltham, MA) in the presence of 25 nm dithiothreitol. For D2 and D3 activity, homogenates were incubated, respectively, with 1 nm T4 or 2 nm T3. Deiodination was determined based on the percentage of labeled iodine released (D2) or the amount of labeled 3,3′-diiodothyronine produced. The latter was determined after separation of reaction products by paper chromatography, as described (32). A factor of 2 was included in the calculation of D2 activity to correct for the chemical equivalence of the outer ring iodine residues and the fact that only one of them is labeled in a given molecule. Protein content in the homogenates was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA).
Statistical analysis
Differences between means were obtained by one- or two-way ANOVA, depending on the experiment, and the Tukey or Bonferroni post hoc tests, respectively. Calculations were done using the GraphPad Prism software (http://www.graphpad.com/prism/). Significance was illustrated graphically by asterisks: ***, P < 0.001; **, P < 0.01, *, P < 0.05.
Results
Selection of genes sensitive to hypothyroidism in the mouse cerebral cortex
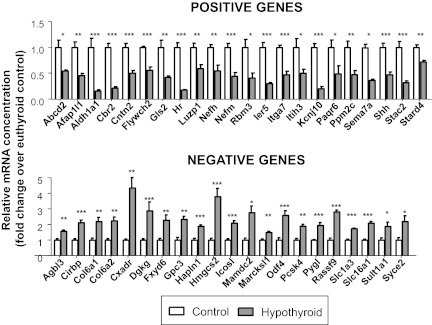
From previous microarray gene expression data from the cerebral cortex of P21 control and hypothyroid mice (27), we selected 43 genes sensitive to thyroid hormone deprivation. Hypothyroidism decreased the expression of 22 of them and increased the expression of the other 21. We refer to these genes as positive or negative genes, respectively, indicating that their expression is likely to be positively or negatively regulated by thyroid hormone. The selection of the gene targets among the annotated probes, was performed taking into account the following two parameters: 1) the expression levels in the microarrays (the A mean value above 6; the A mean value is the log2 intensity values over all samples as a measure of the average expression level) and 2) the response to hypothyroidism (the M value of at least 1.0; M is the log2-fold change between the two conditions). The effects of hypothyroidism on this set of genes was confirmed by TaqMan PCR arrays on microfluidic cards using biological replicates of cerebral cortex RNA from P21 mice (Fig. 1). From the 43-gene set, hypothyroidism decreased the expression of the 22 genes selected as positively regulated by thyroid hormone and increased the expression of the 21 genes selected as negatively regulated by thyroid hormone. The most common effect of hypothyroidism was a 50% decreased expression on the positive genes or a 2-fold increase on the negative genes. Only a few changes in gene expression were 4-fold or higher.
Fig. 1.
Effect of hypothyroidism on gene expression in the cerebral cortex of P21 wild-type mice as measured using TaqMan arrays. Results are mean ± se of six animals for each gene. Statistical analysis was by Student's t test. The set contains genes already studied (27) and additional genes not previously analyzed (Col6a2, Cxadr, Flywch2, Fxyd6, Gls2, Gpc3, Hmgcs2, Kcnj10, Pygl, Rassf9, and Shh). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Effects of Dio3 inactivation on circulating thyroid hormone concentrations before and after treatment with thyroid hormones
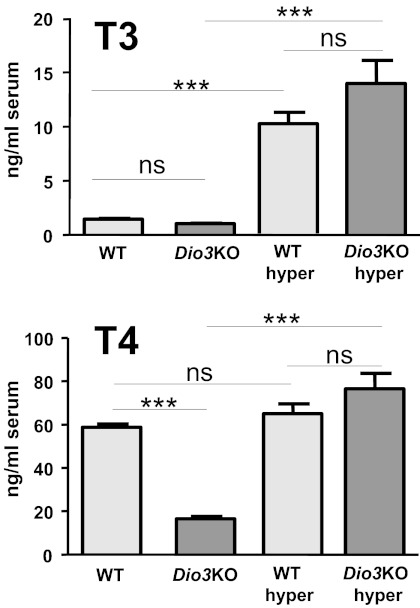
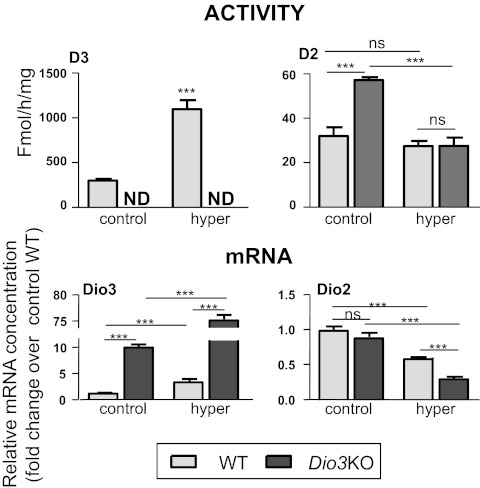
The effects of hyperthyroidism on WT and Dio3KO mice were analyzed by administering high daily doses of T3 in the drinking water from P21 to P30. Serum T3 and T4 concentrations in the untreated and treated animals are shown in Fig. 2. The mean serum concentrations of T3 in the Dio3KO mice (1.03 ± 5.1 ng/ml) were lower than in the WT (1.44 ± 0.07 ng/ml), as previously reported (33). Although the difference was significant (P < 0.001) using the Student's t test, it was not significant using two-way ANOVA in the four-group comparison, which also includes the T3-treated groups. T3 treatment increased serum T3 similarly in WT (10.19 ± 1.07 ng/ml) and Dio3KO (13.96 ± 2.13) mice. Because T3 treatment was expected to block thyroid secretion, the T3-treated mice were also given T4 in physiological amounts to maintain normal circulating levels of T4. T4 concentration in untreated Dio3KO mice (16.5 ± 1.2 ng/ml) was significantly reduced compared with the untreated WT mice (58.7 ± 1.5 ng/ml) as found in previous work (26, 33). In the treated WT and Dio3KO mice, the T4 concentrations were similar to the untreated WT (65.0 ± 4.5 and 76.5 ± 7.1 ng/ml, respectively).
Fig. 2.
Plasma T3 and T4 concentrations in P30 wild-type (WT) and Dio3KO untreated mice and in T4+T3-treated WT and Dio3KO mice (WT hyper and Dio3KO hyper). Results are mean ± se of eight animals in each column. Statistical analysis was two-way ANOVA, factors being genotype and treatment. ***, P < 0.001.
Effects of Dio3 inactivation on cerebrocortical gene expression before and after treatment with thyroid hormones
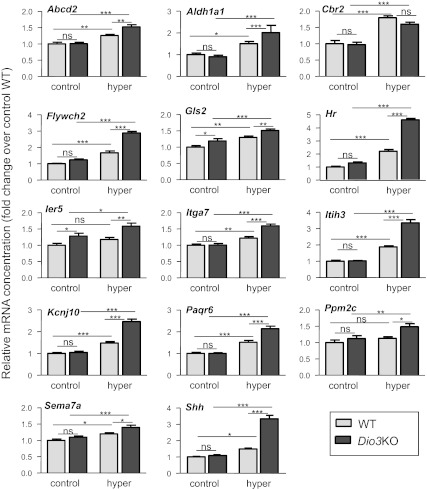
Gene expression levels in the cerebral cortex of untreated and treated animals are shown in Figs. 3 (positive genes) and 4 (negative genes). From the 22 positive genes, 14 showed a difference in at least one of the experimental groups using two reference control RNA, Ppia (cyclophilin) mRNA and 18S RNA (Fig. 3). The other eight positive genes showed no significant changes in any of the experimental situations and are not shown in the figure. D3 inactivation did not affect the basal expression of most genes except for a small increase in two of them, Gls2 and Ier5. Thyroid hormone administration to the WT mice increased the expression of 12 genes, with responses that varied between 1.2-fold (Itga7 and Sema7a) and 2.2-fold (Hr). Two genes (Ppm2c and Ier5) were unresponsive. D3 inactivation enhanced the T3 response for all genes except for Cbr2. The enhancement of the T3 effect was notable on genes such as Aldh1a1, Flywch2, Hr, Itih3, Kcnj10, Paqr6, and Shh, with increments of 2- to 4-fold the basal expression. Taking into account all genes, the effects of T3 treatment on the WT and the Dio3KO were correlated (r = 0.689, P = 0.0003). This indicates that, although D3 inactivation did not significantly influence the basal expression of positive genes, it enhanced the effect of T3 treatment.
Fig. 3.
Effects of D3 inactivation and T4+T3 treatment on positive gene expression in the P30 mouse cerebral cortex. Only the genes showing changes under at least one of the experimental situations are shown. Results are mean ± se of seven animals for each condition. Statistical analysis was by two-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
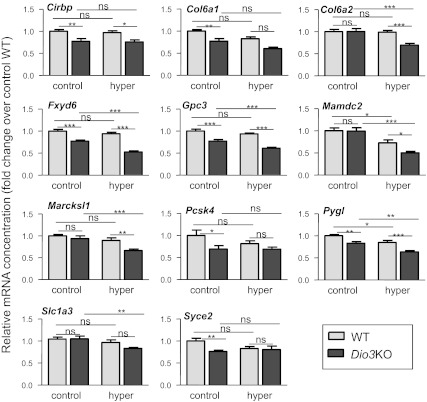
Fig. 4.
Effects of D3 inactivation and T4+T3 treatment on negative gene expression in the P30 mouse cerebral cortex. Only the genes showing changes under at least one of the experimental situations are shown. Results are mean ± se of seven animals for each condition. Statistical analysis was by two-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Of the 21 negative genes analyzed, 10 were insensitive to D3 inactivation and to T3 treatment, and the other 11 showed a significant change in at least one of the experimental situations and are represented in Fig. 4. With the exception of Mamdc2 and Pygl, no effect of hyperthyroidism was observed in the WT mice. However, D3 inactivation significantly reduced the expression of seven genes (Cirbp, Col6a1, Gpc3, Fxyd6, Pcsk4, Pygl, Syce2). Treatment of Dio3KO mice with T3 resulted in further reductions in the expression of seven genes (Col6a2, Fxyd6, Gpc3, Mamdc2, Marcksl1, Pygl, Slc1a3).
We also measured the relative concentrations of Dio2 and Dio3 mRNA as well as D2 and D3 activities in the same groups of animals (Fig. 5). Dio3 is a positively regulated gene, and in WT mice T3 treatment increased Dio3 mRNA by 2.7-fold. D3 activity was also increased by 3.6-fold in the hyperthyroid WT group. Although no D3 activity was present in the Dio3KO mice, Dio3 mRNA could be measured in these mice because inactivation of D3 enzymatic activity was achieved by mutating the codon corresponding to the active site of the D3 protein (34). Therefore, a minimally modified Dio3 mRNA is still expressed in Dio3KO animals. At odds with the response of other positive genes, the effect of D3 inactivation on Dio3 mRNA was stronger (10-fold increase) than the effect of T3 treatment on the WT mice. Furthermore, T3 treatment increased Dio3 mRNA by 60-fold in the Dio3KO mice.
Fig. 5.
Effects of D3 inactivation and T4+T3 treatment on Dio3 and Dio2 expression (D3 and D2 activities, upper panels, and Dio3 and Dio2 mRNA, lower panels) in the cerebral cortex. Results are mean ± se of seven animals for each condition. Statistical analysis was by two-way ANOVA, except for D3 activity in which the t test was used. ND, Not determined; ns, not significant. ***, P < 0.001.
Dio2 mRNA concentration was not altered by D3 inactivation, but was decreased by a factor of 1.7 by T3 treatment in the WT and by 3.3 in the Dio3KO mice. In contrast, D2 activity was increased 1.8-fold in the untreated Dio3KO mice and was unchanged in the hyperthyroid groups. These changes reflect the fact that D2 activity is regulated by T4 concentrations, whereas Dio2 mRNA is sensitive to T3 treatment.
Dio3 expression attenuates T3 action in cultured neuroblastoma cells
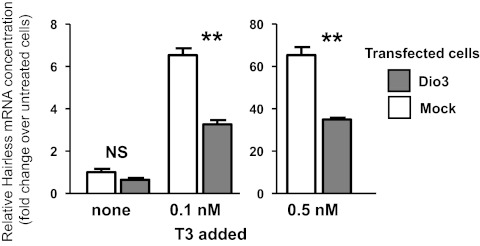
To examine whether cellular changes in D3 activity indeed modified genomic responses to T3, we used a mouse neuroblastoma cell line, N2a, stably expressing the TRα1isoform. These cells do not express the endogenous Dio3 gene (not shown). The cells were transfected with a Dio3 expression vector. Then T3 was added at concentrations of 0.1 and 0.5 nm in the presence of 10% fetal calf serum, and the expression of the endogenous Hr gene was measured by qPCR in RNA isolated from the whole culture (Fig. 6). In the absence of D3, Hr was very sensitive to T3 in these cells. T3 increased Hr expression almost 7-fold at a concentration of 0.1 nm and 65-fold at 0.5 nm. Dio3 expression after transfection interfered with T3 action and decreased T3 activity by about 50% at the two concentrations of T3 tested.
Fig. 6.
Effect of Dio3 expression on the induction of the Hr gene by T3 in a TRα-expressing mouse neuroblastoma N2a cell line. An N2a cell line stably expressing TRα1, but not Dio3, was transfected with an expression plasmid encoding mouse Dio3. T3 was added to the cultures and Hr transcripts were measured by qPCR. The response to T3 was higher in cells not expressing Dio3. **, P < 0.01.
A summary of all gene expression data are shown in Tables 1 and 2. The tables list the gene symbols and names and the effects of the experimental manipulations described. For comparison we also included in the table the effects of Dio2 inactivation on genes previously reported (27) and on additional genes (Col6a2, Cxadr, Flywch2, Fxyd6, Gls2, Gpc3, Hmgcs2, Kcnj10, Pygl, Rassf9, and Shh) not previously analyzed. In addition, we included data on cellular expression of genes enriched at least 3-fold in astrocytes or neurons. These data were calculated from the expression values given by Cahoy et al. (35) for astrocytes and neurons on P16. Nine genes were enriched in astrocytes and 16 in neurons. Given that all genes were sensitive to thyroid hormone deprivation, positive and negative genes differed in their responses to hyperthyroidism and D3 inactivation. More positive than negative genes were sensitive to hyperthyroidism, whereas more negative than positive genes were sensitive to D3 inactivation.
Table 1.
Summary of the effects of hypo- and hyperthyroidism and D3 or D2 deficiencies on the expression of cerebral cortex genes
| Positive genes |
Fold enrichment in cell type |
Fold change |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene symbol | Gene name | Astro | Neuron | Hypo | Hyper | Dio3 KO | Dio3 KO+T3 | Dio2 KO |
| Abcd2 | ATP-binding cassete, subfamily D, member 2 | 18 | −1.9 | 1.3 | nc | 1.5 | nc | |
| Afap1l1 | Actin filament-associated protein 1 | −2.4 | nc | nc | nc | nc | ||
| Aldh1a1 | Aldehyde dehydrogenase family 1, subfamily A1 | 13 | −6.3 | 1.6 | nc | 2.0 | nc | |
| Cbr2 | Carbonyl reductase 2 | −4.8 | 1.8 | nc | 1.5 | nc | ||
| Cntn2 | Contactin 2 (TAG-1) | 5 | −1.9 | nc | nc | nc | nc | |
| Flywch2 | FLYWCH family member 2 | 3 | −1.7 | 1.5 | nc | 2.8 | nc | |
| Gls2 | Glutaminase 2 | 14 | −2.4 | 1.3 | 1.2 | 1.5 | nc | |
| Hr | Hairless | −6.3 | 2.2 | nc | 4.7 | nc | ||
| Luzp1 | Leucine zipper protein 1 | −1.7 | nc | nc | nc | nc | ||
| Nefh | Neurofilament, heavy polypeptide | 37 | −1.9 | nc | nc | nc | nc | |
| Nefm | Neurofilament, medium polypeptide | 83 | −2.4 | nc | nc | nc | nc | |
| Rbm3 | RNA binding motif protein 3 | −2.4 | nc | nc | nc | −2.0 | ||
| Ier5 | Immediate early response 5 | −3.2 | nc | 1.2 | 1.6 | nc | ||
| Itga7 | Integrin α7 | 3 | −2.1 | 1.2 | nc | 1.6 | nc | |
| Itih3 | Inter-α trypsin inhibitor, heavy chain 3 | −2.1 | 2.0 | nc | 3.5 | nc | ||
| Kcnj10 | Potassium inwardly rectifying channel, subfamily J, member 10 | 3 | −4.8 | 1.5 | nc | 2.6 | −1.6 | |
| Paqr6 | Progestin and adipoQ receptor family member VI | 6 | −2.1 | 1.5 | nc | 2.1 | nc | |
| Ppm2c | Pyruvate dehyrogenase phosphatase catalytic subunit 1 | 4 | −2.1 | nc | nc | 1.5 | nc | |
| Sema7a | Semaphorin 7A | 5 | −2.7 | 1.2 | nc | 1.4 | nc | |
| Shh | Sonic hedgehog | 5 | −2.1 | 1.5 | nc | 3.5 | −1.6 | |
| Stac2 | SH3 and cysteine rich domain 2 | −3.2 | nc | nc | nc | nc | ||
| Stard4 | StAR-related lipid transfer (START) domain containing 4 | −2.1 | nc | nc | nc | nc | ||
This is a summary of changes in expression of positive genes as measured by qPCR. Genes of enriched expression more than 3-fold in astrocytes or neurons are indicated. Values for enrichment were obtained from the published database by Cahoy et al. (35). nc, No change.
Table 2.
Summary of the effects of hypo- and hyperthyroidism and D3 or D2 deficiencies on the expression of cerebral cortex genes
| Negative genes |
Fold enrichment in cell type |
Fold change |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene symbol | Gene name | Astro | Neuron | Hypo | Hyper | Dio3 KO | Dio3 KO+T3 | Dio2 KO |
| Agbl3 | ATP/GTP binding protein-like 3 | 1.6 | nc | nc | nc | 2.8 | ||
| Cirbp | Cold inducible RNA binding protein | 2.2 | nc | −1.3 | −1.3 | 2.3 | ||
| Col6a1 | Collagen, type VI, α1 | 32 | 2.4 | nc | −1.3 | −1.3 | 1.9 | |
| Col6a2 | Collagen, type VI, α2 | 2.4 | nc | nc | −1.3 | 2.4 | ||
| Cxadr | Coxsackie virus and adenovirus receptor (CAR) | 40 | 4.5 | nc | nc | nc | 2.3 | |
| Dgkg | Diacylglycerol kinase, γ | 7 | 4.0 | nc | nc | nc | nc | |
| Fxyd6 | FXYD domain-containing ion transport regulator 6 | 6 | 2.4 | nc | −1.3 | −1.9 | nc | |
| Gpc3 | Glypican 3 | 11 | 2.4 | nc | −1.3 | −1.7 | nc | |
| Hapln1 | Hyaluronan and proteoglycan link protein 1 | 8 | 2.0 | nc | nc | nc | nc | |
| Hmgcs2 | 3-hydroxy-3-methylglutaryl-coenzyme A synthase 2 | 5 | 4.0 | nc | nc | nc | 2.0 | |
| Icosl | Icos ligand | 2.2 | nc | nc | nc | nc | ||
| Mamdc2 | MAM domain-containing proteoglycan | 4 | 2.9 | −1.3 | nc | −2.0 | 1.8 | |
| Marcksl1 | MARCKS-like 1 | 6 | 1.5 | nc | nc | −1.5 | 3.6 | |
| Odf4 | Outer dense fiber of sperm tails 4 | 2.7 | nc | nc | nc | 2.2 | ||
| Pcsk4 | Proprotein convertase subtilisin/kexin type 4 | 2.0 | nc | −1.4 | −1.4 | nc | ||
| Pygl | Glycogen phosphorylase | 2.0 | −1.2 | −1.2 | −1.5 | nc | ||
| Rassf9 | Ras association (RalGDS/AF-6) domain family (N terminal) member 9 | 2.9 | nc | nc | nc | nc | ||
| Slc1a3 | Glial high-affinity glutamate transporter | 37 | 1.8 | nc | nc | −1.3 | nc | |
| Slc16a1 | Monocarboxylic acid transporter 1 | 3 | 2.2 | nc | nc | nc | 2.6 | |
| Sult1a1 | Sulfotransferase family 1A, phenol-preferring, member 1 | 7 | 2.0 | nc | nc | nc | 1.9 | |
| Syce2 | Synaptonemal complex central element protein 2 | 2.2 | nc | −1.5 | −1.9 | nc | ||
This is a summary of changes in expression of negative genes as measured by qPCR. Genes of enriched expression more than 3-fold in astrocytes or neurons are indicated. Values for enrichment were obtained from the published database by Cahoy et al. (35). nc, No change.
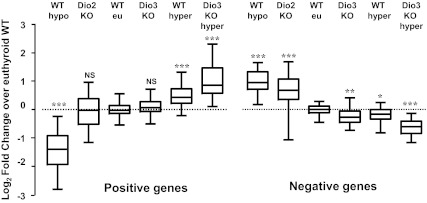
A comprehensive analysis of the gene expression changes in the different experimental situations is illustrated in Fig. 7. This figure was constructed taking the individual data for all the genes shown in the previous tables. It displays the reciprocal changes of expression of positive and negative genes. In contrast to the negative genes that show altered expression in all the experimental situations, the positive genes are remarkably stable in situations of D2 or D3 deficiency and show altered expression only in hypo- or hyperthyroidism.
Fig. 7.
Effects of the different manipulations affecting the supply and metabolism of thyroid hormones on negatively and positively regulated genes. To construct this figure, we used all the single qPCR data from the genes shown in Figs. 3 and 4,to calculate the fold change, relative to the WT values, and plotted the Log2FC (fold change) to make the results quantitatively comparable. The data were represented in a box-and-whiskers (5–95%) plot. Statistical significance between each group and the WT was calculated by one-way ANOVA. For the positive genes, F5, 537 = 272, P < 0.0001. For the negative genes, F5, 400 = 145, P < 0.0001.*, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
The important role of D3 in the control of tissue T3 concentration has been recently demonstrated using genetically modified mice (26, 33). To gain further insight into the role of D3 in modulating T3 action in the brain, we used WT and Dio3KO mice to investigate the expression of a relatively large set of thyroid hormone-sensitive genes identified by microarray analysis of the cerebral cortex of P21 mice (27). After confirming the sensitivity of these genes to thyroid hormone deprivation, we compared the effects of D3 deficiency with those of induced hyperthyroidism and D2 deficiency. The age to perform the present investigation was chosen as the same at which the sensitive genes were identified. This report contains the largest set of brain genes analyzed so far in terms of their dependency to thyroid hormones. A limitation of the study is that the brain of Dio3KO mice manifests abnormal exposure to T3 since early development, and this is maintained throughout life (33). As a result, it might be possible that unintended, permanent changes at the molecular level are influencing the signaling pathways that determine the response to T3 later in life. Although this kind of drawback is generally shared by studies involving germ line gene mutations, it should be kept in mind when extrapolating experimental observations from these mouse models to the normal physiology.
D3 deficiency did not have a marked impact in thyroid hormone-dependent gene expression because it altered the expression of only two positive genes and seven negative genes. This is somewhat surprising, given that D3 expression in the cerebral cortex at this age is comparable with that at an early postnatal age, when significant changes in the expression of the T3-sensitive genes, Hr and Nrgn, have been reported in the Dio3KO mice (26). This does not necessarily mean that D3 is not an important determinant of basal brain T3 action at this age. Although D3 deficiency generally tends to increase brain T3 availability at all developmental stages, we have to take into consideration the suppression exerted on the thyroid axis and on serum levels of T3 and T4. This suppression is most dramatic during the third week of life (26) and may be responsible for the reduced brain expression of the T3-sensitive genes, Nrgn and Hr, on P17 and P21 (33). Hr expression then becomes normalized on P30 as in the present study. Thus, at this age, the Dio3KO brain may still be significantly influenced by the serum hypothyroid status, which would be consistent with previous results (26). However, the Dio3 mRNA concentration in the untreated Dio3KO mice was elevated up to 10-fold, indicating that these animals may actually have an increased T3 availability in the brain despite the lack of effect on most positive genes. A large fraction of brain T3 is generated locally in the brain (36), and although Dio2 mRNA was not increased in the Dio3KO mice, D2 activity in the cortex was increased, in agreement with previous observations (26). Therefore, the concerted action of D2 and D3 activities appears more important for brain homeostasis than the circulating T3 concentrations. Presumably a more robust effect of D3 deficiency on cortical gene expression at this age might be achieved in a tissue specific model of D3 deficiency that leaves the thyroid axis unaffected.
Further evidence of an important role for D3 derives from the results in the animals treated with T3. In Dio3KO mice, T3 treatment led to responses in gene expression that were larger than those in WT mice. It seems unlikely that this was due to differences in the basal concentrations of T3 at the start of treatment, which, if any, would have been overridden by the large dose of T3 administered. The protection afforded by D3 may be largely secondary to increased expression of the Dio3 gene, which is strongly up-regulated by T3 (24). In this regard, we observed an increased D3 activity in the T3-treated WT mice as well as dramatic increases in Dio3 mRNA expression in WT and Dio3KO animals after T3 treatment, demonstrating the great sensitivity of the Dio3 gene to T3. In addition to a direct transcriptional effect of T3, other factors such as increased stability of the mutated mRNA may have contributed to the large induction observed in the untreated and treated Dio3KO mice compared with the respective WT animals. Interestingly, T3 induces the Dio3 gene specifically through the TRα1 isoform (37). Therefore, it is possible that the protecting effect afforded by the increased expression of Dio3 may depend on the relative expression of TRα1 in specific cell types.
A direct role of D3 in the regulation of the genomic responses to T3 was demonstrated in neuroblastoma cells after transfection with a Dio3 expression vector. The Hr response to T3 was attenuated by about 50% in comparison with cells not expressing Dio3. The Hr response was measured in RNA from all cells in the culture, of which 18% were transfected. Therefore, it is likely that Dio3 expression in a fraction of the cells in culture was sufficient to reduce the free T3 concentration in the medium.
Concerning the negative genes, from the original set of 21 genes, seven had a lower expression in the Dio3KO than in the WT. Treatment with excess T3 had a modest effect on the WT, and in some cases the effect of Dio3 inactivation was similar to the effect of T3 administration. The results indicate that some negative genes are very sensitive to local increments of T3 concentration that might have taken place as a result of D3 inactivation. The comparative response of these genes to hyperthyroidism and to D3 inactivation may indicate that the local T3 concentrations attained in D3 deficiency are higher than those resulting from treatment with an excess of T3. However, this conclusion is not supported by the response of the positive genes. We do not have an explanation for this paradox, but it may reflect subtle spatial differences in T3 concentrations affecting only certain subsets of cells.
Another observation worth to explore further is that none of the 10 genes with enriched expression in astrocytes were sensitive to D3 inactivation. In contrast, from the 17 genes, including Dio3, not enriched in astrocytes, 10 were sensitive to D3 inactivation. Because D3 is a neuronal enzyme, this could be explained by a preferential accumulation of T3 in neurons. Because Dio3KO mice manifest increased brain D2 activity due to reduced T4 availability (26), it would be expected that astrocytes provide an increased supply of T3 to neurons. However, the number of genes analyzed in this work is too limited to reach definitive conclusions, and examination of the global expression of neuronal and astrocytic genes in the context of D3 inactivation would be needed.
Although all genes analyzed were highly sensitive to hypothyroidism, it is interesting to note that eight positive genes and 10 negative genes were insensitive to administration of high doses of T3, either to WT or to Dio3KO mice. This indicates a failure of thyroid hormone to increase their expression above the level associated with euthyroid status. This may be due to the high basal occupancy of cortex thyroid hormone receptors, which in adult rats at steady state was calculated to be as high as 90% (36). Individual gene differences in the T3 response may reflect cellular heterogeneity in thyroid hormone receptor occupancy, which should be expected because D2 activity accounts for a large fraction of the total occupancy. Another explanation is that some genes are regulated only in the transition from hypothyroidism to euthyroidism. In hypothyroidism the activity of unliganded receptors (38) may contribute to repression of the positive genes and the induction of the negative genes. Similar differences in the response to hypothyroidism and to excess T3 were already noted for different gene clusters in the liver (39).
Previous data indicated that the negative genes are more sensitive to the absence of D2 than the positive genes (27). As shown in this work, negative genes are also more sensitive to the absence of D3. This may reflect a difference between positive and negative genes in the kinetics of their response to changes in T3 concentrations. Figure 7 shows the reciprocal changes in the expression of the genes that were sensitive to each of the manipulations. However, it is not possible to correlate these changes with T3 concentration in the whole tissue. For example, T3 concentrations in the cerebral cortex of Dio2KO are similar to hypothyroid WT mice (40). But from the expression of the positive genes, it could be predicted that there are no changes of T3 concentration, whereas from the negative genes, one could reach a different conclusion.
In summary, our results demonstrate a role for D3 in the regulation of T3-dependent gene expression in the cerebral cortex. They also suggest that, when thyroid hormone brain status is close to physiological conditions, genes that are negatively regulated by T3 in the cerebral cortex are the most sensitive to subtle changes in local T3. In this scenario, the coordinated expression of D2 and D3 appears to be necessary for maintaining the expression of these genes within narrow limits. These results underscore the fine-tuning of the factors influencing local T3 availability in the central nervous system.
Acknowledgments
We acknowledge Professor Samuel Refetoff and his coworkers for the critical reading of the manuscript and Lauren Keyes and Eulalia Moreno for their technical assistance.
This work was supported by Grants SAF2008-01168 and SAF2008-00429-E from the Ministry of Education and Science of Spain, Grant LSHM-CT-2005-018652 from the European Union Integrated Project CRESCENDO, a grant from the Center for Biomedical Research on Rare Diseases, an initiative of the Instituto de Salud Carlos III, and by Grant NIMH-083220 from the National Institute of Mental Health. A.C. was recipient of a predoctoral fellowship from the Plan Nacional de I+D+i.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BBB
- Blood-brain barrier
- D2
- 5′-deiodinase type 2
- D3
- 5′-deiodinase type 3
- KO
- knockout
- Mct8
- monocarboxylate 8
- P
- postnatal
- qPCR
- quantitative PCR
- TRα1
- thyroid hormone receptor-α1 isoform
- WT
- wild type.
References
- 1. Morreale de Escobar G, Obregon MJ, Escobar del Rey F. 2004. Role of thyroid hormone during early brain development. Eur J Endocrinol 151(Suppl 3):U25–U37 [DOI] [PubMed] [Google Scholar]
- 2. Bernal J. 2007. Thyroid hormone receptors in brain development and function. Nat Clin Pract Endocrinol Metab 3:249–259 [DOI] [PubMed] [Google Scholar]
- 3. Patel J, Landers K, Li H, Mortimer RH, Richard K. 2011. Thyroid hormones and fetal neurological development. J Endocrinol 209:1–8 [DOI] [PubMed] [Google Scholar]
- 4. Cheng SY, Leonard JL, Davis PJ. 2010. Molecular aspects of thyroid hormone actions. Endocr Rev 31:139–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernal J. 2005. Thyroid hormones and brain development. Vitam Horm 71:95–122 [DOI] [PubMed] [Google Scholar]
- 6. Lazar MA. 1993. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev 14:184–193 [DOI] [PubMed] [Google Scholar]
- 7. Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, Simonides WS, Zeöld A, Bianco AC. 2008. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev 29:898–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schneider MJ, Fiering SN, Pallud SE, Parlow AF, St Germain DL, Galton VA. 2001. Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol Endocrinol 15:2137–2148 [DOI] [PubMed] [Google Scholar]
- 9. St Germain DL, Galton VA, Hernandez A. 2009. Minireview: defining the roles of the iodothyronine deiodinases: current concepts and challenges. Endocrinology 150:1097–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Freitas BC, Gereben B, Castillo M, Kalló I, Zeöld A, Egri P, Liposits Z, Zavacki AM, Maciel RM, Jo S, Singru P, Sanchez E, Lechan RM, Bianco AC. 2010. Paracrine signaling by glial cell-derived triiodothyronine activates neuronal gene expression in the rodent brain and human cells. J Clin Invest 120:2206–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bianco AC. 2011. Minireview: cracking the metabolic code for thyroid hormone signaling. Endocrinology 152:3306–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zavacki AM, Arrojo E, Drigo R, Freitas BC, Chung M, Harney JW, Egri P, Wittmann G, Fekete C, Gereben B, Bianco AC. 2009. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol Cell Biol 29:5339–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burmeister LA, Pachucki J, St Germain DL. 1997. Thyroid hormones inhibit type 2 iodothyronine deiodinase in the rat cerebral cortex by both pre- and posttranslational mechanisms. Endocrinology 138:5231–5237 [DOI] [PubMed] [Google Scholar]
- 14. Guadaño-Ferraz A, Obregón MJ, St. Germain DL, Bernal J. 1997. The type 2 iodothyronine deiodinase is expressed primarily in glial cells in the neonatal rat brain. Proc Natl Acad Sci USA 94:10391–10396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tu HM, Kim SW, Salvatore D, Bartha T, Legradi G, Larsen PR, Lechan RM. 1997. Regional distribution of type 2 thyroxine deiodinase messenger ribonucleic acid in rat hypothalamus and pituitary and its regulation by thyroid hormone. Endocrinology 138:3359–3368 [DOI] [PubMed] [Google Scholar]
- 16. Guadaño-Ferraz A, Escámez MJ, Rausell E, Bernal J. 1999. Expression of type 2 iodothyronine deiodinase in hypothyroid rat brain indicates an important role of thyroid hormone in the development of specific primary sensory systems. J Neurosci 19:3430–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Visser WE, Friesema EC, Visser TJ. 2011. Minireview: thyroid hormone transporters: the knowns and the unknowns. Mol Endocrinol 25:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu C, Li JY, Boado RJ, Pardridge WM. 2008. Blood-brain barrier genomics and cloning of a novel organic anion transporter. J Cereb Blood Flow Metab 28:291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roberts LM, Woodford K, Zhou M, Black DS, Haggerty JE, Tate EH, Grindstaff KK, Mengesha W, Raman C, Zerangue N. 2008. Expression of the thyroid hormone transporters monocarboxylate transporter-8 (SLC16A2) and organic ion transporter-14 (SLCO1C1) at the blood-brain barrier. Endocrinology 149:6251–6261 [DOI] [PubMed] [Google Scholar]
- 20. Friesema EC, Ganguly S, Abdalla A, Manning Fox JE, Halestrap AP, Visser TJ. 2003. Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter. J Biol Chem 278:40128–40135 [DOI] [PubMed] [Google Scholar]
- 21. Grijota-Martínez C, Díez D, Morreale de Escobar G, Bernal J, Morte B. 2011. Lack of action of exogenously administered T3 on the fetal rat brain despite expression of the monocarboxylate transporter 8. Endocrinology 152:1713–1721 [DOI] [PubMed] [Google Scholar]
- 22. Hernandez A. 2005. Structure and function of the type 3 deiodinase gene. Thyroid 15:865–874 [DOI] [PubMed] [Google Scholar]
- 23. Galton VA, Martinez E, Hernandez A, St Germain EA, Bates JM, St. Germain DL. 1999. Pregnant rat uterus expresses high levels of the type 3 iodothyronine deiodinase. J Clin Invest 103:979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tu HM, Legradi G, Bartha T, Salvatore D, Lechan RM, Larsen PR. 1999. Regional expression of the type 3 iodothyronine deiodinase messenger ribonucleic acid in the rat central nervous system and its regulation by thyroid hormone. Endocrinology 140:784–790 [DOI] [PubMed] [Google Scholar]
- 25. Escámez MJ, Guadaño-Ferraz A, Cuadrado A, Bernal J. 1999. Type 3 iodothyronine deiodinase is selectively expressed in areas related to sexual differentiation in the newborn rat brain. Endocrinology 140:5443–5446 [DOI] [PubMed] [Google Scholar]
- 26. Hernandez A, Martinez ME, Fiering S, Galton VA, St. Germain D. 2006. Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J Clin Invest 116:476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morte B, Ceballos A, Diez D, Grijota-Martínez C, Dumitrescu AM, Di Cosmo C, Galton VA, Refetoff S, Bernal J. 2010. Thyroid hormone-regulated mouse cerebral cortex genes are differentially dependent on the source of the hormone: a study in monocarboxylate transporter-8- and deiodinase-2-deficient mice. Endocrinology 151:2381–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pastor R, Bernal J, Rodríguez-Peña A. 1994. Unliganded c-erbA/thyroid hormone receptor induces trkB expression in neuroblastoma cells. Oncogene 9:1081–1089 [PubMed] [Google Scholar]
- 29. Samuels HH, Stanley F, Casanova J. 1979. Depletion of l-3,5,3′-triiodothyronine and l-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology 105:80–85 [DOI] [PubMed] [Google Scholar]
- 30. Bates JM, St. Germain DL, Galton VA. 1999. Expression profiles of the three iodothyronine deiodinases, D1, D2, and D3, in the developing rat. Endocrinology 140:844–851 [DOI] [PubMed] [Google Scholar]
- 31. Escobar-Morreale HF, Obregón MJ, Hernandez A, Escobar del Rey F, Morreale de Escobar G. 1997. Regulation of iodothyronine deiodinase activity as studied in thyroidectomized rats infused with thyroxine or triiodothyronine. Endocrinology 138:2559–2568 [DOI] [PubMed] [Google Scholar]
- 32. Galton VA, Hiebert A. 1987. Hepatic iodothyronine 5-deiodinase activity in Rana catesbeiana tadpoles at different stages of the life cycle. Endocrinology 121:42–47 [DOI] [PubMed] [Google Scholar]
- 33. Hernandez A, Quignodon L, Martinez ME, Flamant F, St. Germain DL. 2010. Type 3 deiodinase deficiency causes spatial and temporal alterations in brain T3 signaling that are dissociated from serum thyroid hormone levels. Endocrinology 151:5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hernandez A, Fiering S, Martinez E, Galton VA, St. Germain D. 2002. The gene locus encoding iodothyronine deiodinase type 3 (Dio3) is imprinted in the fetus and expresses antisense transcripts. Endocrinology 143:4483–4486 [DOI] [PubMed] [Google Scholar]
- 35. Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crantz FR, Silva JE, Larsen PR. 1982. An analysis of the sources and quantity of 3,5,3′-triiodothyronine specifically bound to nuclear receptors in rat cerebral cortex and cerebellum. Endocrinology 110:367–375 [DOI] [PubMed] [Google Scholar]
- 37. Barca-Mayo O, Liao XH, Alonso M, Di Cosmo C, Hernandez A, Refetoff S, Weiss RE. 2011. Thyroid hormone receptor α and regulation of type 3 deiodinase. Mol Endocrinol 25:575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morte B, Manzano J, Scanlan T, Vennström B, Bernal J. 2002. Deletion of the thyroid hormone receptor α1 prevents the structural alterations of the cerebellum induced by hypothyroidism. Proc Natl Acad Sci USA 99:3985–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yen PM, Feng X, Flamant F, Chen Y, Walker RL, Weiss RE, Chassande O, Samarut J, Refetoff S, Meltzer PS. 2003. Effects of ligand and thyroid hormone receptor isoforms on hepatic gene expression profiles of thyroid hormone receptor knockout mice. EMBO Rep 4:581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Galton VA, Wood ET, St Germain EA, Withrow CA, Aldrich G, St. Germain GM, Clark AS, St. Germain DL. 2007. Thyroid hormone homeostasis and action in the type 2 deiodinase-deficient rodent brain during development. Endocrinology 148:3080–3088 [DOI] [PubMed] [Google Scholar]