Abstract
Mitochondria are the primary locus for the generation of reactive nitrogen species including peroxynitrite and subsequent protein tyrosine nitration. Protein tyrosine nitration may have important functional and biological consequences such as alteration of enzyme catalytic activity. In the present study, mouse liver mitochondria were incubated with peroxynitrite, and the mitochondrial proteins were separated by 1D and 2D gel electrophoresis. Nitrotyrosinylated proteins were detected with an anti-nitrotyrosine antibody. One of the major proteins nitrated by peroxynitrite was carbamoyl phosphate synthetase 1 (CPS1) as identified by LC-MS protein analysis and Western blotting. The band intensity of nitration normalized to CPS1 was increased in a peroxynitrite concentration-dependent manner. In addition, CPS1 activity was decreased by treatment with peroxynitrite in a peroxynitrite concentration- and time-dependent manner. The decreased CPS1 activity was not recovered by treatment with reduced glutathione, suggesting that the decrease of the CPS1 activity is due to tyrosine nitration rather than cysteine oxidation. LC-MS analysis of in-gel digested samples, and a Popitam-based modification search located 5 out of 36 tyrosine residues in CPS1 that were nitrated. Taken together with previous findings regarding CPS1 structure and function, homology modeling of mouse CPS1 suggested that nitration at Y1450 in an α-helix of allosteric domain prevents activation of CPS1 by its activator, N-acetyl-L-glutamate. In conclusion, this study demonstrated the tyrosine nitration of CPS1 by peroxynitrite and its functional consequence. Since CPS1 is responsible for ammonia removal in the urea cycle, nitration of CPS1 with attenuated function might be involved in some diseases and drug-induced toxicities associated with mitochondrial dysfunction.
Keywords: protein tyrosine nitration, carbamoyl phosphate synthetase 1, peroxynitrite, LC-ESI-MS/MS, homology modeling, mitochondria
Introduction
Protein tyrosine nitration is a post-translational modification which occurs in the presence of reactive nitrogen species [1]. One of the reactive nitrogen species is peroxynitrite (ONOO–) which is formed by the reaction of a superoxide radical anion (O 2–) with nitric oxide (NO) [2]. Tyrosine nitration can alter protein structure due to a shift in pKa from 10.1 to 7.2 of the tyrosine hydroxyl group, as well as the addition of bulkiness to the tyrosine residue [3]. The structural changes may have important functional and biological consequences such as alteration of enzyme catalytic activity, modulation of the tyrosine phosphorylation cascade and induction of immune responses [1]. Peroxynitrite can be formed intramitochondrially through the generation of reactive oxygen species by complexes I – III of the mitochondrial respiratory chain [4]. Due to the rather short-lived nature of these reactive species, it is conceivable that mitochondrial proteins are the primary targets of protein tyrosine nitration. One of the best-studied examples is nitration and inactivation of manganese superoxide dismutase (MnSOD) [5]. A normal function of MnSOD is to decrease peroxynitrite formation by dismutating superoxide to hydrogen peroxide and molecular oxygen. However, recent studies have shown that MnSOD can be inactivated by tyrosine nitration, resulting in increased peroxynitrite formation and further oxidative/nitrative stress. Decreases in MnSOD activity due to nitrative stress in mitochondria has been observed in animal models of renal ischemia/reperfusion [6] and in drug-induced liver injury caused by toxic doses of acetaminophen [7].
Protein tyrosine nitration demonstrates a degree of selectivity for proteins and specific tyrosine residues within those proteins [8, 9]. There is no specific or preferred consensus sequence that promotes tyrosine nitration, however, the protein secondary structure and local environment of the tyrosine residues seem to be important for reactivity and accessibility of reactive nitrogen species to the tyrosine residue [8, 9]. Therefore, the identification of tyrosine nitrated proteins and the sites of nitrated tyrosine residues is key to understanding the link between protein tyrosine nitration and its biological consequences.
In the present study, we have examined tyrosine nitration of mouse liver mitochondrial proteins after treatment with peroxynitrite followed by 1D and 2D SDS gel electrophoresis, Western blot analysis using an anti-nitrotyrosine antibody, and LC-MS/MS analysis of in-gel tryptic digests of the nitrated protein samples. Several algorithms such as Sequest and Popitam were utilized to identify proteins and search post-translational modifications, including tyrosine nitration, based on tandem mass spectrometry data (MS/MS spectra) and peptide sequence databases [10]. On the basis of our initial findings, we focused on the tyrosine nitration of carbamoyl phosphate synthetase 1 (CPS1) in mouse mitochondria. CPS1 is the mitochondrial enzyme that catalyzes the first and rate-limiting step of the urea cycle and responsible for the detoxification of excess ammonia [11]. N-acetyl-L-glutamate (NAG) is an allosteric activator of CPS1, playing a key role in controlling the flow of nitrogen through the urea cycle [12]. Here, we report tyrosine nitration of CPS1 with functional changes by treatment with peroxynitrite and possible involvement of a nitrated tyrosine residue as a functional consequence.
Materials and methods
Chemicals and reagents
Peroxynitrite was purchased from Cayman Chemical (Ann Arbor, MI). Pyruvate kinase, lactate dehydrogenase, phosphoenolpyruvate, NADH, ATP, NAG, glutathione (GSH) and glycylglycine were purchased from Sigma-Aldrich (St. Louis, MO). Mouse anti-nitrotyrosine antibody (clone 2A8.2) was purchased from Millipore Corporation (Temecula, CA). Rabbit anti-CPS1 antibody was obtained from Abcam Inc. (Cambridge, MA). Secondary antibodies were obtained from Li-Cor Biosciences (Lincoln, NE).
Preparation of mouse mitochondrial fractions
Mouse liver samples were collected from male C57BL/6 mice (n =3), which were obtained form Jackson Labs (Bar Harbor, ME). The animal experiments were conducted with approval by the IACUC for the University of Washington. Preparation of mitochondrial fractions was based on a previously published protocol [13].
1D and 2D gel electrophoresis
For 1D gel electrophoresis, the mitochondrial fractions (10 μg protein each) collected from three mice were individually incubated with each concentration of peroxynitrite (0, 0.1 and 1.0 mM) at 37°C for 10 min in an aqueous solution containing 50 mM glycylglycine. Each reaction mixture was resolved using 7% SDS-polyacrylamide gels. For 2D gel electrophoresis, the mitochondrial fractions from three mice were pooled. The reaction mixtures (20 μL) including 100 μg protein of the pooled mitochondrial fractions, 50 mM glycylglycine, and peroxynitrite (0 or 1.0 mM) were incubated at 37°C for 10 min. Each reaction mixture was separated by pH 3 - 10 isoelectric focusing as previously described [14].
Western blot analysis
Proteins were transferred to nitrocellulose membranes, which were blocked in a 1:1 ratio of phosphate buffered saline (pH 7.4) and Li-Cor Blocking Buffer for 1 h at room temperature. Membranes were incubated overnight at 4°C with anti-nitrotyrosine antibody (1:500, mouse IgG) and anti-CPS1 antibody (1:10000, rabbit IgG). Membranes were then incubated with secondary anti-mouse IRDye 800CW antibody (1:5000, goat IgG) and anti-rabbit IRDye 680LT antibody (1:10000, donkey IgG) for 1 h at room temperature. Detection and quantification of the immunofluorescence bands was accomplished using the Odyssey Infrared Imaging System and Software ver. 2.1 (Li-Cor).
CPS1 activity assay
The mitochondrial fractions (5 μg protein, n = 3) were incubated with different concentrations of peroxynitrite at 37°C for 10 min, or incubated with 1.0 mM peroxynitrite at 37°C for different time periods. The assay mixtures contained 5 μg protein of the mitochondrial fractions after peroxynitrite treatment, 2.5 mM phosphoenolpyruvate, 0.2 mM NADH, 10 mM NH4Cl, 100 mM KHCO3, 5 mM ATP, 10 mM MgSO4, 10 mM NAG, 10 U/ml pyruvate kinase, 12.5 U/ml of lactate dehydrogenase, and 50 mM glycylglycine. For (-) NAG assays, NAG was omitted. CPS1 activity was calculated from the initial velocity of NADH oxidation (absorbance at 340 nm). For the reversibility assay, the mitochondrial fractions were incubated with 1.0 mM peroxynitrite at 37°C for 10 min and subsequently incubated with or without 5.0 mM GSH at 37°C for 10 min, followed by the CPS1 activity assay with 10 mM NAG.
Detection and characterization of nitrated peptides by LC-MS/MS
The Coomassie blue-stained 1D gel bands and 2D gel spots that correspond to nitrated CPS1 were excised, and proteins were subjected to in-gel tryptic digestion. The peptides obtained were analyzed using a nano-HPLC (nanoAcquity systems; Waters, Milford, MA) coupled to a hybrid linear ion trap Orbitrap (LTQ-Orbitrap) mass spectrometer (Thermo Scientific, San Jose, CA). Peptide separation was performed with a laser-pulled 75 μm i.d. capillary packed with 15 cm of 100 Å (5 μm) Magic C18 particles (C18AQ; Michrom). Peptides were eluted using an acetonitrile gradient flowing at 250 nL/min in a mobile phase consisting of the following: A, water, 0.1% formic acid; B, acetonitrile, 0.1% formic acid. Peptides were analyzed in the positive ion mode with an MS survey scan acquired intense precursor over a full m/z range of 400-2000 with the Orbitrap at a resolution of 60,000. The seven most ions were sequentially isolated using a data-dependent mode and subjected to collision induced dissociation before a tandem mass spectrum was obtained with the Orbitrap at a resolution of 15,000. The Sequest (version 27) algorithm was applied to search the MS data against the IPI mouse database (ipi.MOUSE.fasta.v3.68). Additionally, the raw data was searched utilizing the open-modification search engine Popitam (freely accessible at www.expasy.ch/tools/popitam/).
Homology modeling of NAG binding domain of mouse CPS1
The crystal structures deposited in the PDB as 1JDB [15] and 2YVQ [16], corresponding to the whole structure of E. Coli CPS and the NAG binding domain of human CPS1 (residues 1343 - 1478), respectively, were used as the templates. Homology modeling of mouse CPS1 was performed in the SWISS-MODEL Workspace [17]. Figures depicting protein structures were prepared with PyMOL (ver. 1.1, DeLano Scientific).
Results and Discussion
Western blots of nitrated proteins in mouse liver mitochondria
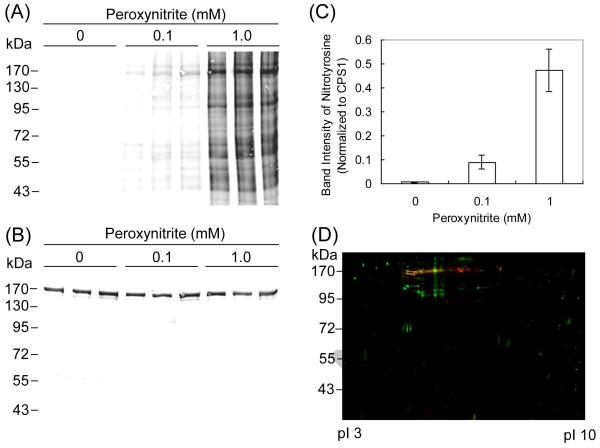
The mitochondrial fractions isolated from mouse liver (n = 3) were incubated with different concentrations of peroxynitrite for 10 min, followed by 1D SDS gel electrophoresis and Western blot analysis using an anti-nitrotyrosine antibody (Figure 1A). Many bands corresponding to nitrated proteins were observed and the most intense band was determined to be that at around 170 kDa by densitometry analysis. This band was subjected to in-gel tryptic digestion, LC-MS/MS analysis and a Sequest search, which showed that the band contained CPS1 (accession number: Q8C196; sequence coverage: 29%; number of unique peptides assigned to the protein: 60). The blot probed with anti-CPS1 antibody represented the bands at around 170 kDa corresponding to CPS1 in all samples (Figure 1B). The band intensity of nitrotyrosine corresponding to CPS1 was normalized to that of CPS1, demonstrating that nitration of CPS1 increases in a peroxynitrite concentration-dependent manner (Figure 1C). However, the bands at 170 kDa corresponding to nitrated CPS1 could include some other nitrated proteins because the 1D Western blots, especially for 1 mM peroxynitrite treatment, showed numerous bands of nitrated proteins without sufficient resolution. To further examine the nitration of CPS1, the mitochondrial fractions after incubation with 1.0 mM peroxynitrite were also subjected to 2D gel electrophoresis, followed by Western blot analysis using anti-nitrotyrosine and anti-CPS1 antibodies (Figure 1D). CPS1 was detected as multiple serial spots, a so-called “charge-train”, at around 170 kDa, which is consistent with previously reported findings [18]. The spots of nitrated proteins at around 170 kDa were largely overlapped with those of CPS1, as represented by yellow spots in Figure 1D. This observation confirms that CPS1 is one of the major targets of tyrosine nitration in mitochondria and the calculation of nitrated CPS1 from 1D Western blot shown in Figure 1C presents the peroxynitrite concentration dependency of CPS1 nitration, even though it is not rigorously quantitative. In the present study, SDS-PAGE analysis was performed on gels containing a relatively low concentration of acrylamide in order to resolve high molecular weight proteins including CPS1. In this case, however, small molecular weight proteins including mitochondrial MnSOD (25 kDa), which is well known to be nitrated, migrated at the buffer-front and, therefore, nitrated MnSOD was not detected. In preliminary experiments on gels containing probably corresponds to nitrated MnSOD (data not shown).
Figure 1.
Western blot analysis of mouse mitochondrial proteins after treatment with peroxynitrite. (A) The mitochondrial fractions collected from three mice were individually incubated with each concentration of peroxynitrite (0, 0.1 and 1.0 mM) at 37°C for 10 min and subjected to 1D SDS gel electrophoresis. The blots were probed with antibodies to nitrotyrosine. (B) The blots were probed with antibodies to CPS1. (C) Densitometry analysis of nitration to CPS1 was performed using the Odyssey Infrared Imaging System. (D) The mitochondrial proteins pooled from three mice were incubated with 1.0 mM peroxynitrite at 37°C for 10 min and subjected to 2D gel electrophoresis. The blots of nitrated proteins (green) were superimposed with those of CPS1 (red). The overlap is represented by yellow.
Effects of peroxynitrite treatment on CPS1 activity
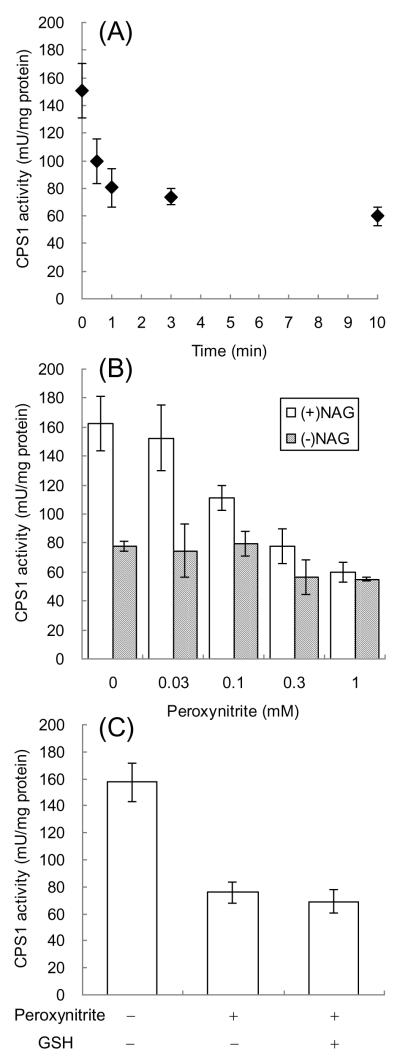
The mitochondrial fractions were incubated with 1.0 mM peroxynitrite for different time periods, followed by measuring their CPS1 activities in the presence of NAG, an allosteric activator of CPS1 (Figure 2A). The CPS1 activity was quickly and significantly decreased by treatment with peroxynitrite in a time-dependent manner (47% decrease in 1 min and 60% decrease in 10 min). The CPS1 activities after 10-min incubations with different concentrations of peroxynitrite were also examined (Figure 2B). In the presence of NAG, CPS1 activity was decreased in a peroxynitrite concentration-dependent fashion. Significant decreases in the activity were observed for the samples treated by 0.1 mM or higher concentrations of peroxynitrite. In the absence of NAG, CPS1 activity without treatment with peroxynitrite was approximately half of that in the presence of NAG, which is consistent with the previous findings that NAG activates CPS1 by increasing the affinities of ATP, Mg2+ and K+ to CPS1 [12]. Treatment with increasing concentrations of peroxynitrite did not have significant effects on CPS1 activity in the absence of NAG as shown in Figure 2B. CPS1 activities after treatment with 1.0 mM peroxynitrite were comparable between in the presence and absence of NAG. These observations suggest that tyrosine nitration decreases CPS1 activity by affecting the interaction of CPS1 with NAG. In addition to tyrosine nitration, peroxynitrite can oxidize thiol groups of cysteines. The contribution of thiol oxidation to the decrease of CPS1 activity can be evaluated because thiol oxidation is reversible in the presence of the reducing agents such as GSH and dithiothreitol [19]. CPS1 activity decreased by peroxynitrite was not recovered by treatment with reduced GSH as shown in Figure 2C, suggesting that the decrease of the CPS1 activity is due to tyrosine nitration rather than cysteine oxidation.
Figure 2.
The effects of peroxynitrite treatment on CPS1 activity in mouse mitochondrial fractions. (A) The mitochondrial fractions were incubated with 1.0 mM peroxynitrite at 37°C for different time periods and subjected to the CPS1 activity assay. (B) The mitochondrial fractions were incubated with different concentrations of peroxynitrite at 37°C for 10 min and subjected to the CPS1 activity assay with or without 10 mM NAG. (C) The mitochondrial fractions were incubated with 1.0 mM peroxynitrite at 37°C for 10 min and subsequently incubated with or without 5.0 mM GSH at 37°C for 10 min, followed by the CPS1 activity assay with 10 mM NAG.
Determination of nitrated tyrosine residues by LC-MS/MS and Popitam analysis
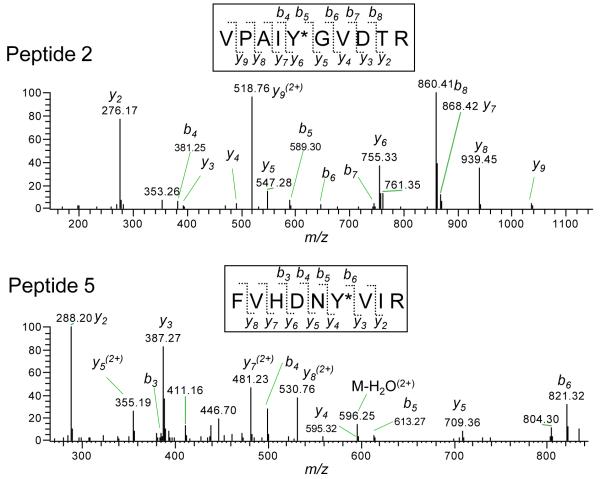
Mouse CPS1 consists of 1,500 amino acids including 36 tyrosine residues (NCBI protein database, Accession No. AAI26970). In order to determine the target tyrosine residues of nitration, the 1D gel bands and 2D gel spots corresponding to nitrated CPS1 were excised, followed by in-gel tryptic digestion of the proteins and LC-MS/MS of the digested peptides. LC-MS/MS spectra were analyzed using Popitam, which is an algorithm designed to identify mutations and modifications to peptides based on MS/MS spectral data. The Popitam-based modification search of the mouse CPS1 sequence identified 5 peptides that could contain nitrotyrosine (Table 1). The observed mass number of the precursor ions was close to the theoretical mass number for all candidate peptides (mass errors < 3.1 ppm). The structures of the 5 nitrated peptides were verified by their MS/MS spectra. Representative MS/MS spectra and assignments of b and y ions are shown in Figure 3. The mass difference of 208 Da corresponding to nitration (+45 Da) of tyrosine (163 Da) was observed in y ions and b ions. Previous studies reported that most nitrated tyrosines are found in loop structures with turn-inducing residues, proline or glycine, in the vicinity (from −5 to +5 residues around nitrated tyrosines) [8]. The proximity of a negative charge of acidic amino acids (i.e., aspartate and glutamate) is also beneficial [8]. All five nitrated tyrosines have acidic amino acid residue(s) in their proximal sequences (Table 2), which is consistent with the previous findings mentioned above. The tyrosines Y120, Y162, Y529 and Y1305 have turn-inducing residue(s) in their proximity, suggesting that they are located in loop structures.
Table 1.
Nitrated peptides of mouse CPS1 detected by LC-MS/MS and Popitam analysis.
| Peptide No. |
Sequence | Calculated m/za |
Observed m/za (mass error: ppm) |
|
|---|---|---|---|---|
| Sample from 1D gel | Sample form 2D gel | |||
| 1 | 113DELGLNKY*M#ESDGIK127 | 886.9045 | 886.9055 (1.1) |
886.9046 (0.1) |
| 2 | 158VPAIY*GVDTR167 | 568.2914 | 568.2917 (0.5) |
568.2917 (0.5) |
| 3 | 524GVLKEY*GVK532 | 519.2855 | 519.2855 (0) |
N.D. |
| 4 | 1293LPTLEQPIIPSDY*VAIK1309 | 971.5309 | 971.5339 (3.1) |
971.5311 (0.2) |
| 5 | 1445FVHDNY*VIR1453 | 604.2969 | 604.2974 (0.8) |
604.2975 (1.0) |
Numbers show m/z of doubly protonated molecular ions. Y*and M# represent nitrotyrosine and oxidized (+ O) methionine, respectively. N.D. represents “not detected”.
Figure 3.
Representative LC-MS/MS spectra and assignments of the sequence-specific fragment ions of the nitrated peptides. The sequence-specific fragment ions are labeled as y and b ions on the spectra. For the peptide 2, the mass difference of 208 detected between b4 and b5 and between y5 and y6 corresponds to nitrated tyrosine residue (163 (Tyr) + 45 (nitration)). For the peptide 5, the mass difference of 208 detected between b5 and b6 and between y3 and y4 corresponds to nitrated tyrosine residue.
Table 2.
Proximal amino acids of nitrated tyrosine residues
| No. | Sequence from −5 to +5 amino acids relative to the nitrated tyrosine |
|---|---|
| 1 | LGLNKY120*MESDG |
| 2 | KVPAIY162*GVDTR |
| 3 | GVLKEY529*GVKVL |
| 4 | IIPSDY1305*VAIKA |
| 5 | FVHDNY1450*VIRRT |
Turn-inducing residues, proline and glycine, are shown in bold. Negatively-charged residues, aspartate and glutamate, are underlined. Y* represents nitrotyrosine.
Homology modeling of mouse CPS1 and location of nitrated tyrosine residues
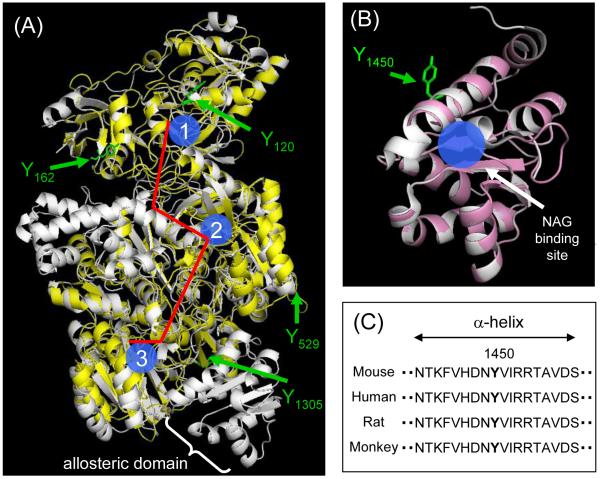
Since a crystal structure of mouse CPS1 has not been solved, we created its model based on E. Coli CPS (PDB file: 1JDB) [15] by homology modeling on SWISS-MODEL Workspace [17]. The created model of mouse CPS1 superimposed with the template structure is shown in Figure 4A. The model mostly covered the sequence of mouse CPS1, whereas some parts of the protein, such as the allosteric domain, were not modeled because of poor homology of the sequence. Four out of five nitrated tyrosine residues (Y120, Y162, Y529 and Y1305) were included in the created model of mouse CPS1 as shown in Figure 4A. They appear to be located in or close to loop structures, which is consistent with the findings in primary sequences. Also, all these nitrated tyrosine residues are positioned on the protein surface, which can be explained by their accessibility to reactive nitrogen species. According to previous studies regarding the crystal structure of CPS from E. Coli, there is a molecular tunnel for the transport of reaction intermediates between remotely located active sites (Figure 4A) [15]. The superimposed image of mouse CPS1 indicates that those four tyrosine residues are located away from active sites and the molecular tunnel, suggesting that they are unlikely to be involved in the nitration-mediated decrease in CPS1 activity. The other tyrosine residue, Y1450, was considered to be located in the allosteric domain which interacts with the activator, NAG. Recently, the crystal structure of the NAG binding domain of human CPS1 (residues 1343 – 1478, PDB file: 2YVQ) has been solved and NAG-interacting residues were characterized [16]. We created the NAG binding domain of mouse CPS1 using the three-dimensional structure of human CPS1 as a template (Figure 4B). Since the sequence of the domain is highly conserved among species including human and mouse, the created model is almost identical to the template. The tyrosine Y1450 is located in an α-helix, which is considered to directly contribute to NAG binding via an electrostatic interaction with N1449 and by hydrophobic interactions with F1445 and I1452 [16]. The primary amino acid sequence of this α-helix, including Y1450, is strictly conserved as shown in Figure 4C. The importance of this α-helical structure for CPS1 activity has also been demonstrated by CPS1 mutations found in CPS1 deficient patients. It was reported that the mutations R1453Q and R1453W, which were found in severe CPS1 deficiency, inactivated CPS1. Since R1453 does not interact with NAG directly, CPS1 inactivation by the mutations to R1453 was considered to be due to distortion of the α-helical structure [20]. Similarly, nitration to Y1450 could elicit a structural change of the α-helix and decrease the binding affinity to NAG, which is a possible mechanism for the decrease in CPS1 activity by peroxynitrite. This interpretation seems to be consistent with the finding that the peroxynitrite-dependent decrease in CPS1 activity was more significant in the presence of NAG than in its absence as shown in Figure 2B. Even though most nitrated tyrosine residues are found in flexible loop regions, as mentioned above, functionally critical tyrosine nitration tends to be found in α-helices, such as Y34 in MnSOD [21] and Y97 and Y74 in cytochrome c [22]. Also, a recent study demonstrated that the content of the α-helical structure of insulin was reduced by nitration to a tyrosine residue in an α-helix [23]. These findings support our results and interpretation. Future studies such as mutation analysis will be needed to clarify a mechanistic link between the nitration to Y 1450 and loss of NAG binding.
Figure 4.
Homology modeling of mouse CPS1. (A) The superimposed image of the created model of mouse CPS1 (yellow) with the template structure for the E. Coli CPS (white, PDB file: 1JDB). Three active sites are shown in blue (1: glutamine binding site, 2: bicarbonate binding site, 3: carbamoyl phosphate binding site) and the molecular tunnel is shown in red. (B) The superimposed image of the created model of the mouse NAG binding domain (pink) with the template structure for the human NAG binding domain (white, PBD file: 2YVQ). (C) The sequences (residues 1442 - 1459) in NAG binding domain of CPS1s from mouse (Mus musculus), human (Homo sapiens), rat (Rattus norvegicus) and monkey (Macaca mulatta), were obtained from the NCBI protein database.
In conclusion, our study for the first time revealed that CPS1 is one of the major targets of tyrosine nitration in mitochondria after treatment with peroxynitrite, and the functional consequence is the decrease in its enzymatic activity in the presence of NAG. Since CPS1 is responsible for the detoxification of excess ammonia, nitration of CPS1 with attenuated function may result in hyperammonemia and subsequent mitochondrial dysfunction in in vivo situations. Future in vivo studies will be required to investigate the relevance of these findings to some diseases and drug-induced toxicities.
Highlights.
CPS1 is one of the major proteins nitrated by peroxynitrite in liver mitochondria.
The enzymatic activity of CPS1 is inactivated by peroxynitrite.
CPS1 activity decreased by peroxynitrite is not recovered by reduced GSH.
Five out of 36 tyrosine residues in CPS1 are selectively nitrated.
Nitration at Y1450 is suggested to be involved in the loss of CPS1 activity.
Acknowledgement
This work was supported in part by National Institutes of Health Grant [GM32165] and support to H.T. from Daiichi Sankyo Co., Ltd. (Tokyo, Japan).
Abbreviations
- CPS1
carbamoyl phosphate synthetase 1
- MnSOD
manganese superoxide dismutase
- NAG
N-acetyl-L-glutamate
- GSH
glutathione
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was performed while H.T. was a visiting scholar at the University of Washington.
References
- [1].Abello N, Kerstjens HA, Postma DS, Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome. Res. 2009;8:3222–3238. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- [2].Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat. Rev. Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- [3].Sokolovsky M, Riordan JF, Vallee BL. Conversion of 3-nitrotyrosine to 3-aminotyrosine in peptides and proteins. Biochem. Biophys. Res. Commun. 1967;27:20–25. doi: 10.1016/s0006-291x(67)80033-0. [DOI] [PubMed] [Google Scholar]
- [4].Radi R, Cassina A, Hodara R, Quijano C, Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radic. Biol. Med. 2002;33:1451–1464. doi: 10.1016/s0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- [5].MacMillan-Crow LA, Crow JP, Kerby JD, Beckman JS, Thompson JA. Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allografts. Proc. Natl. Acad. Sci. USA. 1996;93:11853–11858. doi: 10.1073/pnas.93.21.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cruthirds DL, Novak L, Akhi KM, Sanders PW, Thompson JA, MacMillan-Crow LA. Mitochondrial targets of oxidative stress during renal ischemia/reperfusion. Arch. Biochem. Biophys. 2003;412:27–33. doi: 10.1016/s0003-9861(03)00039-0. [DOI] [PubMed] [Google Scholar]
- [7].Agarwal R, MacMillan-Crow LA, Rafferty TM, Saba H, Roberts DW, Fifer EK, James LP, Hinson JA. Acetaminophen-induced hepatotoxicity in mice occurs with inhibition of activity and nitration of mitochondrial manganese superoxide dismutase. J. Pharmacol. Exp. Ther. 2011;337:110–116. doi: 10.1124/jpet.110.176321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. Factors determining the selectivity of protein tyrosine nitration. Arch. Biochem. Biophys. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- [9].Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- [10].Hernandez P, Gras R, Frey J, Appel RD. Popitam: towards new heuristic strategies to improve protein identification from tandem mass spectrometry data. Proteomics. 2003;3:870–878. doi: 10.1002/pmic.200300402. [DOI] [PubMed] [Google Scholar]
- [11].Haussinger D. Nitrogen metabolism in liver: structural and functional organization and physiological relevance. Biochem. J. 1999;267:281–290. doi: 10.1042/bj2670281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rubio V, Britton HG, Grisolia S. Mitochondrial carbamoyl phosphate synthetase activity in the absence of N-acetyl-L-glutamate. Mechanism of activation by this cofactor. Eur. J. Biochem. 1983;134:337–343. doi: 10.1111/j.1432-1033.1983.tb07572.x. [DOI] [PubMed] [Google Scholar]
- [13].Zhao P, Kalhorn TF, Slattery JT. Selective mitochondrial glutathione depletion by ethanol enhances acetaminophen toxicity in rat liver. Hepatology. 2002;36:326–335. doi: 10.1053/jhep.2002.34943. [DOI] [PubMed] [Google Scholar]
- [14].O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- [15].Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I. Structure of Carbamoyl Phosphate Synthetase: A Journey of 96 Å from Substrate to Product. Biochemistry. 1997;36:6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- [16].Pekkala S, Martinez AI, Barcelona B, Gallego J, Bendala E, Yefimenko I, Rubio V, Cervera J. Structural insight on the control of urea synthesis: identification of the binding site for N-acetyl-L-glutamate, the essential allosteric activator of mitochondrial carbamoyl phosphate synthetase. Biochem. J. 2009;424:211–220. doi: 10.1042/BJ20090888. [DOI] [PubMed] [Google Scholar]
- [17].Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modeling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- [18].Craig A, Sidaway J, Holmes E, Orton T, Jackson D, Rowlinson R, Nickson J, Tonge R, Wilson I, Nicholson J. Systems toxicology: integrated genomic, proteomic and metabonomic analysis of methapyrilene induced hepatotoxicity in the rat. J. Proteome Res. 2006;5:1586–1601. doi: 10.1021/pr0503376. [DOI] [PubMed] [Google Scholar]
- [19].Murray J, Taylor SW, Zhang B, Ghosh SS, Capaldi RA. Oxidative damage to mitochondrial complex I due to peroxynitrite: identification of reactive tyrosines by mass spectrometry. J. Biol. Chem. 2003;278:37223–37230. doi: 10.1074/jbc.M305694200. [DOI] [PubMed] [Google Scholar]
- [20].Pekkala S, Martónez AI, Barcelona B, Yefimenko I, Finckh U, Rubio V, Cervera J. Understanding carbamoyl-phosphate synthetase I (CPS1) deficiency by using expression studies and structure-based analysis. Hum. Mutat. 2010;3:801–808. doi: 10.1002/humu.21272. [DOI] [PubMed] [Google Scholar]
- [21].Yamakura F, Taka H, Fujimura T, Murayama K. Inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J. Biol. Chem. 1998;273:14085–14089. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]
- [22].Batthyany C, Souza JM, Duran R, Cassina A, Cervenansky C, Radi R. Time course and site(s) of cytochrome c tyrosine nitration by peroxynitrite. Biochemistry. 2005;44:8038–8046. doi: 10.1021/bi0474620. [DOI] [PubMed] [Google Scholar]
- [23].Wang Y, Luo Y, Zhong R, Gao D, Cui S. Identification of site(s) of insulin nitration by peroxynitrite and characterization of its structural change. Protein Pept. Lett. 2008;15:1063–1067. doi: 10.2174/092986608786071111. [DOI] [PubMed] [Google Scholar]