Abstract
DEK is a biochemically distinct protein that is generally found in the nucleus, where it is vital to global heterochromatin integrity. However, DEK is also secreted by cells (eg, macrophages) and influences other adjacent cells (eg, acts as a chemoattractant for certain mature blood cells). We hypothesized that DEK may modulate functions of hematopoietic stem (HSCs) and progenitor (HPCs) cells. C57Bl/6 mice were used to demonstrate that absolute numbers and cycling status of HPCs (colony forming unit-granulocyte macrophage [CFU-GM], burst forming unit-erythroid [BFU-E], and colony forming unit-granulocyte erythroid macrophage megakaryocyte [CFU-GEMM]) in bone marrow (BM) and spleen were significantly enhanced in DEK −/− as compared with wild-type (WT) control mice. Moreover, purified recombinant DEK protein inhibited colony formation in vitro by CFU-GM, BFU-E, and CFU-GEMM from WT BM cells and human cord blood (CB) cells in a dose-dependent fashion, demonstrating that DEK plays a negative role in HPC proliferation in vitro and in vivo. Suppression was direct acting as determined by inhibition of proliferation of single isolated CD34+ CB cells in vitro. In contrast, DEK −/− BM cells significantly demonstrated reduced long term competitive and secondary mouse repopulating HSC capacity compared with WT BM cells, demonstrating that DEK positively regulates engrafting capability of self-renewing HSCs. This demonstrates that DEK has potent effects on HSCs, HPCs, and hematopoiesis, information of biological and potential clinical interest.
Introduction
Hematopoiesis is regulated by cell-cell and cytokine-cell interactions on hematopoietic stem (HSCs) and progenitor (HPCs) cells [1]. Extracellular and intracellular players involved in this regulation continue to be identified, and understanding these factors is crucial to modulating hematopoiesis for clinical benefit.
In our continuing efforts to elucidate players involved in regulation of HSC and HPC growth [1], we focused on DEK, an abundant and unusual protein found in multicellular organisms [2]. DEK has 2 DNA binding modules and has some affinity for specific DNA sequences, but primarily recognizes and binds to superhelical and cruciform DNA and induces positive supercoiling. DEK manifests multiple cellular activities that include transcriptional repression and activation, mRNA processing, and chromatin architectural functions [2]. We recently demonstrated that DEK modulates global heterochromatin integrity in vivo [3]. Interestingly, DEK, an autoantigen in juvenile idiopathic arthritis (JIA), can leave the cell and act as a chemoattractant for CD8+T cells and natural killer cells [4]. Its secretion from macrophages is modulated by casein kinase 2 and interleukin (IL)-8, while being inhibited by dexamethasone and cyclosporine A [5]. Further, DEK is present in synovial fluid and in immune complexes of patients with JIA, and the chemotactic activity of DEK suggest that DEK may contribute to joint inflammation [4]. DEK autoantigenicity is augmented by acetylation. DEK is an oncogene that is overexpressed in multiple different malignancies [6,7], and is involved in melanoma proliferation and chemoresistence [6,7], promotion of epithelial transformation in vitro and in vivo [8], and in the pathogenesis of breast cancer [9].
Being intrigued that a nuclear protein was able to be secreted by hematopoietic cells, and act on other hematopoietic cells, we hypothesized that DEK might play a role in HSC/HPC function and hematopoiesis. Towards this possibility, we utilized recombinant (r) DEK protein, and DEK −/− mice, to demonstrate that DEK is a positive regulator of long-term repopulating HSC proliferation and engraftment, and a negative regulator of HPC proliferation.
Materials and Methods
Recombinant human His-DEK
Recombinant human His-DEK (rhu DEK) was purified from insect cells essentially as described [10].
Three days postinfection with a high-titer virus stock, HighFive cells were harvested and washed 3 times with phosphate-buffered saline prior to lysis with 2 mL of lysis buffer per 175-cm2 flask (100 mM Tris-Cl (pH 7.5), 150 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 1% NP-40, 5 mM imidazole). The lysate was further treated with 1.3 M NaCl for 20 min at room temperature, cleared (100,000 g, 10 min), and diluted with lysis buffer to a final concentration of 700 mM NaCl. After incubation for 1 h at 4°C with Ni-nitrilotriacetic acid-agarose (Qiagen), the beads were washed 3 times with 10 volumes of buffer 1 [50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 50 mM imidazole], 3 times with 10 volumes of buffer 2 [50 mM Tris-Cl (pH 7.5), 300 mM NaCl, and 50 mM imidazole], and again with 10 volumes of buffer 1. Elution was performed with 50 mM Tris-Cl (pH 7.5)-150 mM NaCl-500 mM imidazole. Aliquots of recombinant protein were stored at −80°C.
DEK −/− mice
These mice (C57BI/6/129/SVEV) were obtained from Dr. Gerard Grosveld, St Jude Children's Hospital, Memphis, TN [8], and were generated at the University of Michigan to a C57Bl/6 background by back-breeding the −/− mice with WT C57BI/6 mice for 9 generations. Mice were housed in specific pathogen-free conditions at the Animal Maintenance Facility of the University of Michigan Medical Center and used at Indiana University for experiments at 10–13 weeks of age. The University of Michigan and Indiana University Committees on Use and Care of Animals reviewed and approved the animal protocols.
HPC assay
C57Bl/6 mouse bone marrow (BM) and spleen cells were plated respectively at 5×104 and 5×105 cells/mL in 1% methylcellulose culture medium in the presence of hemin 0.1 mM, 30% fetal bovine serum (FBS; Hyclone), and the following growth factors, unless otherwise noted: [1 U/mL rhu erythropoietin (EPO; Amgen), 50 ng/mL recombinant murine (rmu) stem cell factor (SCF; R&D Systems), and 5% vol/vol pokeweed mitogen mouse spleen cell conditioned medium (PWMSCM)] [11]. Colonies were scored after 7 days incubation at 5% C02 and lowered (5%) 02 in a humidified chamber and granulocyte macrophage (CFU-GM), erythroid (BFU-E), and multipotent (CFU-GEMM) progenitors distinguished. When other growth factor combinations were used for stimulation of CFU-GM-derived colonies, no hemin was added. Human cord blood (CB) cells were separated into a low density fraction, and plated at 5×104 cells/mL in 1% methylcellulose culture media with 30% FBS, and various rhu cytokines (EPO, 1 U/mL; IL-3, 10 ng/mL; granulocyte macrophage-colony stimulating factor (GM-CSF), 10 ng/mL; SCF, 50 ng/mL) as noted. rmu and rhu GM-CSF, rmu macrophage (M)-CSF, rmu, and rhu IL-3 and rmu and rhu SCF were from R&D Systems [11]. Before sorting, the CD34+ population of hu CB was enriched via MACS (Miltenyi Biotec). After enrichment via MACS, cells were stained with FITC-conjugated anti-CD34 antibodies (Miltenyi Biotec) until sorted by FACSAria (BD Biosciences Immunocytometry Systems). Single CD34+ cells were directly sorted into 1 well of a 96-well microtiter plate containing 100 μL methylcellulose (1%) culture medium, containing Iscove's modified Dulbecco's medium (IMDM), 30% FBS, SCF, IL-3, GM-CSF, and EPO [12].
HSC engrafting studies
All mice used were on a C57Bl/6 strain background. BM cells from DEK −/− or wild type (WT) control mice (CD45.2+) served as donor cells and were used at a 1:1 (2.5×105 to 2.5×105) ratio with BoyJ congenic (CD45.1) competitor cells; they were intravenously transplanted together into primary F1 (dual CD45.1/CD45.2) mice that were given a lethal dose (950 cGy) of gamma irradiation [13]. For secondary transplants BM cells from primary mice engrafted for 6 months were transplanted at 5×105 cells into secondary lethally irradiated F1 mice in a noncompetitive transplant assay [13]. Results are given as percent CD45.2+ donor cell chimerism in the peripheral blood or BM at the times noted.
Statistical analysis
In vitro and in vivo differences between WT and DEK −/− mice were assessed by double-tailed student's t-test. Significance for single cell assays was determined by Fischer's exact test. P values of at least P<0.05 were considered significant.
Results and Discussion
HPC proliferation in DEK −/− mice
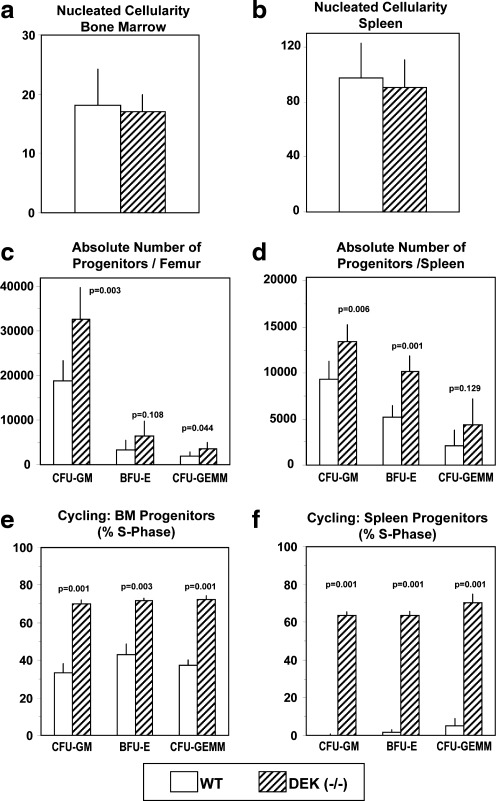
To determine whether DEK had an effect on steady state hematopoiesis, BM and spleen cells from DEK −/− mice were compared with that of WT control mice for absolute numbers and cycling status of CFU-GM, BFU-E, and CFU-GEMM. Although there were no differences in BM (Fig. 1a) or spleen (Fig. 1b) nucleated cellularity between DEK −/− and WT mice, the absolute numbers of CFU-GM, BFU-E, and CFU-GEMM per femur (Fig. 1c) and per spleen (Fig. 1d) were increased in DEK −/− mice with significant increases in BM CFU-GM and CFU-GEMM, and in splenic CFU-GM and BFU-E. Increases in DEK −/− progenitors in BM and spleen were consistent with significantly increased percentages of CFU-GM, BFU-E, and CFU-GEMM in S-Phase of the cell cycle in DEK −/− BM (Fig. 1e) and spleen (Fig. 1f). Increased numbers and cycling of HPCs in DEK −/− BM and spleen suggest that DEK acts as a negative regulator of HPC proliferation in vivo.
FIG. 1.
Hematopoietic progenitor cell numbers and cycling characteristics (% in S-Phase of the cell cycle) in bone marrow (femur) and spleens of DEK −/− and WT control mice. Results (mean±1 SEM) are based on 6 individually assessed mice per group for each progenitor cell population in a total of 2 separate experiments. P values compare DEK −/− with WT mice. WT, wild type; SEM, standard error of the mean; CFU-GM, colony forming unit-granulocyte macrophage; BFU-E, burst forming unit-erythroid; CFU-GEMM, colony forming unit-granulocyte erythroid macrophage megakarocyte.
Influence of rhu DEK on colony formation in vitro by HPCs
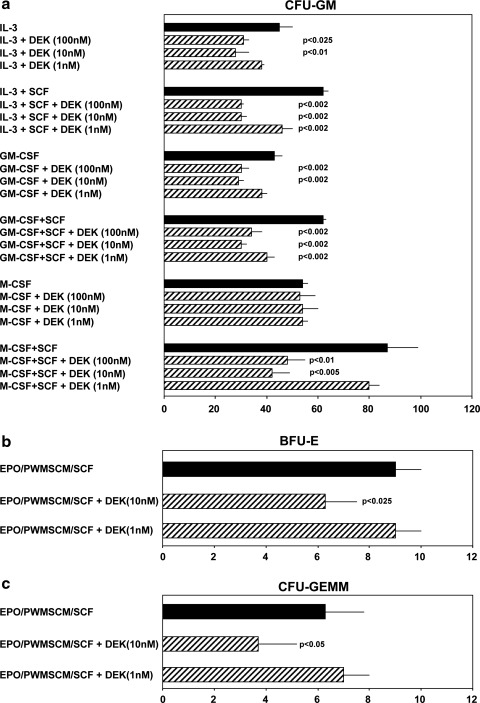
To assess this negative role for DEK further, rhu DEK was tested for effects on HPC proliferation using unseparated mouse BM (Fig. 2) and low density hu CB (Fig. 3) cells. DEK, dose-dependently suppressed colony formation by mouse BM CFU-GM stimulated by either IL-3 or GM-CSF, each alone; it did not influence colony formation stimulated by M-CSF alone (Fig. 2a). However, it dose-dependently inhibited CFU-GM colony formation by either IL-3, GM-CSF, or M-CSF when these cytokines were combined with the potent co-stimulating cytokine SCF. In fact, inhibition by DEK was greater on CFU-GM stimulated by the combination of IL-3, GM-CSF, or M-CSF, each in the presence of SCF, compared with CFU-GM stimulated by IL-3, GM-CSF, or M-CSF each alone in terms of percent inhibition, and the amount of DEK required to inhibit colony formation. Although the lowest amount of DEK that could inhibit colony formation of CFU-GM stimulated by IL-3 or GM-CSF alone was 10 nM, concentrations as low as 1 nM DEK could inhibit colony formation stimulated by IL-3 plus SCF, or GM-CSF plus SCF. Although DEK did not inhibit colony formation of CFU-GM stimulated by M-CSF at up to 100 nM, it was active at concentrations as low as 10 nM in suppressing M-CSF plus SCF stimulated colony formation. DEK at concentrations of 10 nM also inhibited colony formation by BFU-E (Fig. 2b) and by CFU-GEMM (Fig. 2c) each stimulated by a combination of growth factors (EPO, PWMSCM, and SCF). It is known that HPCs stimulated by a single CSF (eg, IL-3, GM-CSF, or M-CSF) represent a more mature subset of HPCs than those stimulated by a combination of cytokines (such as a CSF plus SCF) [1], suggesting that immature subsets of HPCs are more sensitive in vitro to the suppressive effects of DEK than are the more mature HPCs.
FIG. 2.
Influence of rhu DEK on colony formation CFU-GM (a), BFU-E (b), and CFU-GEMM (c) in 5×104 unseparated WT control mouse bone marrow cells per milliliter. Results (mean±1 SEM) shown are 1 of 3 reproducible experiments. P values are compared with control values without added DEK. Rhu, recombinant human.
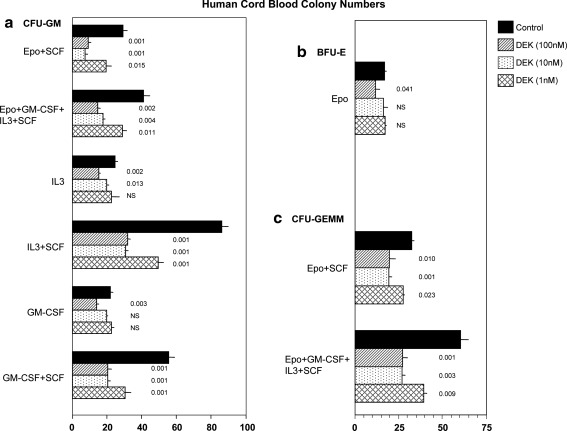
FIG. 3.
Influence of rhu DEK on colony formation by CFU-GM (a), BFU-E (b), and CFU-GEMM (c) per milliliter in low density human cord blood. Results (mean±1 SEM) shown are 1 of 2 reproducible experiments. P values are compared with control values without added DEK.
Similar results were noted for HPCs present in hu CB (Fig. 3). DEK was an inhibitor of CFU-GM (Fig. 3a) and this inhibition was greater and apparent at lower concentrations of DEK when colonies formed from CFU-GM were stimulated by combinations of cytokines, compared with those stimulated by a single cytokine. Colony formation by BFU-E (Fig. 3b) and CFU-GEMM (Fig. 3c) in hu CB were also suppressed by DEK.
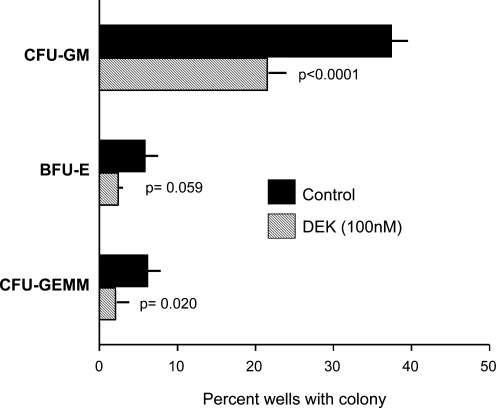
The activity of DEK in vitro in inhibiting colony formation by CFU-GM, BFU-E, and CFU-GEMM (Figs. 2 and 3) is consistent with the increased numbers and cycling of these progenitors in DEK −/− compared with WT BM and spleen (Fig. 1c–f), confirming the negative regulatory effects of DEK on proliferation of HPCs. Since the HPCs in Figs. 2 and 3 were present in a relatively unseparated population of mouse BM and low density cord blood cells, the suppressive effects of DEK on colony formation could have been due to direct effects on the HPCs, or indirectly mediated by effects on accessory cells. To determine if the DEK effects were directly or indirectly manifesting on the HPCs, DEK was assessed for effects on single isolated CD34+ cord blood cells [10], each in a single well stimulated by EPO, GM-CSF, IL-3, and SCF. As seen in Fig. 4, DEK significantly decreased the number of wells containing a CFU-GM-, BFU-E-, or CFU-GEM- colony. The BFU-E effects were borderline significant, but scoring the numbers of wells with either a BFU-E- or CFU-GEMM-colony together resulted in inhibition that attained a P value of P<0.0013. This demonstrates that DEK directly initiates its suppressive effect on HPC. This however, does not rule out the possibility that DEK may also act on other, possibly accessory, cells to influence HPC proliferation.
FIG. 4.
Influence of rhu DEK directly on hematopoietic progenitor cells. DEK was assessed on colony formation by single isolated CD34+ human cord blood cells each in a single well. Results (mean±1 standard deviation) of each experimental point are based on analysis of 288 wells.
Engrafting capability of DEK −/− mouse BM cells
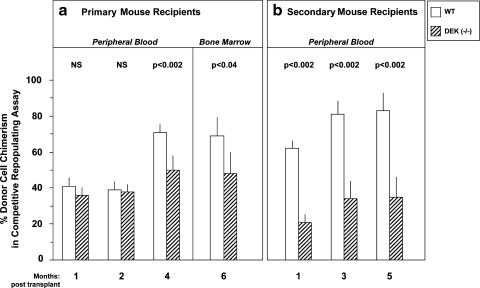
The competitive repopulating assay in vivo allows assessment of the short- and longer-term repopulating HSC, and transplantation of BM cells from primary to secondary lethally-irradiated recipients in a noncompetitive assay describes the longer-term repopulating HSC [1,14], and can offer information on the self-renewal capacity of this population of HSCs. As shown in Fig. 5a, although there was no difference in the HSC repopulating capacity of DEK −/− and WT shorter-term repopulating cells (months 1 and 2 for blood chimerism), there was a significant decrease in DEK −/− compared with WT BM cell repopulation at month 4 in the blood, and month 6 in the BM. This decreased repopulation of DEK −/− compared with WT HSC was even more apparent in secondary mouse recipients (Fig. 5b) suggesting that DEK played a positive role in engraftment of longer-term repopulating HSCs, and perhaps in the self-renewal capacity of mouse BM HSCs. Exactly how DEK acts to influence long term repopulating self-renewing HSCs remains to be determined. The effects may be direct acting on the HSCs, or indirectly mediated by accessory cells in the mouse.
FIG. 5.
Primary and secondary engrafting capability of DEK −/− and WT control donor hematopoietic stem cells. Experiments were designed as noted in the Materials and Methods section. Results are given as donor cell (CD45.2+) chimerism in blood and bone marrow of lethally-irradiated (F1; dual CD45.2+/CD45.1+) recipients. Primary engraftment was with competitor cells (BoyJ; CD45.1+); secondary transplants did not use additional competitor cells. Results are expressed as mean±1 SEM. For primary transplants, bone marrow cells from 2 WT and 2 DEK −/− mice were used. Cells from each mouse were combined with competitor cells and intravenously infused into 5 lethally irradiated recipients each for WT cells, and into 4 and 5 recipients each for DEK −/− cells cells, for a total of 10 primary recipients for WT and 9 primary recipients for DEK −/−. Bone marrow from each group of primary mice, 6 months after transplant were separately pooled (10 for WT; 9 for DEK −/−) and these pools injected without additional competitor cells into 10 (WT) and 9 (DEK −/−) recipients.
The studies shown in Figs. 1–5 demonstrate a here-to-fore unknown role for DEK in the regulation of HPCs, HSCs, and hematopoiesis. DEK could have separate effects on HPC and HSC, as suggested by the direct acting effects of DEK on single HPC. Alternatively, DEK may alone or in addition allow HSC to favor a self-renewal, versus a differentiation pathway to HPC. Exactly how DEK mediates this regulation in the cell is not yet known. This may entail transcriptional effects. DEK is heavily posttranslationally modified, and is acetylated in vivo at lysine residues in the first 70 N-terminal amino acids [15]. Both acelylation and phosphorylation of DEK decrease its affinity for DNA elements in transcriptional promoter regions, consistent with its involvement in transcriptional modulation [3,6,10]. Our recent observation that DEK is vital to global heterochromatin integrity in vivo suggests that this activity may also play a role in the way DEK affects hematopoiesis [3].
It is possible that DEK may be of potential clinical value for enhancing HSC activity/proliferation in vivo, or in an ex-vivo situation. Either of these possibilities would have clinical applicability and relevance, and further assessment of these activities are warranted.
Acknowledgments
These studies were supported by U.S. Public Health Service Grants R01 HL056416 and R01 HL067284 from the NIH to H.E.B., and R01 AI087128 to D.M.M. J.K. was sequentially funded by NIH T32 grants: HL07910 and DK07519 to H.E.B. N.M.-V. was funded by NIH K01 AR055620 and R03 AR056748. F.K. received research support from an Arthritis Foundation Postdoctoral Fellowship and by the START-Program of the Faculty of Medicine, RWTH Aachen. The authors wish to thank Gerard Grosveld for the gift of the DEK knockout mice.
Author Disclosure Statement
The authors have no conflicts of interest to disclose.
References
- 1.Shaheen M. Broxmeyer HE. Shattil SJ. Furie B. Silberstein LE. McGlave P. Heslop H. Anastasi J. The humoral regulation of hematopoiesis. In: Hoffman R, editor; Benz EJ Jr, editor; Hematology: Basic Principles and Practice. 5th. Elsevier Churchill Livingston; Philadelphia, PA: 2009. pp. 253–275. Part III, Chapter 24. [Google Scholar]
- 2.Waldmann T. Scholten I. Kappes F. Hu HG. Knippers R. The DEK protein–an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 3.Kappes F. T Waldmann T. Mathew V. Yu J. Zhang L. Khodadoust MS. Chinnaiyan AM. Luger K. Erhardt S. Schneider R. Markovitz DM. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 2011;25:673–678. doi: 10.1101/gad.2036411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mor-Vaknin N. Punturieri A. Sitwala K. Faulkner N. Legendre M. Khodadoust MS. Kappes F. Ruth JH. Koch A, et al. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol Cell Biol. 2006;26:9484–9496. doi: 10.1128/MCB.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mor-Vaknin N. Kappes F. Dick AE. Legendre M. Damoc C. Teitz-Tennenbaum S. Kwok R. Ferrando-May E. Adams BS. Markovitz DM. DEK in the synovium of patients with juvenile idiopathic arthritis. Arthritis Rheum. 2011;63:556–567. doi: 10.1002/art.30138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khodadoust MS. Verhaegen M. Kappes F. Riveiro-Falkenbach E. Cigudosa JC. Kim DSL. Chinnaiyan AM. Markovitz DM. Soengas MS. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riveiro-Falkenbach E. Soengas MS. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin Cancer Res. 2010;16:2932–2938. doi: 10.1158/1078-0432.CCR-09-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wise-Draper TM. Mintz-Cole RA. Morris TA. Simpson DS. Wikenheiser-Brokamp KA. Currier MA. Cripe TP. Grosveld GC. Wells SI. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Privette Vinnedge LM. R McClaine R. Wagh PK. Wikenheiser-Brokamp KA. Waltz SE. Wells SI. The human DEK oncogene stimulates beta-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30:2741–2752. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kappes F. Damoc C. Knippers R. Przybylski M. Pinna LA. Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK Mol. Cell Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broxmeyer HE. Orschell CM. Clapp DW. Hangoc G. Cooper S. Plett PA. Liles W. Li X. Graham-Evans B, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J. Exp. Med. 2005;201:1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinzfogl J. Hangoc G. Broxmeyer HE. Neurexin iα/Neurexophilin 1 function as a myelosuppressive axis in human cord blood and murine bone marrow hematopoiesis. Blood. 2011;118:565–575. doi: 10.1182/blood-2010-12-325381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell TB. Basu S. Hangoc G. Tao W. Broxmeyer HE. Overexpression of Rheb2 enhances mouse hematopoietic progenitor cell growth while impairing stem cell repopulation. Blood. 2009;114:3392–3401. doi: 10.1182/blood-2008-12-195214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christopherson KW., II Hangoc G. Mantel C. Broxmeyer HE. Modulation of hematopoietic stem cell homing and engraftment by CD26. Science. 2004;305:1000–1003. doi: 10.1126/science.1097071. [DOI] [PubMed] [Google Scholar]
- 15.Cleary J. Sitwala KV. Khodadoust MS. Kwok RPS. Mor-Vaknin N. Cebrat M. Cole PA. Markovitz DM. P300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280:31760–317673. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]