Abstract
Stromal cells and mesenchymal stem cells (MSCs), 2 important cell populations within the hematopoietic microenvironment, may play an important role in the development of hematopoietic stem/progenitor cells. We have successfully cultured human umbilical cord blood-derived stromal cells (hUCBDSCs). It has been demonstrated that MSCs also exist in hUCB. However, we have not found any reports on the distinct characteristics of hUCBDSCs and human umbilical cord blood-derived mesenchymal stem cells (hUCBDMSCs). In this study, hUCBDSCs and hUCBDMSCs were isolated from the cord blood of full-term infants using the same density gradient centrifugation and cultured in the appropriate medium. Some biological characteristics and hematopoietic supportive functions were compared in vitro. hUCBDSCs were distinct from hUCBDMSCs in morphology, proliferation, cell cycle, passage, immunophenotype, and the capacity for classical tri-lineage differentiation. Finally, quantitative real-time polymerase chain reaction analysis revealed that granulocyte colony-stimulating factor (G-CSF) gene expression was higher in hUCBDSCs than that in hUCBDMSCs. Enzyme-linked immunosorbent assay revealed that the secretion of G-CSF, thrombopoietin (TPO), and granulocyte macrophage colony-stimulating factor (GM-CSF) by hUCBDSCs was higher than that by hUCBDMSCs. After coculture, the granulocyte/macrophage colony-forming units (CFU-GM) of hematopoietic cells from the hUCBDSC feeder layer was more than that from the hUCBDMSC feeder layer. Flow cytometry was used to detect CD34+ hematopoietic stem/progenitor cell committed differentiation during 14 days of coculture; the results demonstrated that CD14 and CD33 expression in hUCBDSCs was significantly higher than their expression in hUCBDMSCs. This observation was also true for the granulocyte lineage marker, CD15. This marker was expressed beginning at day 7 in hUCBDSCs. It was expressed earlier and at a higher level in hUCBDSCs compared with hUCBDMSCs. In conclusion, hUCBDSCs are different from hUCBDMSCs. hUCBDSCs are superior to hUCBDMSCs in supporting hematopoiesis stem/progenitor cells differentiation into myeloid lineage cells at an early stage in vitro.
Introduction
As an alternative and attractive source for cellular or gene therapy, human umbilical cord blood (hUCB) is abundant, easy to collect, and has a low probability of pathophoresis [1]. Umbilical cord blood also has a low contaminate rate of malignant cells and low immunogenicity [2,3].
Human umbilical cord blood-derived stromal cells (hUCBDSCs), which were successfully isolated and cultured in our laboratory, are mainly composed of macrophage-like cells, fibroblast-like cells, and small-sphere-like cells [4]. Data from our previous research shows that hUCBDSCs secrete thrombopoietin (TPO), stem cell factor (SCF), and granulocyte macrophage colony-stimulating factor (GM-CSF), which support the development of hematopoietic stem cells (HSCs). In particular, hUCBDSCs are better than human bone marrow stromal cells in the secretion of TPO and the expansion of CFU megakaryocytes both in vitro and in vivo [4,5].
Several research groups have demonstrated that mesenchymal stem cells (MSCs), the precursor cells of stromal cells [6–8], also exist in hUCB [2,9–14]. Human umbilical cord blood-derived mesenchymal stem cells (hUCBDMSCs) obtained from colony-forming unit fibroblasts (CFU-F) display a homogeneous morphology and express some CD molecules that are characteristic of bone marrow-derived MSCs [12]. These cells also have the potential to support hematopoietic stem/progenitor cell proliferation and differentiation in vitro [10,15–17] and have the capacity to differentiate into the 3 classical connective tissue lineages in induction medium [2,12,18,19].
As 2 important cell populations in the hematopoietic microenviroment, stromal cells and MSCs may play an important role in the development of hematopoietic stem/progenitor cells. Thus far, the differences between the biological characteristics of hUCBDSCs and hUCBDMSCs have not been identified. In particular, the use of hUCBDSCs and hUCBDMSCs to support hematopoiesis still needs to be evaluated. The current study focuses on the biological characteristics of hUCBDSCs and hUCBDMSCs and their capacity to support hematopoiesis in vitro. This study supports the potential utility of hUCBDSCs in cell-based therapies.
Materials and Methods
Cell separation of hUCB
hUCB was obtained from consenting parturients who delivered full-term infants at the Department of Obstetrics of South-west Hospital, Chongqing, China. The volume of umbilical cord blood obtained was between 50 and 100 mL (mean volume 84 mL; n=12). The average cell number in hUCB was 2.3×109 cells/mL. Cell separation was performed as previously described [4]. Briefly, the majority of the red blood cells were depleted using the 6% gelatin sedimentation method. The leukocyte-rich fraction was collected in four 50 mL centrifuge tubes and centrifuged at 400×g for 10 min at 4°C. The supernatant was removed, and the pellet was resuspended in 4 mL of Ca2+–Mg2+-free phosphate-buffered saline (PBS). This suspension was then carefully loaded onto Percoll density gradient fractionation columns (density=1.077 g/L; Pharmacia Biotech) of the same volume. Cells were centrifuged at 2,000 rpm for 20 min at 4°C. The mononuclear cells (MNCs) at the interface were centrifuged at 1,000 rpm for 3 min at room temperature after being collected and washed with PBS. The number of hUCB MNCs ranged from 0.5×107 to 12.5×107 cells (mean 7.0×105 MNCs per mL of cord blood). All of these manipulations were completed within 4 h.
Establishment of hUCBDSCs
hUCB MNCs (4×107 cells, 5.3×105 cells/cm2) were cultured in a modified Dexter system to obtain hUCBDSCs. MNCs were resuspended in Dulbecco's modified Eagle medium (DMEM)/F12 medium (Gibco) containing 12.5% fetal bovine serum (Hyclone), 12.5% horse serum (Gibco), and 10 μM hydrocortisone and supplemented with streptomycin (100 ng/mL) and penicillin (100 U/mL). Culture medium was replaced with fresh medium after 48 h. Adherent cells were left undisturbed, and half of the medium was then replaced weekly with fresh medium. When the density of the adherent cells was greater than 80%, hUCBDSCs were subcultured at a 1:2 ratio using the same culture medium and conditions.
Establishment of hUCBDMSCs
hUCB MNCs (4×107 cells, 5.3×105 cells/cm2) were cultured in mesenchymal stem cell medium (MSCM) (ScienCell; Cat. No. 7501) to obtain hUCBDMSCs. MNCs were resuspended in MSCM, and the culture medium was replaced with fresh medium after 48 h. Adherent cells were demidepopulated weekly, and fresh medium was added. When the density of the adherent cells was greater than 80%, hUCBDMSCs were subcultured at a 1:3 ratio using the same culture medium and conditions.
CFU-F assay
The frequency of CFU-F was measured using the following method. A total of 1×107 MNCs isolated from hUCB (n=12) were seeded in DMEM/F12 medium (Gibco) or MSCM (ScienCell; Cat. No. 7501) in T-25 flasks and incubated in a humidified atmosphere with 5% CO2 at 37°C. The medium was demidepopulated every week. The fibroblast-like colonies were counted on day 10 of culture with an inverted microscope. Cell clusters containing >50 cells were scored as CFU-F colonies.
Detecting the cell cycle stage of hUCBDSCs and hUCBDMSCs by flow cytometry
Passage 1 cells were collected after digestion with 0.25% trypsin and prepared by standard methods. Briefly, the harvested cells were washed twice with ice-cold PBS, fixed with 70% cold ethanol, washed again with ice-cold PBS, and incubated with 10 μg/mL RNase A (Sigma). DNA was subsequently stained with 50 μg/mL propidium iodide (Sigma). The cell cycle phase was analyzed by FACS flow cytometry. The cell cycle distribution (G0/G1, S and G2 + M) was quantified using the Cell Fit 2.0 and 2.1 software (Becton Dickinson).
Cells proliferation assay
Cell proliferation assays were performed using the Cell Counting Kit-8 (Dojindo Japan). One hundred microliters of a 5×104 cells/ml single-cell suspension from passage 1 cells were added into a 96-well plate with triplicate wells for each group. Medium was used as a control. The cells were cultured at 37°C with 5% CO2 for 24 h. At the indicated time points, the cell numbers were measured in triplicate by absorbance (450 nm) every day for the next 7 or 10 days.
Phenotypic analysis
Cells from hUCBDSCs (n=4, P0 to P2, P refers to passage) and hUCBDMSCs (n=5, P2 to P6) were stained with phycoerythrin (PE)-conjugated antibodies against CD106 (vascular cell adhesion molecule 1, VCAM-1/CD106) or CD34 (the 2 antibodies were purchased from eBioscience) or with fluorescein isothiocyanate (FITC)-conjugated antibodies against fibronectin (Fn), laminin (Lm) (the 2 antibodies were purchased from Southern-Biotech), CD45, CD44, CD29 (the 3 antibodies were purchased from eBioscience), or Stro-1 (Biolegend). Relative mouse isotypic antibodies were used as the control. Cells were stained with a single label and then analyzed by flow cytometry with an FACScan cytometer (Becton Dickinson).
Multilineage differentiation assays
The potency of hUCBDSC and hUCBDMSC differentiation into various cell types including osteoblasts, adipocytes, and chondrocytes was examined using the Stempro Osteogenesis, Adipogenesis, and Chondrogenesis Differentiation Kits (Gibco Cat. No. A10072-01, A10071-01 and A10070-01, respectively) according to the manufacturer's recommended methods. After induction, the multilineage potential was evaluated by Alizarin Red staining for osteoblasts, accumulation of lipid-rich vacuoles and Oil Red O staining for adipocytes, and by Alcian Blue staining for chondrocytes on day 21. Cells maintained in regular growth medium served as the control group.
Quantitative real-time polymerase chain reaction
Total RNA from hUCBDSCs or hUCBDMSCs was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. One milliliter of TRIzol was used for every 5×106 cells in an RNase free environment. Chloroform was then added (200 μL for every 1 mL of TRIzol), and the samples were centrifuged at 12,000 g for 15 min at 4°C. The aqueous layer was then transferred to a new tube, and the RNA was precipitated with isopropanol followed by 1 wash with 75% ethanol. The RNA precipitate was then dissolved in 20 μL of RNase-free water and analyzed for quantity and quality using a spectrophotometer. The integrity of the total RNA was examined by 1% agarose gel electrophoresis, the quantity was determined based on the absorbance at 260 nm (A260), and the purity was analyzed based on the absorbance ratio at 260 and 280 nm (A260/280) (Beckman; spectrophotometer DU 640). The cDNA was synthesized from 1 μg of total RNA using the PrimeScript™ RT reagent Kit (Takara). Five genes were tested by real-time polymerase chain reaction (RT-PCR) and included granulocyte colony-stimulating factor (G-CSF), SCF, TPO, stromal cell-derived factor 1 (SDF-1), and GM-CSF (primer information is provided in Table 1). Quantitative RT-PCR was performed using an ABI 7500 RT-PCR system (Applied Biosystems) according to the manufacturer's recommendations. We used SYBR Green (Takara) for all the tested genes. The PCR cycling program was as follows: 94°C for 5 min; 30 cycles at 94°C for 10 s, 57°C for 30 s, and 72°C for 30 s; and 72°C for 10 min. Glyceraldehyde phosphate dehydrogenase (GAPDH) served as an internal control. The data analysis was performed using the Sequence Detector System software (ABI), and the results were expressed as a fold change in relative mRNA expression level. This level was calculated using the ΔΔCt method with GAPDH as the reference gene and the hUCBDSCs as the baseline. Each evaluation was performed in triplicate in independent experiments.
Table 1.
Primers for Quantity Real-Time Polymerase Chain Reaction
| Primer | Sequence | Product (bp) |
|---|---|---|
| G-CSF | F: 5′-GCCCCCTTACCCACTACACCATC-3′ | 275 |
| R: 5′-GAGCCAGGCAGTTCCACAGAG-3′ | ||
| TPO | F: 5′-TCGCTGGCTGCTCTGCTGATC-3′ | 215 |
| R: 5′-CGGCCTTCCATCCCAGCACT-3′ | ||
| GM-CSF | F: 5′-TGGGGGTTAGGGCTTGGAAG-3′ | 251 |
| R: 5′-CCACTTGGGCCGTTAACTTCTCT-3′ | ||
| SCF | F: 5′-TTGGGGCACGTTTTCCTACTC-3′ | 222 |
| R: 5′-GGCTCCGGATATCTTTTGCAACA-3′ | ||
| SDF-1 | F: 5′-GGGCGCAGAGAAGAAAAAGTGG-3′ | 139 |
| R: 5′-GCGCCGAGAGCAAGTGAACTG-3′ | ||
| GAPDH | F: 5′-ACCCATCACCATCTTCCAGGAG-3′ | 159 |
| R: 5′-GAAGGGGCGGAGATGATGAC-3′ |
GAPDH, glyceraldehyde phosphate dehydrogenase; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte macrophage colony-stimulating factor; SCF, stem cell factor; SDF-1, stromal cell-derived factor 1; TPO, thrombopoietin.
Enzyme-linked immunosorbent assay
An enzyme-linked immunosorbent assay (ELISA) was performed according to the manufacturer's instructions (R&D Systems) to analyze the concentration of G-CSF, GM-CSF, SCF, SDF-1, and TPO in the supernatants of hUCBDSCs or hUCBDMSCs. After a γ-irradiation dose of 12 Gy for each group of cells (80% confluency) and coculture with an equal amount of hematopoietic cells (from cord blood MNCs), we collected supernatants and analyzed the cytokine concentrations on days 0, 7, and 14.
Coculture of hematopoietic cells colony assays
MNCs were isolated from hUCB using Percoll density gradient fractionation (density=1.077 g/L; Pharmacia Biotech). After culture, the CD34+ fraction of suspended MNCs was harvested. After a γ-irradiation dose of 12 Gy, 2.0×105 hUCBDSCs or 2.0×105 hUCBDMSCs were seeded in 24-well plates as feeder layer cells. After 24 h, 1×106 MNCs were seeded. On the fifth day, nonadherent expanded cells were inoculated in methylcellulose-based semisolid cultures (Stem Cell Technologies, Inc.). Cultures were maintained at 37°C and in 5% CO2 humidified air. After 5–14 days of culture, cells with clonogenic potential were assessed by erythroid colony-forming units (CFU-E), granulocyte/macrophage colony-forming units (CFU-GM) and granulocyte, erythrocyte, macrophage, and megakaryocyte colony-forming units (CFU-GEMM) using an inverted microscope. The noncoculture CD34+ fraction in suspended MNCs was used as a control. Colonies were counted and evaluated according to the manufacturer's instructions.
Coculture of hematopoietic stem/progenitor cell phenotypic analysis
hUCB MNCs were CD34+-enriched using immunomagnetic cell sorting (human CD34+ Selection Kit; Catalog 18056; Stemcell Technologies), according to the manufacturer's instructions. After analyzing the percentage of CD34+ cells by flow cytometry, hematopoietic stem/progenitor cells (7.5×104 cells/ml) were seeded in T-25 flasks, which contained confluent, growth-inactivated hUCBDSCs or hUCBDMSCs as a feeder layer. These cultures were grown at 37°C in 5% CO2 humidified air for 14 days.
Hematopoietic stem/progenitor cells were analyzed by flow cytometry (FACSCalibur equipment, Becton Dickinson) on days 7 and 14 using a panel of monoclonal antibodies (FITC- or PE-conjugated; purchased from eBioscience) including CD34 for stem/progenitor cells, CD14, CD15, and CD33 for myeloid lineage cells, CD3 and CD19 for lymphocytes, CD41a for megakaryocytes, and CD71 for erythrocytes. Isotype controls were also prepared for every experiment.
Statistical analysis
The data are presented as the mean±SD. Statistical comparisons were performed using a Student's t-test. P<0.05 was considered significant.
Results
Dynamic observation of hUCBDSCs and hUCBDMSCs
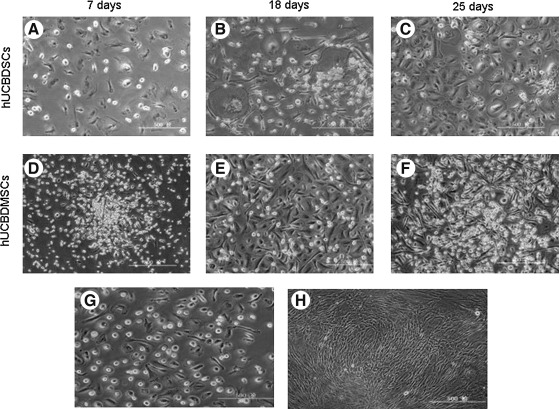
hUCB MNCs were cultured in a modified Dexter system. After 3–4 days, the morphology of the adherent cells varied between round, triangular, fusiform, and irregular. After 7 days, the cells were mainly composed of macrophage-like cells, fibroblast-like cells, and small-sphere-like cells as observed using an inverted microscope. Cells in different visual fields of the same magnification were sorted and counted. The percentages of the 3 types of cells were ∼48% macrophage-like cells, 44% fibroblast-like cells, and 8% small-sphere-like cells (Fig. 1A). Stromal cells did not form colonies during culture (Fig. 1A–C). When the cell density was ∼80% (Fig. 1C), hUCBDSCs were subcultured at a 1:2 ratio. hUCBDSCs can be subcultured for up to 3 generations. During this limited number of passages, cell morphology remained heterogeneous (Fig. 1G). As the number of generations increased, the time to confluence was delayed. Every donor samples was successfully isolated and cultured into hUCBDSCs.
FIG. 1.
Morphology of primary (A–F) and passage (G, H) cultures of hUCBDSCs and hUCBDMSCs. (A), (B) and (C) represent phase-contrast images of primary hUCBDSCs on day 7, 18, 25 respectively. (D), (E) and (F) represent phase-contrast images of primary hUCBDMSCs on day 7, 18, and 25, respectively. (G) represent passage 1 hUCBDSCs on day 10. (H) represent passage 5 hUCBDMSCs on day 5. Original magnification, 100×. Scale bar indicates 500 μm. hUCBDSCs, human umbilical cord blood-derived stromal cells; hUCBDMSCs, human umbilical cord blood-derived mesenchymal stem cells.
hUCB MNCs were cultured in MSCM. The morphology of adherent cells was small fusiform, triangular, or spherical cells at approximately day 7 (Fig. 1D). After 10 days, a few fibrous spindle cells developed into dense colonies and began to grow rapidly (Fig. 1D). However, the growth of cells with other morphologies decreased greatly (Fig. 1E). Cell morphology tended to be homogeneous. At ∼28 days, hUCBDMSCs increased significantly and filled the culture flask (Fig. 1F). The cells were then subcultured at a 1:3 ratio. hUCBDMSCs could be readily expanded in vitro by 12 serial passages with no visible change in morphology (Fig. 1H). With the increase in the number of generations, the time to confluence was gradually delayed. In MSCM, 8 of 12 hUCB MNCs formed fibroblast-like colonies. The mean number of adherent fibroblast-like colonies was 5 per 1×108 MNCs.
Cell cycle and growth curves
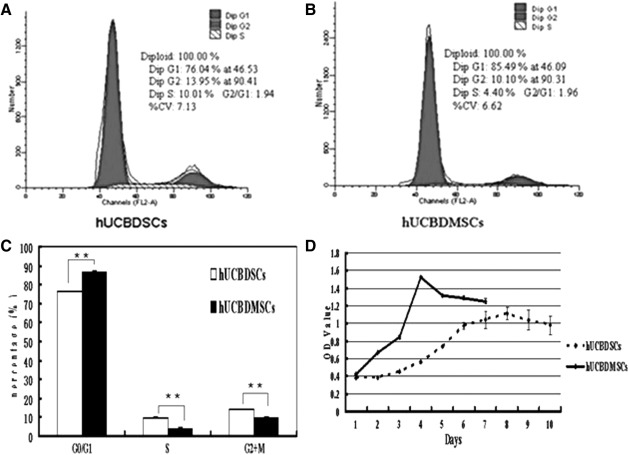
The analysis of the hUCBDSCs cycle indicated that 76.18% of cells were in the G0/G1 phase, 9.87% cells were in the S phase and 13.94% cells were in the G2 + M phase. The analysis of the hUCBDMSCs cycle demonstrated that 86.34% cells were in the G0/G1 phase, 4.14% cells were in the S phase, and 9.61% cells were in the G2 + M phase. Figure 2A and B indicates the representative cell cycle spots. As seen in Fig. 2C, significant differences in the cell cycles of hUCBDSCs and hUCBDMSCs were observed.
FIG. 2.
Cell cycle and growth curves of hUCBDSCs and hUCBDMSCs. (A, B) representative cell cycle plots from hUCBDSCs or hUCBDMSCs. (C) shows the statistical difference of different phases of hUCBDSCs and hUCBDMSCs. (D) shows the growth curves of hUCBDSCs and hUCBDMSCs. The bars represent the mean±SD (n=3), **P<0.01.
The growth curve (Fig. 2D) demonstrated that hUCBDSCs were in the dormancy and adaptation phase during the first 4 days of culture. The cells then began a logarithm growth phase, reaching their peak growth rate at day 8. Finally, the cell growth decreased slightly and entered a stable period. The hUCBDMSCs were in the dormancy and adaptation phase during the first 2 days of culture. They then began a logarithmic growth phase and reached their peak growth rate at day 4. Finally, the cell growths decreased slightly and entered a stable period.
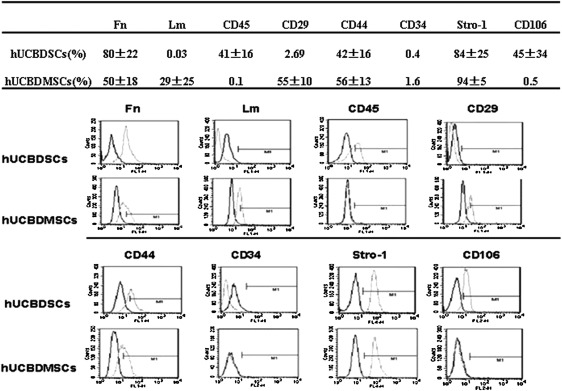
Immunophenotype of hUCBDSCs and MSCs
Figure 3 illustrates that hUCBDSCs shared some of their immunophenotypes with hUCBDMSCs. Both were positive for Fn, CD44, and Stro-1 and negative for CD34. Lm and CD29 were expressed in hUCBDMSCs but not in hUCBDSCs. CD45 and CD106 were expressed in hUCBDSCs but not in hUCBDMSCs.
FIG. 3.
Immunophenotype of hUCBDSCs and hUCBDMSCs. Flow cytometric analysis of cultured hUCBDSCs and hUCBDMSCs with monoclonal antibodies, no differences were found in the expression of Fn, CD44 and Stro-1 between hUCBDSCs and hUCBDMSCs by t-test (P=0.137, 0.293, and 0.565, respectively). Black line histogram indicates isotype contrast; gray line histogram indicates positive reactivity. Fn, fibronectin; Lm, laminin.
Multilineage differentiation capacities
hUCBDSCs and hUCBDMSCs from passage 2–5 (n=3) were used to assess tri-lineage differentiation.
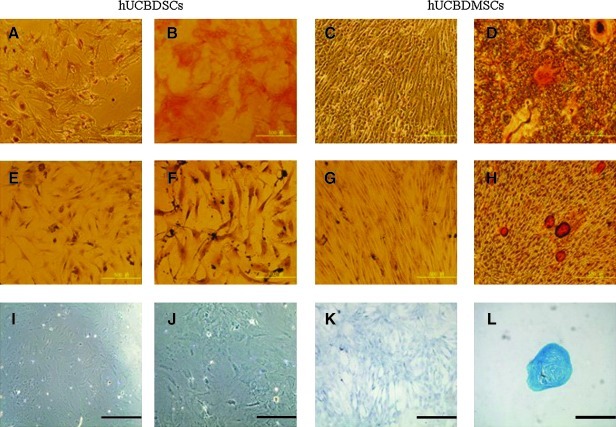
Osteogenic differentiation was detected by matrix calcification after Alizarin Red staining (Fig. 4A–D). The hUCBDMSCs formed mineralized matrix (Fig. 4D). However, the cells were maintained in regular growth medium (Fig. 4A, C), and the osteogenically induced hUCBDSCs (Fig. 4B) did not form mineralized matrix.
FIG. 4.
Osteogenic, chondrogenic and adipogenic differentiation from hUCBDSCs and hUCBDMSCs. (A–D) Osteogenic differentiation was assayed by Alizarin red. (E–H) Adipocytic differentiation was evidenced by the formation of lipid vacuoles by oil-red staining. (I–L) Chondrogenic differentiation was evidenced by Alcian Blue staining. (A, E, I) the control group of hUCBDSCs maintained in the regular medium. (B, F, J) the group of hUCBDSCs in induction condition. (C, G, K) the control group of hUCBDMSCs maintained in the regular medium. (D, H, L) the group of hUCBDMSCs in the induction condition. Original magnification, 100×. Scale bar indicates 500 μm.
Adipogenic differentiation was evidenced by the formation of lipid vacuoles indicated by Oil Red O staining (Fig. 4E–H). The hUCBDMSCs formed lipid vacuoles (Fig. 4H). No lipid vacuoles were found in cells that were maintained in the regular medium (Fig. 4E, G) or in adipogenically induced hUCBDSCs (Fig. 4F).
High-density cells cultured under chondrogenic conditions produced slightly larger pellets. After 3 weeks, the deposition of sulfated glycosaminoglycans was assayed by Alcian Blue staining (Fig. 4I–L). Staining by Alcian Blue was only observed in chondrogenically induced hUCBDMSCs (Fig. 4L). No Alcian Blue were observed in either cells maintained in the regular medium (Fig. 4I, K) or in chondrogenically induced hUCBDSCs (Fig. 4J). All the representative samples are shown at 100×magnification.
Gene expression of growth factors by hUCBDSCs and hUCBDMSCs
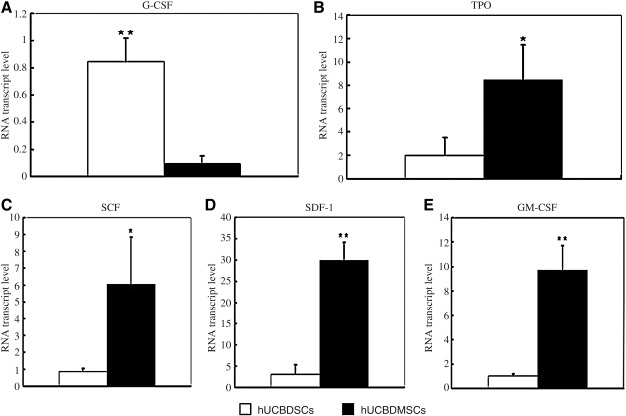
The quantitative RT-PCR results demonstrated that hUCBDSCs and hUCBDMSCs expressed G-CSF, TPO, SCF, SDF-1, and GM-CSF mRNA (Fig. 5A–E). These results also showed that hUCBDSCs expressed lower levels of TPO, SCF, SDF-1, and GM-CSF mRNA (Fig. 5B–E) but higher levels of G-CSF mRNA compared with hUCBDMSCs (Fig. 5A).
FIG. 5.
Gene expression of growth factors by hUCBDSCs and hUCBDMSCs. mRNA was extracted and reversed transcribed to cDNA for real-time polymerase chain reaction analysis. (A) G-CSF gene expression. (B) TPO gene expression. (C) SCF gene expression. (D) SDF-1 gene expression. (E) GM-CSF gene expression. Bars represent the mean±SD of n=3 donor sample; **P<0.01; *P<0.05. G-CSF, granulocyte colony-stimulating factor; SCF, stem cell factor; TPO, thrombopoietin; SDF-1, stromal cell-derived factor 1; GM-CSF, granulocyte macrophage colony-stimulating factor.
Cytokine secretion by hUCBDSCs and hUCBDMSCs
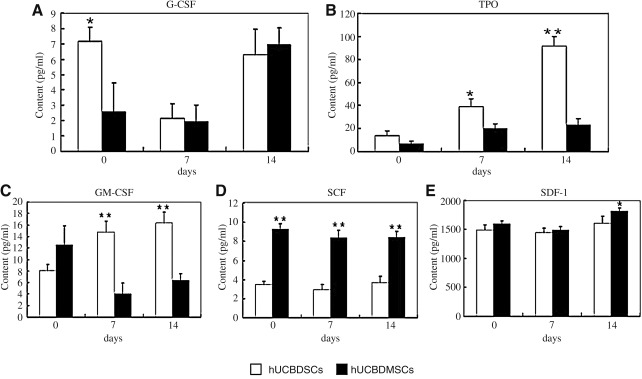
Analysis by ELISA indicated that hUCBDSCs and hUCBDMSCs dynamically secreted G-CSF, TPO GM-CSF, SCF, and SDF-1 when cocultured with hematopoietic cells (Fig. 6A–E). On day 0, the level of G-CSF was higher in the hUCBDSC group than in the hUCBDMSC group (Fig. 6A); the level of SCF was lower in the hUCBDSC group than in the hUCBDMSC group (Fig. 6D). The levels of TPO, GM-CSF and SDF-1 were comparable (Fig. 6B, C, E). The levels of G-CSF, SCF, and SDF-1 decreased on day 7, then increased in each group on day 14 (Fig. 6A, D, E). The level of TPO in each groups demonstrated a marked increase during the 14 days of growth, and the TPO level of the hUCBDSC group was higher than that of the hUCBDMSC group at the 3 time points (Fig. 6B). The level of GM-CSF in the hUCBDMSC group showed a decrease on day 7 and an increase on day 14, whereas the GM-CSF level increased gradually in the hUCBDSC group. Additionally, the level of GM-CSF in the hUCBDSC group was significantly higher than in the hUCBDMSC group at the last 2 time points (Fig. 6C).
FIG. 6.
Cytokine secretion of hUCBDSCs and hUCBDMSCs analysis by enzyme-linked immunosorbent assay. (A) G-CSF secretion. (B) TPO secretion. (C) GM-CSF secretion. (D) SCF secretion. (E) SDF-1 secretion. Bars represent the mean±SD of n=3 donor sample; **P<0.01; *P<0.05.
Expansion of hUCB-derived hematopoietic cells on a hUCBDSC feeder layer or a hUCBDMSC feeder layer
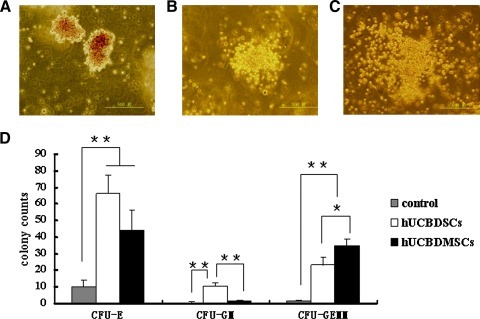
Both the hUCBDSCs and hUCBDMSCs could be used for the expansion of a nonadherent CD34+ fraction in suspended MNCs. Our data showed a significant difference in the quantity of CFU-E, CFU-GM, and CFU-GEMM after the expansion of hematopoietic cells under different culture conditions. Figure 7A–C show the morphology of CFU-E, CFU-GM, and CFU-GEMM, respectively. After 5 days of coculture with hematopoietic cells, the CFU-GM count of hematopoietic cells was greater with the hUCBDSC feeder layer than with the hUCBDMSC feeder layer (Fig. 7D); the CFU-GEMM count from the hUCBDMSC feeder layer was greater than that from the hUCBDSC feeder layer (Fig. 7D). The control group almost did not form colonies (Fig. 7D). The hUCBDSCs were better than hUCBDMSCs at enhancing the expansion of CFU-GM (P<0.01).
FIG. 7.
Expansion of hUCB-derived hematopoietic cells on cell feeder layers. (A) The morphology of CFU-E was composed of more mature erythroid progenitors. Erythroblasts reach maturity by 5–7 days and could be distinguished by the reddish color displayed. (B) The morphology of CFU-GM. Progenitors developed colorless colonies containing macrophages and granulocytes. Individual granulocytes were smaller in size than macrophages. (C) The morphology of CFU-GEMM. Multilineage progenitors developed colonies containing granulocytes, erythrocytes, macrophages, and megakaryocytes. It could be distinguished by reddish cells mixed with colorless cells. (D) hUCBDSCs was superior to hUCBDMSCs on the expansion of CFU-GM, but not on the expansion of CFU-GEMM. Bars represent the mean±SD (n=3); **P<0.01; *P<0.05. CFU-E, erythroid colony-forming units; CFU-GM, granulocyte/macrophage colony-forming units; CFU-GEMM, granulocyte, erythrocyte, macrophage, and megakaryocyte colony-forming units.
HSC committed differentiation
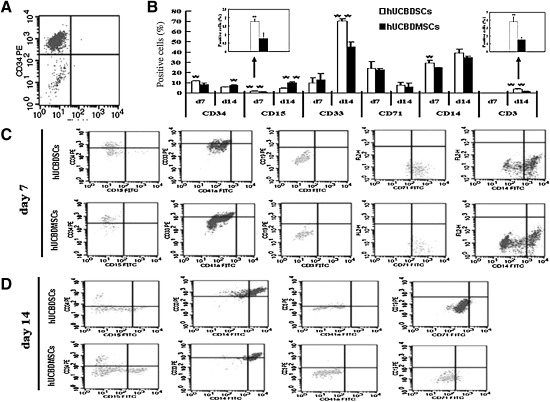
The average percentage of CD34+ HSCs in the CD34+-enriched cell population was determined by flow cytometry analysis to be 90.2% (Fig. 8A).
FIG. 8.
Differentiation potential of expanding cells with hUCBDSCs or hUCBDMSCs. No-adherent cells were harvested periodically and analyzed by flow cytometry with CD34 PE, CD33 PE, CD19 PE, CD3 PE/FITC, CD15 FITC, CD41a FITC, CD71 FITC, and CD14 FITC antibodies. Isotype control antibodies were used to determine the level of nonspecific binding. (A) The percentage of sorting CD34+ hematopoietic stem cells (90.2%). (B) Positive committed cells on day 7 and 14. Bars represent the mean±SD (n=3); **P<0.01; *P<0.05. (C) The significant represent histogram of flow cytometric on day 7. (D) The significant represent histogram of flow cytometric on day 14. FITC, fluorescein isothiocyanate; PE, phycoerythrin.
As shown in Fig. 8B and C, the percentage of CD34+ cells on day 7 was decreased in different groups. The expression of CD19, CD3, and CD41a was negative, whereas the expression of CD33, CD71, and CD14 was positive. The expression of CD15 was positive in the hUCBDSCs group and negative in the hUCBDMSC group (P=0.008). The level of CD14 expression in the hUCBDSC group was higher than in the hUCBDMSC group (P=0.025). The levels of CD33 and CD71 in hUCBDMSCs were comparable with those in hUCBDSCs (P=0.539 and P=0.813, respectively).
As shown in Fig. 8B and D, the percentage of CD34+ cells was further decreased on day 14. The expression of CD19 and CD41a was still negative, whereas the expression of CD15, CD3, CD33, CD71, and CD14 was positive. The levels of CD33 and CD71 significantly increased and decreased, respectively. The CD15 expression level increased in the hUCBDSC group, but was still lower than the level in the hUCBDMSC group (P=0.0005). The hUCBDSC group was better than the hUCBDMSC group at inducing the differentiation of CD34+ HSCs into CD33+ myelocytes and CD3+ lymphocytes (P=0.001 and P=0.004, respectively). During this phase, the expression of the erythrocyte maker CD71 decreased significantly.
Discussion
We successfully cultured hUCBDSCs and hUCBDMSCs from hUCB and compared their characteristics by analyzing cell morphology, CFU-F frequency, cell cycle, cell growth kinetics, immunophenotype, multilineage differentiation potentials, hematopoietic cytokines, and hematopoietic-supportive function. Unlike previously reported methods, we isolated MNCs from umbilical cord blood. The hUCBDMSCs and hUCBDSCs were isolated for culture using the same density gradient centrifugation method (Percoll density=1.077 g/mL). We found that different methods of density gradient centrifugation were used in other studies of MSCs culture. All these results were positive. Haynesworth and coworkers [20] selected a 1.073 g/mL Percoll solution, Lee et al. [12] and Barachini et al. [2] selected a 1.077 g/mL Ficoll-Paque solution, and Jang et al. [15] used a 1.077 g/mL Histopaque. It appears that the type and density of the gradient centrifugation solution were not important factors. However, the CFU-F frequency and MSC proliferation/expansion are dependent on the optimal medium [21,22]. It is well known that the frequency of hUCBDMSCs is deficient [10,23–25], and the successful rate of culture ranges from 23.1% [26] to 63% [10,27]. To increase the achievement ratio of culturing hUCBDMSCs, we used MSCM from ScienCell. While the frequency of fibroblast-like colony formation per 1×106 MNCs from hUCB was less than 3.5±0.7 in the study by Wang et al. [9], ∼8 of the 12 cord bloods samples (66.7%) were successfully isolated and cultured into hUCBDMSCs in the current study. This success rate might be due to the use of optimal culture medium and the fact that sample collection and the subsequent steps never exceeded 4 h. Compared with hUCBDMSCs, hUCBDSCs could be easily isolated from every umbilical cord blood sample and were less likely to form fibroblast-like colonies in a modified Dexter system. The hUCBDMSCs were a homogenous population of fibroblast-like cells that were distinct from hUCBDSCs and were comprised of macrophage-like cells, fibroblast-like cells, and small-sphere-like cells, which were similar to the MSCs and stromal cells isolated from bone marrow [16].
The cell-cycle results demonstrated that more hUCBDMSCs than hUCBDSCs were in the G0/G1 phase (P<0.01). The doubling time of the hUCBDMSC population was shorter than that of the hUCBDSC population. Additionally, the morphological characteristics of the hUCBDMSC population did not change until 12 passages, which is in agreement with the results reported by Wang et al. [9]. Meanwhile, the population doubling time of hUCBDSCs was longer, and they retained their morphology for 3 generations. These results suggest that hUCBDMSCs possess a higher proliferation capacity than hUCBDSCs. Additionally, hUCBDSCs expressed Fn, CD45, CD44, Stro-1, and CD106 but did not express Lm, CD29, and CD34. hUCBDMSCs expressed Fn, Lm, CD29, CD44, and Stro-1 but did not express CD45, CD34, and CD106. Some of these molecules are important in supporting hematopoiesis, hematopoietic cell migration and homing [28,29]. These results suggest that Lm, CD29, CD45, and CD106 may be used as specific markers to identify hUCBDSCs and hUCBDMSCs. Moreover, cord blood cells have low immunogenicity [2,3]. The expression of HLA-DR might be lower in hUCBDMSCs [21,30,31], and the expression of HLA-II, CD80, CD86, and CD40 is lower in hUCBDSCs [32]. All these characteristics may favor the use of hUCBDSCs and hUCBDMSCs in allogeneic cell therapy.
Our results also demonstrated that hUCBDMSCs possessed the capacity to differentiate into osteoblasts, adipocytes, and chondrocytes under induction conditions in vitro. This result was in agreement with previous reports [2,10,12]. In contrast, hUCBDSCs had no capacity for differentiation. This result demonstrated that hUCBDMSCs have more stem cell characteristics than hUCBDSCs according to the clarification of MSCs by International Society of Cell Therapy [33].
To further examine the hematopoietic-supportive function of hUCBDSCs and hUCBDMSCs, we analyzed several common hematopoietic cytokine genes by quantitative RT-PCR and determined the concentration of hematopoietic cytokines in the supernatant by ELISA. The quantitative RT-PCR results showed that the SCF, TPO, SDF-1, and GM-CSF genes were expressed in both hUCBDSCs and hUCBDMSCs. However, the expression was lower in hUCBDSCs. In contrast, G-CSF gene expression in hUCBDSCs was higher than in hUCBDMSCs (P=0.001). Previous research has shown that MSCs and stromal cells isolated from bone marrow do not expressed the G-CSF and GM-CSF genes unless induced by IL-1α. However, their analysis method in this study was not quantitative; qualitative RT-PCR was the major analysis method [16]. In our study, we did not use IL-1α to induce hUCBDSCs and hUCBDMSCs. Lu et al. [34] confirmed that MSCs from bone marrow did not express G-CSF and GM-CSF, but MSCs from umbilical cord did express the 2 genes. The ELISA results showed that hUCBDSCs and hUCBDMSCs secreted G-CSF, GM-CSF, SCF, SDF-1, and TPO. The secretion of G-CSF, TPO, and GM-CSF by hUCBDSCs was greater than that by hUCBDMSCs. In cocultures of hUCBDSCs or hUCBDMSCs and hematopoiesis cells, the dynamic analysis of cytokines showed that G-CSF, SCF, and SDF-1 secretion decreased on day 7 and increased on day 14. The same change in GM-CSF was observed in the hUCBDMSC group. However, GM-CSF expression increased gradually at the last 2 time points in the hUCBDSC group. This result suggests that hUCBDSCs display increased secretion of GM-CSF when cocultured with hematopoietic cells. The level of TPO in each group increased markedly during the 14 days of culture and at the relevant time points. It was higher in the hUCBDSC group than in the hUCBDMSC group. These results suggest that TPO was not the key cytokine for hematopoietic cells differentiation at early stages in vitro. The increase and accumulation of TPO might contribute to poststage hematopoietic cell differentiation. However, these results also suggest that hUCBDSCs might be better than hUCBDMSCs at promoting thrombocytopoiesis.
The hUCBDSCs group was better than the hUCBDMSC group at enhancing the expansion of CFU-GM in methylcellulose-based semisolid cultures (P=0.002). The hUCBDMSC group was better than the hUCBDSC group at enhancing the expansion of CFU-GEMM (P=0.037). These results suggest that there were more lineage-committed progenitor cells and fewer multipotential progenitor cells in suspended MNCs after a short length of time in coculture with hUCBDSCs than when in coculture with hUCBDMSCs. Hematopoietic cells proliferation and differentiation correlated with hematopoietic growth factors and cytokines [35]. The difference in the colony formation of hematopoietic cells between hUCBDSCs and hUCBDMSCs was due to different levels of cytokines in each cell type. It appeared that hUCBDSCs were better than hUCBDMSCs at inducing hematopoietic cells differentiation into a myeloid lineage in vitro. Subsequent assays of hematopoietic stem/progenitor cells-committed differentiation further confirmed this observation. On day 7, the expression of CD15 was positive in the hUCBDSC group and negative in the hUCBDMSCs group. This result suggests that hematopoietic stem/progenitor cell differentiation into granulocytes was induced earlier by hUCBDSCs than by hUCBDMSCs. On day 14, despite the lower level of CD15 in the hUCBDSC group, the levels of CD33 and CD14 increased significantly more than in the hUCBDMSC group. CD14, CD15, and CD33 were all myeloid lineage cell markers. We also observed that under hUCBDSC and hUCBDMSC coculture conditions, the erythropoiesis marker CD71 demonstrated a transient increase at approximately day 7. We should point out that the assays for hematopoietic stem/progenitor cell-committed differentiation were performed at only 2 time points due to the low amounts of CD34+ cell sorting.
In summary, hUCBDSCs were distinguished from hUCBDMSCs, hUCBDSCs were obtained easily, secreted higher levels of G-CSF, TPO, and GM-CSF, and induced HSCs differentiation into myeloid lineage cells, especially differentiation into the granulocyte lineage at an earlier coculture stage in vitro. Furthermore, hUCBDSCs possessed strengthen potency in promoting thrombocytopoiesis. All these characteristics support the clinic use of hUCBDSCs. Of course, these results should be further confirmed in vivo.
Acknowledgments
The authors thank Mr. Mao-Hua Hu (Central Laboratory, Third Military Medical University, Chongqing, China) and Mr. Ping Wang (Laboratory, Department of Hematology, Second Affiliated Hospital, Third Military Medical University, Chongqing, China) for technical assistance with flow cytometry. This work was funded by grants awarded to Dr. Chen from the National Natural Science Foundation (No. 30971109) and the Chongqing Natural Science Foundation (CSTC2009BA5011).
Author Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Barker JN. Wagner JE. Umbilical cord blood transplantation: current practice and future innovations. Crit Rev Oncol Hematol. 2003;48:35–43. doi: 10.1016/s1040-8428(03)00092-1. [DOI] [PubMed] [Google Scholar]
- 2.Barachini S. Trombi L. Danti S. D'Alessandro D. Battolla B. Legitimo A. Nesti C. Mucci I. D'Acunto M. Cascone MG. Lazzeri L. Mattii L. Consolini R. Petrini M. Morpho-functional characterization of human mesenchymal stem cells from umbilical cord blood for potential uses in regenerative medicine. Stem Cells Dev. 2009;18:293–305. doi: 10.1089/scd.2008.0017. [DOI] [PubMed] [Google Scholar]
- 3.Park KS. Lee YS. Kang KS. In vitro neuronal and osteogenic differentiation of mesenchymal stem cells from human umbilical cord blood. J Vet Sci. 2006;7:343–348. doi: 10.4142/jvs.2006.7.4.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao L. Chen XH. Zhang X. Liu Y. Kong PY. Peng XG. Liu L. Liu H. Zeng D. Human umbilical cord blood-derived stromal cell, a new resource of feeder layer to expand human umbilical cord blood CD34+ cells in vitro. Blood Cells Mol Dis. 2006;36:322–328. doi: 10.1016/j.bcmd.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 5.Gao L. Chen XH. Feng YM. Zhang X. Yu SC. Gao L. Zhang C. Liang X. Peng XG. Wang QY. Human umbilical cord blood-derived stromal cells Multifaceted regulators of megakaryocytopoiesis. Cell Cycle. 2010;9:1342–1353. doi: 10.4161/cc.9.7.11146. [DOI] [PubMed] [Google Scholar]
- 6.Short B. Brouard N. Occhiodoro-Scott T. Ramakrishnan A. Simmons PJ. Mesenchymal stem cells. Arch Med Res. 2003;34:565–571. doi: 10.1016/j.arcmed.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J. Niu C. Ye L. Huang H. He X. Tong WG. Ross J. Haug J. Johnson T, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 8.Erices A. Conget P. Minguell JJ. Mesenchymal progenitor cells in human umbilical cord blood. Br J Haematol. 2000;109:235–242. doi: 10.1046/j.1365-2141.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang JF. Wang LJ. Wu YF. Xiang Y. Xie CG. Jia BB. Harrington J. McNiece IK. Mesenchymal stem/progenitor cells in human umbilical cord blood as support for ex vivo expansion of CD34(+) hematopoietic stem cells and for chondrogenic differentiation. Haematologica. 2004;89:837–844. [PubMed] [Google Scholar]
- 10.Bieback K. Kern S. Kluter H. Eichler H. Critical parameters for the isolation of mesenchymal stem cells from umbilical cord blood. Stem Cells. 2004;22:625–634. doi: 10.1634/stemcells.22-4-625. [DOI] [PubMed] [Google Scholar]
- 11.Kogler G. Sensken S. Airey JA. Trapp T. Muschen M. Feldhahn N. Liedtke S. Sorg RV. Fischer J, et al. A new human somatic stem cell from placental cord blood with intrinsic pluripotent differentiation potential. J Exp Med. 2004;200:123–135. doi: 10.1084/jem.20040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee OK. Kuo TK. Chen WM. Lee KD. Hsieh SL. Chen TH. Isolation of multipotent mesenchymal stem cells from umbilical cord blood. Blood. 2004;103:1669–1675. doi: 10.1182/blood-2003-05-1670. [DOI] [PubMed] [Google Scholar]
- 13.Markov V. Kusumi K. Tadesse MG. William DA. Hall DM. Lounev V. Carlton A. Leonard J. Cohen RI. Rappaport EF. Saitta B. Identification of cord blood-derived mesenchymal stem/stromal cell populations with distinct growth kinetics, differentiation potentials, and gene expression profiles. Stem Cells Dev. 2007;16:53–73. doi: 10.1089/scd.2006.0660. [DOI] [PubMed] [Google Scholar]
- 14.Wang TT. Tio M. Lee W. Beerheide W. Udolph G. Neural differentiation of mesenchymal-like stem cells from cord blood is mediated by PKA. Biochem Biophys Res Commun. 2007;357:1021–1027. doi: 10.1016/j.bbrc.2007.04.046. [DOI] [PubMed] [Google Scholar]
- 15.Jang YK. Jung DH. Jung MH. Kim DH. Yoo KH. Sung KW. Koo HH. Oh WN. Yang YS. Yang SE. Mesenchymal stem cells feeder layer from human umbilical cord blood for ex vivo expanded growth and proliferation of hematopoietic progenitor cells. Ann Hematol. 2006;85:212–225. doi: 10.1007/s00277-005-0047-3. [DOI] [PubMed] [Google Scholar]
- 16.Majumdar MK. Thiede MA. Mosca JD. Moorman M. Gerson SL. Phenotypic and functional comparison of cultures of marrow-derived mesenchymal stem cells (MSCs) and stromal cells. J Cell Physiol. 1998;176:57–66. doi: 10.1002/(SICI)1097-4652(199807)176:1<57::AID-JCP7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Pittenger MF. Mackay AM. Beck SC. Jaiswal RK. Douglas R. Mosca JD. Moorman MA. Simonetti DW. Craig S. Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 18.Ye ZQ. Burkholder JK. Qiu P. Schultz JC. Shahidi NT. Yang NS. Establishment of an adherent cell feeder layer from human umbilical cord blood for support of long-term hematopoietic progenitor cell growth. Proc Natl Acad Sci U S A. 1994;91:12140–12144. doi: 10.1073/pnas.91.25.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin HS. Bicknese AR. Chien SN. Bogucki BD. Quinn CO. Wall DA. Multilineage differentiation activity by cells isolated from umbilical cord blood: expression of bone, fat, and neural markers. Biol Blood Marrow Transplant. 2001;7:581–588. doi: 10.1053/bbmt.2001.v7.pm11760145. [DOI] [PubMed] [Google Scholar]
- 20.Le Blanc K. Tammik L. Sundberg B. Haynesworth SE. Ringden O. Mesenchymal stem cells inhibit and stimulate mixed lymphocyte cultures and mitogenic responses independently of the major histocompatibility complex. Scand J Immunol. 2003;57:11–20. doi: 10.1046/j.1365-3083.2003.01176.x. [DOI] [PubMed] [Google Scholar]
- 21.Fan XB. Liu TQ. Liu Y. Ma XH. Cui ZF. Optimization of primary culture condition for mesenchymal stem cells derived from umbilical cord blood with factorial design. Biotechn Prog. 2009;25:499–507. doi: 10.1002/btpr.68. [DOI] [PubMed] [Google Scholar]
- 22.Motaln H. Schichor C. Lah TT. Human mesenchymal stem cells and their use in cell-based therapies. Cancer. 2010;116:2519–2530. doi: 10.1002/cncr.25056. [DOI] [PubMed] [Google Scholar]
- 23.Wexler SA. Donaldson C. Denning-Kendall P. Rice C. Bradley B. Hows JM. Adult bone marrow is a rich source of human mesenchymal ‘stem’ cells but umbilical cord and mobilized adult blood are not. Br J Haematol. 2003;121:368–374. doi: 10.1046/j.1365-2141.2003.04284.x. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez-Rodriguez M. Reyes-Maldonado E. Mayani H. Characterization of the adherent cells developed in Dexter-type long-term cultures from human umbilical cord blood. Stem Cells. 2000;18:46–52. doi: 10.1634/stemcells.18-1-46. [DOI] [PubMed] [Google Scholar]
- 25.Rebelatto CK. Aguiar AM. Moretao MP. Senegaglia AC. Hansen P. Barchiki F. Oliveira J. Martins J. Kuligovski C, et al. Dissimilar differentiation of mesenchymal stem cells from bone marrow, umbilical cord blood, and adipose tissue. Exp Biol Med (Maywood) 2008;233:901–913. doi: 10.3181/0712-RM-356. [DOI] [PubMed] [Google Scholar]
- 26.Yang SE. Ha CW. Jung M. Jin HJ. Lee M. Song H. Choi S. Oh W. Yang YS. Mesenchymal stem/progenitor cells developed in cultures from UC blood. Cytotherapy. 2004;6:476–486. doi: 10.1080/14653240410005041. [DOI] [PubMed] [Google Scholar]
- 27.Kern S. Eichler H. Stoeve J. Kluter H. Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 28.Turner ML. Masek LC. Hardy CL. Parker AC. Sweetenham JW. Comparative adhesion of human haemopoietic cell lines to extracellular matrix components, bone marrow stromal and endothelial cultures. Br J Haematol. 1998;100:112–122. doi: 10.1046/j.1365-2141.1998.00543.x. [DOI] [PubMed] [Google Scholar]
- 29.Ghaffari S. Dougherty GJ. Eaves AC. Eaves CJ. Diverse effects of anti-CD44 antibodies on the stromal cell-mediated support of normal but not leukaemic (CML) haemopoiesis in vitro. Br J Haematol. 1997;97:22–28. doi: 10.1046/j.1365-2141.1997.d01-2139.x. [DOI] [PubMed] [Google Scholar]
- 30.Oh W. Kim DS. Yang YS. Lee JK. Immunological properties of umbilical cord blood-derived mesenchymal stromal cells. Cell Immunol. 2008;251:116–123. doi: 10.1016/j.cellimm.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Wagner W. Wein F. Seckinger A. Frankhauser M. Wirkner U. Krause U. Blake J. Schwager C. Eckstein V. Ansorge W. Ho AD. Comparative characteristics of mesenchymal stem cells from human bone marrow, adipose tissue, and umbilical cord blood. Exp Hematol. 2005;33:1402–1416. doi: 10.1016/j.exphem.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Hao L. Zhang C. Chen XH. Zou ZM. Zhang X. Kong PY. Liang X. Gao L. Peng XG. Sun AH. Wang QY. Human umbilical cord blood-derived stromal cells suppress xenogeneic immune cell response in vitro. Croat Med J. 2009;50:351–360. doi: 10.3325/cmj.2009.50.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz EM. Le Blanc K. Dominici M. Mueller I. Slaper-Cortenbach I. Marini FC. Deans RJ. Krause DS. Keating A. Clarification of the nomenclature for MSC: the international society for cellular therapy position statement. Cytotherapy. 2005;7:393–395. doi: 10.1080/14653240500319234. [DOI] [PubMed] [Google Scholar]
- 34.Lu LL. Liu YJ. Yang SG. Zhao QJ. Wang X. Gong W. Han ZB. Xu ZS. Lu YX, et al. Isolation and characterization of human umbilical cord mesenchymal stem cells with hematopoiesis-supportive function and other potentials. Hematol J. 2006;91:1017–1026. [PubMed] [Google Scholar]
- 35.Kemp KC. Hows J. Donaldson C. Bone marrow-derived mesenchymal stem cells. Leuk Lymphoma. 2005;46:1531–1544. doi: 10.1080/10428190500215076. [DOI] [PubMed] [Google Scholar]