Abstract
Epidermal growth factor receptor (EGFR) is one of the most commonly altered genes in human cancer by way of over-expression, amplification, and mutation. Targeted inhibition of EGFR activity suppresses signal transduction pathways which control tumor cell growth, proliferation, and resistance to apoptosis. Small molecule tyrosine kinase inhibitors and monoclonal antibodies are among the most common EGFR-targeting agents and have been used clinically for treating various malignancies. This review discusses the successes and challenges of targeting EGFR in human cancer. The genetic alterations of EGFR tend to occur more often in some solid tumors than others, as do the mechanisms of resistance to targeted inhibition. The clinical and basic science experiences with these agents thus far have important implications for the future of therapeutic targeting of EGFR.
Keywords: EGFR, targeting, therapeutic
Introduction
Epidermal growth factor receptor (EGFR), the founding member of a family of four ErbB receptor tyrosine kinases, has been a focus of intense research since its initial purification in 1982[1]. It was first implicated in cancer in 1984, when it was identified as a cellular homolog of the v-erbB Oncogene of avian erythroblastosis virus[2] and found to be amplified in A431 human carcinoma cells[3],[4]. EGFR-mediated intracellular signaling controls many of the functions required for cell growth, migration, and proliferation[5]. Not surprisingly, therefore, EGFR expression is a poor prognostic factor for cancer patients. EGFR is frequently over-expressed and/or mutated in human cancer; in fact, gain-of-function genetic alterations in EGFR are observed in up to 30% of solid tumors [6]. Indeed, certain tumor cells are dependent on EGFR signaling and thus possess an “Oncogene addiction”, which makes this receptor an attractive target for therapy[7]. These features have prompted the development of a number of drugs targeted at EGFR (Table 1), several of which are approved by the US Food and Drug Administration (FDA) and widely used, or are currently being tested for the treatment of specific malignancies[8]–[19].
Table 1. Epidermal growth factor receptor (EGFR) status and mechanisms of resistance to targeting agents.
| Tumor type | EGFR mutation | EGFR expression changes | EGFR targeting agents (FDA-approved for clinical use) | Resistance mechanisms |
| Non-small cell lung cancer | Kinase domain deletions (exon 19), point mutations (exon 21)[41] | Gene amplification[29] | Erlotinib[10], Gefitinib[11] | T790M gatekeeper mutation (50%)[71],[93], elevated c-Met/HGF expression (20%)[72]–[74] |
| Colorectal cancer (metastatic) | Rare[30],[42] | Overexpression, copy number increase[30] | Cetuximab[12], Panitumumab[13] | K-ras[75]–[77], B-raf[79], PIK3CA[78], PTEN[80] mutations |
| Head and neck squamous cell carcinoma | ΔEGFR (42%)[32],[43] | Transcriptional up-regulation[31], copy number increase[32] | Cetuximab[14], Nimotuzumab[15] | Increased EGFR stability, co-activation of HER2[81] |
| Nasopharyngeal cancer | Not detected[45] | Overexpression[35] | Nimotuzumab[16] | Not determined |
| Glioblastoma | Deletions and truncations (most commonly ΔEGFR)[39],[47],[48] | Focal gene amplification[39], chromosome 7 trisomy[34] | Nimotuzumab[17] | PTEN loss[82],[83], RTK co-activation[85] |
| Pancreatic cancer | Rare[44] | Over-expression of EGFR and EGF and/or TGFα[33] | Erlotinib[18] | EGFR-independent activation of downstream signaling[44] |
| Breast cancer (HER2-amplified metastatic) | Rare[26] | Gene overexpression (40%)[25], amplification (6%)[26] | Lapatinib[19] | PIK3CA mutation[86], increased estrogen receptor signaling[87] |
Unfortunately, it has become increasingly apparent that effective targeting of EGFR to achieve significant clinical benefit is not a straightforward matter, as many tumors harbor inherent or acquired resistance to receptor inhibition. Moreover, some of the molecular and genetic alterations that predict response to EGFR inhibitors appear to be unique to specific tumor types. Elucidation of the mechanisms of resistance to EGFR-targeted therapies and an increased understanding of the biology of EGFR in response to these agents are clearly required to improve their efficacy in cancer patients.
EGFR: A Driver of Oncogenesis
Ligand-dependent activation of EGFR kinase causes trans-phosphorylation of tyrosines in the intracellular domain of the wild-type receptor, which creates docking sites for adaptor proteins that mediate downstream signaling processes (Figure 1) [20],[21]. The PI3K/Akt pathway promotes cell growth, survival, and migration as well as resistance to apoptosis in response to EGFR-mediated activation[22]. EGFR also transduces oncogenic signaling through binding of adapter proteins such as Grb2/Sos and Shc to specific tyrosine residues in the intracellular domain, resulting in activation of the Ras/MAPK signaling cascade and a profound increase in cell proliferation and migration[23],[24].
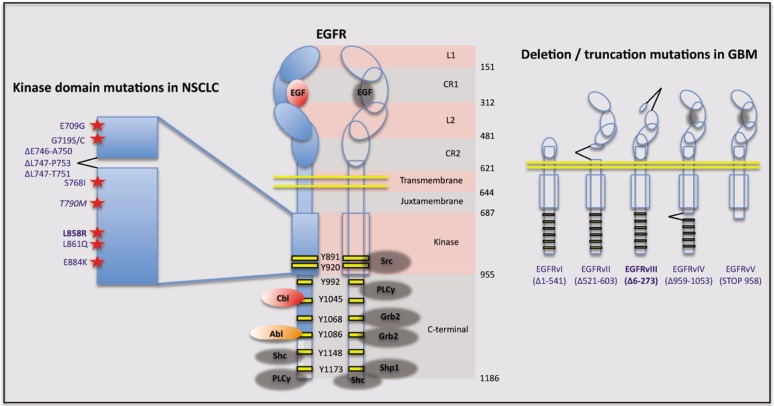
Figure 1. Structural organization, signaling properties, and cancer-associated mutations of epidermal growth factor receptor (EGFR). The domain structure of EGFR is shown, together with the locations of the domain boundaries: L1 and L2, ligand-binding domains 1 and 2; CR1 and CR2, cysteine-rich domains 1 and 2[62]. The major autophosphorylation sites on EGFR, together with the docking proteins and enzymes that are known to associate with these sites to nucleate downstream signaling pathways are shown[62],[63]. Activation of PI3K/Akt signaling by EGFR homodimers is largely driven by recruitment of the p85 regulatory subunit to the Gab1 adaptor protein that binds to Grb2. Along with Shc, Grb2 also mediates activation of Ras signaling by recruitment of the guanine nucleotide exchange factor, SOS. The kinase domain mutations documented in non-small cell lung cancer (NSCLC) and deletion mutations found in glioblastoma (GBM) are detailed, with the most frequent alterations (L858R and ΔEGFR/EGFRvlll respectively) shown in bold [64],[65]. The T790M “gatekeeper mutation” is associated with acquired resistance to Erlotinib in NSCLC (see text for details and further references).
EGFR is expressed at elevated levels in many solid tumors, most often as a result of focal gene amplification or genomic copy number gain[25]–[35]. In some cases, however, over-expression is observed at the protein level in the absence of gene amplification [36]. Overexpression and activation of EGFR is intimately linked to its role in driving tumorigenesis. Activation of EGFR in tumors is often achieved in a ligand-independent manner through somatic mutation of the receptor, and in some cases, these mutations predict response to EGFR-targeted therapies[37],[38]. These mutations (summarized in Figure 1) impart constitutive tyrosine kinase activity to the mutant receptor and result in persistent activation of the downstream oncogenic pathways[39]–[41].
EGFR mutations are tumor-type specific
Although EGFR plays a critical role in the biology of many different tumors, its specific genetic alterations vary depending on tumor type[30],[32],[42]–[45]. More specifically, certain mutations occur at a very high frequency in some tumors but are rare in others. Somatic mutations in the kinase domain, for example, are commonly found in non-small cell lung cancer (NSCLC) while being quite rare in others, such as glioblastoma multiforme (GBM)[53]. These kinase domain mutations typically occur in exons 18–21 and include single base substitutions in exon 18, in-frame deletions in exon 19, insertions in exon 20, and a single base substitution causing a lysine to arginine amino acid change in exon 21 (L858R) (Figure 1)[41]. Exon 19 and 21 mutations, which are the most commonly observed kinase domain mutations in NSCLC, confer tumor cell dependency on EGFR signaling and are predictive of NSCLC response to EGFR tyrosine kinase inhibitors (TKIs). In GBM, the most common EGFR alteration is a deletion of exons 2–7 in the extracellular domain to form the constitutively active ΔEGFR (also referred to as EGFRvlll or EGFR*)[47],[48]. Tumors, therefore, appear to take different, yet specific, means of altering EGFR to reach the common end of activating receptor-regulated oncogenic signaling pathways and processes. By extension, one could argue that although activation of oncogenic signaling is a common end, the tendency to mutate EGFR in distinct ways is reflective of the tumor's preference for specific downstream signaling pathways, such as Ras/MAPK versus PI3K/AKT[49],[50]. Notably, in solid tumors like NSCLC, EGFR mutations that drive Ras activation are more frequent than those that selectively stimulate PI3K/AKT activation[51], while the opposite is true in GBM[52]. Importantly, such mutations have a profound impact on the effectiveness and clinical use of EGFR-targeted therapies.
Therapeutic Targeting of EGFR
The frequent somatic alteration and/or overexpression of EGFR in malignant cells compared to normal cells provides a therapeutic window for targeting the receptor. Targeting approaches vary and often exploit the cancer-associated expression of mutant receptors. Some of these include ΔEGFR peptide vaccines [53], EGFR-mediated delivery of cytotoxic agents to intracellular compartments[54], or EGFR kinase inhibition using antibodies or small molecules which block its essential role in promoting tumor growth and survival. The oncogenic pathways driven by EGFR are interconnected in a complex network involving both negative and positive feedback loops that regulate the activity of pathway components in response to stimuli. Thus, while there are many potential targets within these pathways, there is a significant possibility that direct inhibition of one protein will affect activation of another protein and/or pathway and counteract the growth-suppressive effect caused by inhibition of the targeted pathway. For example, c-Raf, a component of the Ras-MAPK pathway, is negatively regulated by activated AKT, such that PI3K/AKT inhibitors could inadvertently drive activation of MAPK signaling [55],[56]. Moreover, analogs of rapamycin show only modest anti-cancer activity because inhibition of mTOR results in activation of AKT through the relief of negative feedback signaling normally mediated by the mTOR effector p70S6K[57]. Thus, individual targeting of the downstream effectors of EGFR is challenging and may require a combinatorial approach targeting multiple points within these pathways. EGFR is situated at the apex of this complex signaling network, which is an advantage of directly targeting the function of the receptor itself rather than components of the pathways it activates. Furthermore, because it is a cell-surface receptor and is a druggable kinase that is over-expressed and often genetically altered in cancer cells, EGFR is a very attractive therapeutic target for cancer therapy.
Small molecule EGFR tyrosine kinase inhibitors
Since the intrinsic kinase activity of EGFR is essential for many of its oncogenic functions, it is amenable to targeting with small molecule TKIs (Table 1)[58]. Gefitinib (Iressa®) and erlotinib (Tarceva® ) are type I ATP-competitive reversible inhibitors and are both based on a 4-anilinoquinazoline backbone structure[59]. In 2005, a phase III clinical trial comparing gefitinib and placebo in advanced recurrent NSCLC failed to demonstrate increased survival[11], prompting the US FDA to restrict the use of gefinitib to patients who had previously benefited or continue to benefit from it. Nevertheless, gefitinib is approved for use in many other countries, in some cases as a first-line therapy. Erlotinib is FDA-approved for treatment of patients with locally advanced or metastatic NSCLC, either following failure of at least one prior chemotherapy or, more recently, as maintenance therapy for patients whose disease has not progressed after four cycles of platinum-based chemotherapy. Lapatinib is a dual EGFR/ErbB2 inhibitor that is used clinically to treat HER-2 positive breast cancer patients who have failed anti-ErbB2/HER2 therapy. Targeting strategies combining lapatinib with the HER2-targeting antibody, trastuzumab, are now being tested [60]. Several irreversible small molecule inhibitors such as HKI-272 and BIBW 2992, which covalently bind to cys797 in the EGFR kinase domain, are currently being tested for various malignancies[61],[62].
Sensitivity to gefitinib and erlotinib was initially predicted by the presence of EGFR protein expression and/or genomic copy number gain/amplification [29]. In many studies, however, presence and/or activation of EGFR protein by immunohistochemistry and FISH do not appear to correlate with drug response in patients[63]. Presence of the gain-of-function somatic mutations in exons 19 and 21 appear to be a more accurate predictor of response to treatment with EGFR TKIs[64]. In fact, the likelihood that a tumor would respond to treatment with gefitinib alone was 60%–80% in patients pre-screened and identified to have an exon 19 or 21 mutation, and was only 1% for patients with no mutation[65],[66].
Monoclonal antibodies directed at EGFR
Unlike intracellular kinases, as a cell surface receptor, EGFR is also amenable to targeting with antibodies specific to its extracellular ligand-binding domain. Several EGFR-specific antibodies have been developed for clinical use (Table 1). Cetuximab, a chimeric monoclonal antibody and panitumumab, a fully humanized antibody, are both FDA-approved for the treatment of colorectal cancer[12],[13]. Nimotuzumab, another humanized antibody, is used clinically for treatment of head and neck squamous cell carcinoma [15] and glioblastoma[17]. Another chimeric antibody, ch806, binds specifically to the activated form of wild-type EGFR and, with even greater affinity, to ΔEGFR and is thus selective for tumors and has no effect on normal tissue[67],[68]. These antibodies function as direct inhibitors of EGFR signaling by binding and locking the receptor in an inactive conformation. However, they may also be used as highly specific vehicles for the delivery of agents such as oncolytic viruses or cytotoxins to tumor cells in which EGFR is overexpressed or mutated[69].
Mechanisms of Resistance to EGFR Inhibition
A number of patients with NSCLC display a dramatic response to erlotinib[10], and treatment with cetuximab significantly prolongs survival in some patients with colorectal cancer[12]. Unfortunately, EGFR inhibitors often fail to elicit a clinical response, even in instances where the tumor expresses high levels of activated receptor, such as GBM. In some cases, an initial response is achieved as measured by decreased tumor burden, but patients often have tumor relapse and tumors quickly grow back because of acquired resistance to receptor inhibition. In others, the patients never clinically respond to the targeting agent and thus possess inherent, or up-front, resistance. One possible explanation for the latter is insufficient inhibition of the target due to poor penetration of the tumor with drug. Alternatively, it is possible that some tumors are intrinsically resistant because they are not dependent on EGFR for maintenance and progression. In this case, inhibition of the receptor at the later stages when tumors are usually diagnosed would fail to affect tumor growth. This scenario would also offer an explanation for those cases where high levels of EGFR expression/activity are noted in the tumor without clinical response to drug. It has, however, been experimentally demonstrated that the mutant ΔEGFR is required for both initiation and maintenance of GBM growth in vivo, suggesting that the receptor is, indeed, a suitable target for therapeutic intervention[70]. Thus, other mechanisms of both inherent and acquired resistance are likely responsible for the inability of EGFR TKIs to elicit a significant and/or lasting clinical response (Table 1) [44],[61],[71]–[87].
Inherent resistance
The inherent ability to circumvent the effects of EGFR inhibition is an issue that limits the clinical usefulness of these agents for a number of solid tumors. The mutational/genetic background of some tumors simply precludes a function for EGFR inhibition since cells are not dependent on this receptor for oncogenic signaling. For instance, the presence of mutant K-Ras is predictive of failure of response to cetuximab in patients with colorectal cancer and to EGFR TKIs in patients with NSCLC[88]. Somatic mutations in K-Ras and EGFR are mutually exclusive in NSCLC[89], suggesting that gain-of-function alterations are non-redundant and often occur at one node of the signaling cascade. Interestingly, despite that they are both oncogenic, K-Ras mutation predicts resistance to EGFR-targeted therapy and renders independency of cells on EGFR, whereas some EGFR mutations render dependency of cells on EGFR and predict response.
In GBM, failure to clinically respond to EGFR TKIs has been correlated with the lack of functional PTEN, a negative regulator of PI3K activity that frequently undergoes loss-of-function mutation or is lost by deletion in human tumors[82]–[84]. The resultant high levels of PI3K/A kt/mTOR pathway activity induced by PTEN loss persist despite EGFR inhibition. Nevertheless, response to EGFR TKIs in GBM does not often correlate with PTEN or even with expression/amplification/mutation of EGFR [63]. It is possible that the inherent intratumoral heterogeneity of GBM contributes to the up-front resistance that many of these tumors possess.
Interestingly, a recent study approached the question of resistance by considering the full complexity of EGFR signaling networks and identified distinct groups of proteins which, when targeted, increased the intrinsic efficacy of EGFR inhibitors on cell lines[90]. One of the most prominent findings of this study was that co-inhibition of EGFR and Aurora kinase A synergistically induced apoptosis and suppressed the activity of intracellular signaling mediators compared to either inhibitor alone. Thus, tumors may possess up-front resistance based on their genetic make-up and resultant lack of EGFR dependency, or may be EGFR-dependent and undergo additional alterations, intrinsic or acquired, that render EGFR dispensable for tumor survival[82].
Acquired resistance
One of the most direct avenues for acquired resistance is the up-regulation and engagement of efflux pumps to effectively remove the drug from its site of activity. Although gefitinib and erlotinib are substrates for the ABCG2 efflux transporter, they inhibit the activity of these pumps and other proteins involved in drug efflux, such as P-glycoprotein, at high concentrations[91],[92]. Another mechanism that directly interferes with the ability of the drug to access its site of activity is the gatekeeper mutation T790M, which is found in approximately 50% of patients with NSCLC that develop drug resistance[71],[93]. This mutation alters the specificity of the ATP-binding pocket such that the affinity for ATP is greatly increased, thereby interfering with the ability of drugs to compete for binding to the receptor[94]. Studies have shown that a small population of circulating lung cancer cells harbor this mutation[95], so it is likely that these cells are selectively expanded during therapy.
Alternatively, co-activation of multiple receptor tyrosine kinases (RTKs) can reduce or diminish the dependence of tumor cells on EGFR-mediated signaling[85], thereby negating the effects of EGFR inhibition on tumor growth. Depending on the tumor type, expression and/or co-activation of some RTKs likely exists before treatment and could cause intrinsic resistance, but some receptors are specifically up-regulated and activated in response to continued EGFR inhibition and appear to directly compensate for EGFR. Up-regulation of c-Met and/or increase in production of the c-Met ligand, hepatocyte growth factor (HGF), occurs in 20% of NSCLCs and is responsible for maintaining oncogenic signaling despite EGFR inhibition by driving ERBB3-dependent PI3K/AKT activation[72]–[74]. In head and neck squamous cell carcinoma, acquired resistance to cetuximab was caused by dys-regulation of EGFR internalization and degradation, resulting in increased EGFR stability, as well as binding to and activation of HER2 [81]. In addition to these well-studied mechanisms of resistance, there are additional mechanisms that are in the early phase of investigation, such as the up-regulation of novel resistance genes responsible for escape from dependence on ΔEGFR in GBM[70].
Perspectives on resistance
Similar to the EGFR-activating genetic alterations, some of the specific mechanisms that cause resistance to EGFR inhibition appear to be dictated by tumor type. The most striking example of this is evident in NSCLC and GBM. The gatekeeper mutation T790M is perhaps the most common mechanism of resistance in NSCLC, but is rarely, if ever, found in GBM, despite that GBM is often refractory to TKI treatment. Intriguingly, erlotinib-treated NSCLC that metastasized to the liver and the central nervous system (CNS) acquired the T790M mutation in the primary tumor and liver metastases, conferring drug resistance to these tumors, whereas the CNS tumors that did not acquire this mutation remained sensitive to erlotinib[96]. In addition, somatic mutations in K-Ras are very rare in GBM but occur at a high frequency in NSCLC. This pattern may indicate a situation in which tumors are dependent on EGFR in both settings, but perhaps in unique and different ways that are dictated by the specific tumor type, genetic context, and/or microenvironment in which they must adapt. Due to the extremely high level of heterogeneity in GBM tumors, for example, there may be a population of cells within the tumor that is EGFR-independent and responsible for resistance in the event of receptor inhibition. Other solid tumors that possess greater homogeneity in their molecular profile and dependence on EGFR may respond more favorably up-front to EGFR inhibition, but later acquire resistance through secondary mutations such as T790M. Further elucidation of these mechanisms and other aspects of EGFR biology combined with efforts in genome sequencing may uncover novel mutations/alterations in EGFR that are predictive of response in tumors with specific origins or genetic backgrounds.
Conclusion
Targeted EGFR therapy has progressed from the identification of the receptor as a driving factor in oncogenesis to the widespread clinical use of EGFR inhibitors. It is now clear that the complex signaling network that EGFR regulates and the distinct differences in this network in specific tumor types must be considered when predicting which patients are most likely to respond to tailored therapies. These differences dictate how tumors respond to EGFR-targeted therapies, as well as how they become resistant. The future success of drugs and other approaches targeting this receptor lies in our ability to identify biologically vulnerable tumors based on their specific genetic alterations. A greater understanding and appreciation of all of these factors will allow for more rational design of novel therapies and new combinations of existing targeted therapies directed against this receptor.
Acknowledgments
This work was supported by an award from the Goldhirsh Foundation; NIH Grant P01-CA95616; NIH (NINDS) Fellowship Award F32NS066519; Fellow Award from the National Foundation for Cancer Research.
References
- 1.Cohen S, Fava RA, Sawyer ST. Purification and characterization of epidermal growth factor receptor/protein kinase from normal mouse liver [J] Proc Natl Acad Sci USA. 1982;79(20):6237–6241. doi: 10.1073/pnas.79.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downward J, Yarden Y, Mayes E, et al. Close similarity of epidermal growth factor receptor and v-erb-b Oncogene protein sequences [J] Nature. 1984;307(5951):521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 3.Ullrich A, Coussens L, Hayflick JS, et al. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells [J] Nature. 1984;309(5967):418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- 4.Merlino G, Xu Y, Ishii S, et al. Amplification and enhanced expression of the epidermal growth factor receptor gene in A431 human carcinoma cells [J] Science. 1984;224(4647):417–419. doi: 10.1126/science.6200934. [DOI] [PubMed] [Google Scholar]
- 5.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy [J] N Engl J Med. 2005;353(2):172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 6.Salomon DS, Brandt R, Ciardiello F, et al. Epidermal growth factor-related peptides and their receptors in human malignancies [J] Crit Rev Oncol Hematol. 1995;19(3):183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein IB. Cancer. Addiction to oncogenes—the achilles heal of cancer [J] Science. 2002;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 8.Giaccone G. Epidermal growth factor receptor inhibitors in the treatment of non–small-cell lung cancer [J] J Clin Oncol. 2005;23(14):3235–3242. doi: 10.1200/JCO.2005.08.409. [DOI] [PubMed] [Google Scholar]
- 9.Ye F, Gao Q, Cai MJ. Therapeutic targeting of EGFR in malignant gliomas [J] Expert Opin Ther Targets. 2010;14(3):303–316. doi: 10.1517/14728221003598948. [DOI] [PubMed] [Google Scholar]
- 10.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non–small-cell lung cancer [J] N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 11.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non– small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (iressa survival evaluation in lung cancer) [J] Lancet. 2005;366(9496):1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer [J] N Engl J Med. 2004;351(4):337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 13.Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer [J] J Clin Oncol. 2007;25(13):1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 14.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck [J] N Engl J Med. 2006;354(6):567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 15.Crombet T, Osorio M, Cruz T, et al. Use of the humanized anti-epidermal growth factor receptor monoclonal antibody h-R3 in combination with radiotherapy in the treatment of locally advanced head and neck cancer patients [J] J Clin Oncol. 2004;22(9):1646–1654. doi: 10.1200/JCO.2004.03.089. [DOI] [PubMed] [Google Scholar]
- 16.Huang XD, Yi JL, Gao L, et al. Multi-center phase II clinical trial of humanized anti-epidermal factor receptor monoclonal antibody h-R3 combined with radiotherapy for locoregionally advanced nasopharyngeal carcinoma [J] Zhonghua Zhong Liu Za Zhi. 2007;29(3):197–201. [in Chinese] [PubMed] [Google Scholar]
- 17.Ramos TC, Figueredo J, Catala M, et al. Treatment of high-grade glioma patients with the humanized anti-epidermal growth factor receptor (EGFR) antibody h-R3: report from a phase I/II trial [J] Cancer Biol Ther. 2006;5(4):375–379. doi: 10.4161/cbt.5.4.2522. [DOI] [PubMed] [Google Scholar]
- 18.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group [J] J Clin Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 19.Johnston S, Pippen J, Pivot X, et al. Lapatinib combined with letrozole versus letrozole and placebo as first-line therapy for postmenopausal hormone receptor-positive metastatic breast cancer [J] J Clin Oncol. 2009;27(33):5538–5546. doi: 10.1200/JCO.2009.23.3734. [DOI] [PubMed] [Google Scholar]
- 20.Jorissen RN, Walker F, Pouliot N, et al. Epidermal growth factor receptor: mechanisms of activation and signalling [J] Exp Cell Res. 2003;284(1):31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 21.Hynes NE, Lane HA. Erbb receptors and cancer: the complexity of targeted inhibitors [J] Nat Rev Cancer. 2005;5(5):341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 22.Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase-akt pathway in human cancer [J] Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 23.Batzer AG, Rotin D, Urena JM, et al. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor [J] Mol Cell Biol. 1994;14(8):5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowenstein EJ, Daly RJ, Batzer AG, et al. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling [J] Cell. 1992;70(3):431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 25.Toi M, Osaki A, Yamada H, et al. Epidermal growth factor receptor expression as a prognostic indicator in breast cancer [J] Eur J Cancer. 1991;27(8):977–980. doi: 10.1016/0277-5379(91)90262-c. [DOI] [PubMed] [Google Scholar]
- 26.Bhargava R, Gerald WL, Li AR, et al. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mrna and protein expression and her-2 status and absence of EGFR-activating mutations [J] Mod Pathol. 2005;18(8):1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 27.Schlegel J, Merdes A, Stumm G, et al. Amplification of the epidermal-growth-factor-receptor gene correlates with different growth behaviour in human glioblastoma [J] Int J Cancer. 1994;56(1):72–77. doi: 10.1002/ijc.2910560114. [DOI] [PubMed] [Google Scholar]
- 28.Wong AJ, Bigner SH, Bigner DD, et al. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification [J] Proc Natl Acad Sci USA. 1987;84(19):6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non–small-cell lung cancer [J] J Nat Cancer Inst. 2005;97(9):643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 30.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study [J] Lancet Oncol. 2005;6(5):279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 31.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of Carcinogenesis in head and neck cancer [J] Cancer Res. 1993;53(15):3579–3584. [PubMed] [Google Scholar]
- 32.Sharafinski ME, Ferris RL, Ferrone S, et al. Epidermal growth factor receptor targeted therapy of squamous cell carcinoma of the head and neck [J] Head Neck. 2010;32(10):1412–1421. doi: 10.1002/hed.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papageorgio C, Perry MC. Epidermal growth factor receptor-targeted therapy for pancreatic cancer [J] Cancer Invest. 2007;25(7):647–657. doi: 10.1080/07357900701522653. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Gines C, Cerda-Nicolas M, Gil-Benso R, et al. Association of chromosome 7, chromosome 10 and EGFR gene amplification in glioblastoma multiforme [J] Clin Neuropathol. 2005;24(5):209–218. [PubMed] [Google Scholar]
- 35.Zheng X, Hu L, Chen F, et al. Expression of Ki67 antigen, epidermal growth factor receptor and Epstein-Barr virus-encoded latent membrane protein (LMP1) in nasopharyngeal carcinoma [J] Eur J Cancer B Oral Oncol. 1994;30(5):290–295. doi: 10.1016/0964-1955(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 36.Toth J, Egervari K, Klekner A, et al. Analysis of EGFR gene amplification, protein over-expression, and tyrosine kinase domain mutation in recurrent glioblastoma [J] Pathol Oncol Res. 2009;15(2):225–229. doi: 10.1007/s12253-008-9082-4. [DOI] [PubMed] [Google Scholar]
- 37.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib [J] N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 38.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy [J] Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 39.Frederick L, Wang XY, Eley G, et al. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas [J] Cancer Res. 2000;60(5):1383–1387. [PubMed] [Google Scholar]
- 40.Imai K, Takaoka A. Comparing antibody and small-molecule therapies for cancer [J] Nat Rev Cancer. 2006;6(9):714–727. doi: 10.1038/nrc1913. [DOI] [PubMed] [Google Scholar]
- 41.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers [J] Int J Cancer. 2006;118(2):257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 42.Barber TD, Vogelstein B, Kinzler KW, et al. Somatic mutations of EGFR in colorectal cancers and glioblastomas [J] N Engl J Med. 2004;351(27):2883–2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 43.Sok JC, Coppelli FM, Thomas SM, et al. Mutant epidermal growth factor receptor (EGFRvlll) contributes to head and neck cancer growth and resistance to EGFR targeting [J] Clin Cancer Res. 2006;12(17):5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 44.Faller BA, Burtness B. Treatment of pancreatic cancer with epidermal growth factor receptor-targeted therapy [J] Biologics. 2009;3:419–428. [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SC, Lim SG, Soo R, et al. Lack of somatic mutations in EGFR tyrosine kinase domain in hepatocellular and nasopharyngeal carcinoma [J] Pharmacogenet Genomics. 2006;16(1):73–74. doi: 10.1097/01.fpc.0000184959.82903.02. [DOI] [PubMed] [Google Scholar]
- 46.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment [J] Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 47.Sugawa N, Ekstrand AJ, James CD, et al. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas [J] Proc Natl Acad Sci USA. 1990;87(21):8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moscatello DK, Holgado-Madruga M, Godwin AK, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors [J] Cancer Res. 1995;55(23):5536–5539. [PubMed] [Google Scholar]
- 49.Huang PH, Mukasa A, Bonavia R, et al. Quantitative analysis of EGFRvlll cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma [J] Proc Natl Acad Sci U S A. 2007;104(31):12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moscatello DK, Holgado-Madruga M, Emlet DR, et al. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor [J] J Biol Chem. 1998;273(1):200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 51.Sanders HR, Albitar M. Somatic mutations of signaling genes in non–small-cell lung cancer [J] Cancer Genet Cytogenet. 2010;203(1):7–15. doi: 10.1016/j.cancergencyto.2010.07.134. [DOI] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways [J] Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sampson JH, Heimberger AB, Archer GE, et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma [J] J Clin Oncol. 2010;28(31):4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai A, Oh S, Chen H, et al. A novel bispecific ligand-directed toxin designed to simultaneously target EGFR on human glioblastoma cells and uPAR on tumor neovasculature [J] J Neurooncol. 2010 Sep 10; doi: 10.1007/s11060-010-0392-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mabuchi S, Ohmichi M, Kimura A, et al. Inhibition of phosphorylation of BAD and Raf-1 by Akt sensitizes human ovarian cancer cells to paclitaxel [J] J Biol Chem. 2002;277(36):33490–33500. doi: 10.1074/jbc.M204042200. [DOI] [PubMed] [Google Scholar]
- 56.Hatakeyama M, Kimura S, Naka T, et al. A computational model on the modulation of mitogen-activated protein kinase (MAPK) and AKT pathways in heregulin-induced ErbB signalling [J] Biochem J. 2003;373(2):451–463. doi: 10.1042/BJ20021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabatini DM. mTOR and cancer: insights into a complex relationship [J] Nat Rev Cancer. 2006;6(9):729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 58.Albanell J, Gascon P. Small molecules with EGFR-TK inhibitor activity [J] Curr Drug Targets. 2005;6(3):259–274. doi: 10.2174/1389450053765888. [DOI] [PubMed] [Google Scholar]
- 59.Rukazenkov Y, Speake G, Marshall G, et al. Epidermal growth factor receptor tyrosine kinase inhibitors: similar but different? [J] Anticancer Drugs. 2009;20(10):856–866. doi: 10.1097/CAD.0b013e32833034e1. [DOI] [PubMed] [Google Scholar]
- 60.Brandes AA, Franceschi E, Tosoni A, et al. Trastuzumab and lapatinib beyond trastuzumab progression for metastatic breast cancer: strategies and pitfalls [J] Exp Rev Anticancer Ther. 2010;10(2):179–184. doi: 10.1586/era.09.156. [DOI] [PubMed] [Google Scholar]
- 61.Engelman JA, Zejnullahu K, Gale CM, et al. Pf00299804, an irreversible pan-ErbB inhibitor, is effective in lung cancer models with EGFR and ErbB2 mutations that are resistant to gefitinib [J] Cancer Res. 2007;67(24):11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- 62.Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non–small-cell lung cancer [J] J Clin Oncol. 2010;28(18):3076–3083. doi: 10.1200/JCO.2009.27.9414. [DOI] [PubMed] [Google Scholar]
- 63.Yung WKA, Vredenburgh JJ, Cloughesy TF, et al. Safety and efficacy of erlotinib in first-relapse glioblastoma: a phase II open-label study [J] Neurooncol. 2010;12(10):1061–1070. doi: 10.1093/neuonc/noq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dahabreh IJ, Linardou H, Siannis F, et al. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non–small cell lung cancer [J] Clin Cancer Res. 2010;16(1):291–303. doi: 10.1158/1078-0432.CCR-09-1660. [DOI] [PubMed] [Google Scholar]
- 65.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma [J] N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 66.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer [J] N Engl J Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 67.Johns TG, Stockert E, Ritter G, et al. Novel monoclonal antibody specific for the de2-7 epidermal growth factor receptor (EGFR) that also recognizes the EGFR expressed in cells containing amplification of the EGFR gene [J] Int J Cancer. 2002;98(3):398–408. doi: 10.1002/ijc.10189. [DOI] [PubMed] [Google Scholar]
- 68.Luwor RB, Johns TG, Murone C, et al. Monoclonal antibody 806 inhibits the growth of tumor xenografts expressing either the de2-7 or amplified epidermal growth factor receptor (EGFR) but not Wildtype EGFR [J] Cancer Res. 2001;61(14):5355–5361. [PubMed] [Google Scholar]
- 69.Allen C, Vongpunsawad S, Nakamura T, et al. Retargeted oncolytic measles strains entering via the EGFRvlll receptor maintain significant antitumor activity against gliomas with increased tumor specificity [J] Cancer Res. 2006;66(24):11840–11850. doi: 10.1158/0008-5472.CAN-06-1200. [DOI] [PubMed] [Google Scholar]
- 70.Mukasa A, Wykosky J, Ligon KL, et al. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence [J] Proc Natl Acad Sci USA. 2010;107(6):2616–2621. doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kosaka T, Yatabe Y, Endoh H, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non–small cell lung cancer and acquired resistance to gefitinib [J] Clin Cancer Res. 2006;12(19):5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 72.Bean J, Brennan C, Shih JY, et al. Met amplification occurs with or without t790m mutations in egfr mutant lung tumors with acquired resistance to gefitinib or erlotinib [J] Proc Natl Acad Sci USA. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non– small cell lung cancer [J] Clin Cancer Res. 2008;14(10):2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 74.Engelman JA, Zejnullahu K, Mitsudomi T, et al. Met amplification leads to gefitinib resistance in lung cancer by activating ErbB3 signaling [J] Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 75.Amado RG, Wolf M, Peeters M, et al. Wild-type kras is required for panitumumab efficacy in patients with metastatic colorectal cancer [J] J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 76.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis [J] Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 77.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer [J] Anticancer Res. 2008;28(6A):3729–3732. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 78.Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies [J] Cancer Res. 2009;69(5):1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 79.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer [J] J Clin Oncol. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 80.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer [J] J Clin Oncol. 2009;27(35):5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 81.Wheeler DL, Huang S, Kruser TJ, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members [J] Oncogene. 2008;27(28):3944–3956. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mellinghoff IK, Cloughesy TF, Mischel PS. PTEN-mediated resistance to epidermal growth factor receptor kinase inhibitors [J] Clin Cancer Res. 2007;13(2):378–381. doi: 10.1158/1078-0432.CCR-06-1992. [DOI] [PubMed] [Google Scholar]
- 83.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors [J] N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 84.Haas-Kogan DA, Prados MD, Tihan T, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib [J] J Nat Cancer Inst. 2005;97(12):880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 85.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies [J] Science. 2007;318(5848):287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 86.Eichhorn PJA, Gili Mi, Scaltriti M, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235 [J] Cancer Res. 2008;68(22):9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia W, Bacus S, Hegde P, et al. A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer [J] Proc Natl Acad Sci USA. 2006;103(20):7795–7800. doi: 10.1073/pnas.0602468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-ras mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non–small-cell lung cancer and metastatic colorectal cancer [J] Lancet Oncol. 2008;9(10):962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 89.Linardou H, Dahabreh IJ, Bafaloukos D, et al. Somatic EGFR mutations and efficacy of tyrosine kinase inhibitors in NSCLC [J] Nat Rev Clin Oncol. 2009;6(6):352–366. doi: 10.1038/nrclinonc.2009.62. [DOI] [PubMed] [Google Scholar]
- 90.Astsaturov I, Ratushny V, Sukhanova A, et al. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies [J] Sci Signal. 2010;3(140):ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kitazaki T, Oka M, Nakamura Y, et al. Gefitinib, an EGFR tyrosine kinase inhibitor, directly inhibits the function of p-glycoprotein in multidrug resistant cancer cells [J] Lung Cancer. 2005;49(3):337–343. doi: 10.1016/j.lungcan.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 92.Li J, Cusatis G, Brahmer J, et al. Association of variant ABCG2 and the pharmacokinetics of epidermal growth factor receptor tyrosine kinase inhibitors in cancer patients [J] Cancer Biol Ther. 2007;6(3):432–438. doi: 10.4161/cbt.6.3.3763. [DOI] [PubMed] [Google Scholar]
- 93.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib [J] N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 94.Yun CH, Mengwasser KE, Toms AV, et al. The t790m mutation in egfr kinase causes drug resistance by increasing the affinity for ATP [J] Proc Natl Acad Sci USA. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung cancer cells [J] N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruppert AM, Beau-Faller M, Neuville A, et al. EGFR-TKI and lung adenocarcinoma with CNS relapse: interest of molecular follow-up [J] Eur Respir J. 2009;33(2):436–440. doi: 10.1183/09031936.00162307. [DOI] [PubMed] [Google Scholar]