Abstract
Penicillium menonorum is described as a new monoverticillate, non-vesiculate species that resembles P. restrictum and P. pimiteouiense. On the basis of phylogenetic analysis of DNA sequences from four loci, P. menonorum occurs in a clade with P. pimiteouiense, P. vinaceum, P. guttulosum, P. rubidurum, and P. parvum. Genealogical concordance analysis was applied to P. pimiteouiense and P. parvum, substantiating the phenotypically defined species. The species P. rubidurum, P. guttulosum, and P. menonorum were on distinct branches statistically excluded from inclusion in other species and have distinct phenotypes.
Keywords: monoverticillate, fungal systematics, congruence analysis, Penicillium
INTRODUCTION
In the course of a screening program to find useful fungi for conversion of organic matter into high-value products such as lipid precursors to biofuels and animal feed formulations, a Penicillium isolated from garden soil in southern California was obtained that could not be placed with confidence in any described species. Sequencing of the ITS region was performed, with sequence analysis showing that this isolate is phylogenetically related to P. pimiteouiense. DNA distance from P. pimiteouiense suggested that it might be an undescribed species.
Additional gene loci (β-tubulin, calmodulin, and DNA replication licensing factor Mcm7) were amplified and sequenced for this isolate and for phylogenetically and phenotypically similar species. On the basis of the phenotypic and phylogenetic distinctions, this isolate is described as a new species.
MATERIALS AND METHODS
Cultures (Table 1) may be obtained from the Agricultural Research Service Culture Collection (NRRL), Peoria, IL (http://nrrl.ncaur.usda.gov). The P. menonorum culture ex-type is available from the Agricultural Research Service Patent Culture Collection (http://nrrl.ncaur.usda.gov). Cultures were maintained on potato-dextrose agar (PDA) during the course of this study. Colony descriptions were based on 7 d growth of cultures on Czapek’s yeast autolysate agar (CYA), malt extract agar (MEA), and glycerol nitrate agar (G25N) at 25 °C, and on CYA at 5 °C and 37 °C as detailed by Pitt (1980). Some color names are taken from Ridgway (1912) and are designated with an upper case R and a plate number.
Table 1.
Provenance of isolates used in this study.
| Species | NRRL Accession No. | Origin |
|---|---|---|
| Penicillium erubescens MB335726a (syn. Eupenicillium erubescens) | 6223 | South Africa: Pretoria: isolated from nursery soil, 1967, culture ex-type |
| Penicillium guttulosum MB266689 | 907 | USA: Utah: isolated from soil, 1927, culture ex-type |
| Penicillium menonorum MB519297 | 50410 | USA: California: isolated from garden soil, 2009, culture ex-type |
| Penicillium parvum MB289101 (syn. Eupenicillium parvum) | 2095 | Nicaragua: isolated from soil, July 1945, A.G. Kevorkian, culture ex-type |
| 6032 | Papua-New Guinea: isolated from soil, ca. 1973, S. Udagawa, culture ex-type of P. papuanum MB319290 | |
| 35488 | Ghana: Tafo: isolated from soil, ca. 1949 | |
| 35492 | Venezuela: isolated from soil, ca. 1976, D.T. Wicklow | |
| Penicillium pimiteouiense MB460126 | 2063 | New Guinea: isolated from tent cloth, ca. 1944, G.W. Martin |
| 25542 | USA: Illinois: Peoria: isolated from human kidney cell culture plate, April 1996, J.T. Hjelle, culture ex-type | |
| 26932 | USA: Illinois: Peoria: isolated from human kidney cell culture plate, November 1997, M.A. Miller-Hjelle | |
| 26933 | USA: Illinois: Peoria: isolated from human kidney cell culture plate, November 1997, M.A. Miller-Hjelle | |
| 28602 | USA: Illinois: Peoria: isolated from human kidney cell culture plate, July 1998, J.T. Hjelle | |
| Penicillium rubidurum MB319295 (syn. Eupenicillium rubidurum) | 6033 | Papua-New Guinea: isolated from soil, 1975, culture ex-type |
| Penicillium vinaceum MB281754 | 739 | USA: Utah: isolated from soil, 1927, culture ex-type |
| 740 | Unknown: obtained from M.B. Morrow, 1936 |
aMB=MycoBank (http://www.mycobank.org/).
Microscope slides were made by teasing apart bits of mycelium in a drop of lactic acid with cotton blue. A Zeiss axioscope with DIC optics was used for microscopic observations. Photomicrographs were taken with a Kodak 14n digital camera attached to the microscope. Micro- and macro-photographs were sized and placed in a plate using Adobe Photoshop v. 6.0.1.
Biomass for DNA extraction was grown in 125 mL flasks containing 25 mL malt extract (ME) broth incubated at 25 °C on a rotary platform (200 rpm). Biomass ca. 0.5 g wet weight was collected by vacuum filtration, placed in micro centrifuge tubes, and freeze-dried. Freeze-dried mycelium was ground to a powder with a sterile pipette tip and DNA was extracted from the powdered biomass using the CTAB method. Purified DNA was stored in TE buffer (Tris 10 mM, EDTA 1 mM, pH 8.0) at -20 °C until needed. DNA was amplified using the primers and conditions detailed in Peterson et al. (2010). Amplified DNA was prepared for sequencing using ExoSAP-IT (www.usbweb.com). DNA sequences were produced using DyeDeoxy v. 3.1 reagents and an ABI 3730 DNA sequencer (www.appliedbiosystems.com). Complementary strand sequences were assembled and corrected using Sequencher (www.genecodes.com). Finished sequences were aligned using CLUSTALW (Chenna et al. 2003), and maximum parsimony trees and bootstrap proportions were calculated using PAUP v. 4.0b10 (Swofford 2003). MrBayes v. 3.12 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003) was used to calculate Bayesian posterior probabilities. DNA sequences used in this study are deposited in GenBank (www.ncbi.nlm.nih.gov) with accession numbers HQ646566–HQ646603, AF033460–AF033462, AF033464, AF037431, and AF037434. Data sets and tree diagrams are deposited at TREEBASE (www.treebase.org).
The initial search to find phylogenetically related species was performed by BLAST searches of GenBank using the ITS sequence from the new species.
RESULTS
Penicillium menonorum S.W. Peterson sp. nov.
MycoBank MB519297
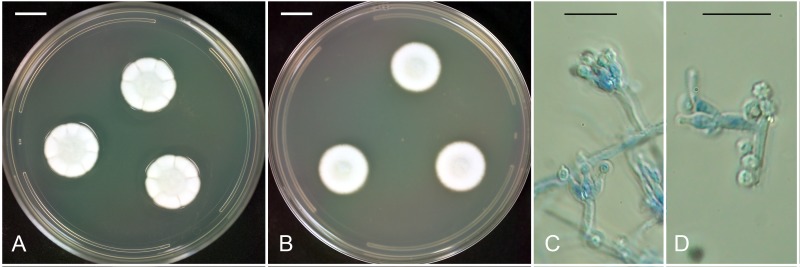
Fig. 1.
Penicillium menonorum NRRL 50410. A. Colonies grown 7 d at 25 °C on CYA showing the radial sulcation and faint blue-gray central color characteristic of the species. Bar = 1 cm. B. Colonies grown 7 d at 25 °C on MEA having wooly consistency and darkened central area where the fungus is sporulating. Bar = 1 cm. C. Conidiophores, phialides and conidia. Bar = 10 μm. D. Roughened conidia. Bar = 10 μm.
Etymology: Named for Menon & Associates whose scientists isolated the fungus.
A speciebus aliis conidiophoris brevibus, conidiis scaberulis, colore in substrato nutritorio CYA pallide caesio atque augmento in temperatura 37 °C distinguendum.
Typus: USA: California: isolated from garden soil, 2009 (BPI 881018 – holotypus; culture ex-holotype NRRL 50410).
Colonies on CYA (Fig. 1A) attaining 17–20 mm diam after 7 d growth at 25 °C, velutinous-silky, radially sulcate peripherally, centrally raised ca. 2–3 mm, sporulation moderate, central region pale bluish gray (court gray R-47), peripheral area white; no exudate or soluble pigments; no sclerotia or ascomata; reverse yellowish brown centrally (buckthorn brown R-15) to pale brownish-yellow (warm buff, R-15) peripherally. On MEA (Fig. 1B) attaining 17–19 mm diam after 7 d growth at 25 °C, mycelium loosely woven, wooly, umbonate 3–4 mm deep centrally, sporulation moderate, white peripherally, court gray (R-47) centrally; no exudate or soluble pigments; no sclerotia or ascomata; reverse yellowish brown centrally to brownish yellow peripherally. On G25N attaining 8–10 mm diam after 7 d growth at 25 °C, umbonate, wooly 1–2 mm deep, white to court gray; no exudate or soluble pigment; no sclerotia or ascomata; reverse white to buff. Incubation for 7 d on CYA at 5 °C produced no growth or germination of conidia. Incubation for 7 d on CYA at 37 °C produced colonies of 29–32 mm diam, resembling growth on CYA at 25 °C, but clear exudate moderately abundant, the reverse color is a darker, more uniform shade of brown. Conidiophores (Fig. 1C) smooth-walled, hyaline, 5–15(–20) × 1.5–2.0 μm , non-vesiculate, with an apical whorl of (1–)2–5 phialides 5–7(–9) × 2.5–3.5 μm, conidia spherical to subspherical, (2–)2.5–3.5μm (Fig. 1D), with roughened to rugose surface.
DNA sequences from the β-tubulin locus included all or part of 4 exon and 4 intron regions. After alignment the data set included 703 base positions. The calmodulin data included all or part of 4 exon and 3 intron regions and aligned with 726 base positions. The ID regions included the ITS1, ITS2, 5.8S rDNA, and ca. 650 bases from the 28S rDNA in an alignment of 1141 bases. DNA replication licensing protein (Mcm7) was composed of an amino acid coding region of 616 bp length. Penicillium erubescens was chosen as the out-group on the basis of phylogenetic trees previously published (Peterson et al. 1999, Peterson 2000).
The most parsimonious trees, bootstrap proportion and Bayesian posterior probabilities for individual data sets were determined and the trees were compared for strongly supported contradictory branch points. Strongly supported nodes are those with > 90 % of the bootstrap sample and a Bayesian posterior probability of > 0.90. The individual locus trees contained no strongly supported contradictions that would preclude combining the data. The data from the four loci were combined to calculate a single phylogenetic tree (Fig. 2).
Fig. 2.
Phylogenetic tree calculated using maximum parsimony criterion for the concatenated data set composed of beta-tubulin, calmodulin, ITS and 28S rDNA, and DNA replication licensing protein (Mcm7). Bootstrap proportions/Bayesian posterior probabilities are placed on internodes.
The five isolates of P. pimiteouiense occur on a single strongly supported branch; three isolates of P. parvum and the single isolate of P. papuanum occur on a different strongly supported branch, and the two P. vinaceum isolates occur on another strongly supported branch. Penicillium rubidurum and P. guttulosum are most closely related to each other and form a sibling group to P. pimiteouiense, while P. menonorum is positioned basal in the tree to this three species branch.
DISCUSSION
Penicillium menonorum is similar phenotypically to P. pimiteouiense, P. restrictum, P. striatisporum, P. vinaceum, P. rubidurum, P. erubescens, and P. parvum. Penicillium restrictum, P. malacaense, P. kurssanovii, P. griseolum, and P. striatisporum, which phenotypically resemble P. menonorum, are phylogenetically positioned in different clades (Peterson & Horn 2009). Other species bearing some resemblance to P. menonorum are either not represented by extant ex-type cultures or the type cultures are not readily available. Penicillium menonorum differs from P. pimiteouiense by producing conidiophores in a basal layer rather than from aerial hyphae and a bluish gray (Court gray R-47) color on CYA versus white in P. pimiteouiense. Additionally, P. pimiteouiense produces yellow exudate and a brown soluble pigment, neither of which appear in P. menonorum after 7 d incubation. On different media (e.g., yeast extract malt agar incubated at 25 °C) or after extended incubation, a clear to rosy exudate often appears in P. menonorum. Penicillium restrictum produces somewhat longer conidiophores (up to 60 μm) and has smaller colonies (< 10 mm diam) at 37 °C than P. menonorum (29–32 mm diam). Penicillium striatisporum produces rosy colored colonies on Czapek’s agar and has striate conidia. Penicillium vinaceum produces copious exudate in yellow to vinaceous colors, yellow to brown soluble pigments, and a dark brown colony reverse on CYA, and colonies grown at 37 °C are somewhat smaller (8–20 mm diam) than those of P. menonorum. Penicillium parvum typically has mycelium that varies from white to yellow to red in color, while the P. menonorum mycelium is uniformly white. Penicillium parvum usually makes brown or purple-brown exudate, a brown soluble pigment, and has a colony reverse that is deep reddish-brown versus P. menonorum, which has no exudate or soluble pigments and a yellow brown colony reverse after 7 d incubation. Penicillium rubidurum produces white to orange or rosy-buff mycelium, red-brown exudate, a dark brown colony reverse, and produces conidia on M40Y medium but not on CYA. Penicillium menonorum produces no exudate or soluble pigment and has a yellow brown reverse and has abundant conidiogenesis on CYA. Penicillium erubescens produces white, pink or flesh color mycelium, reddish-brown exudate, and gray-red to magenta to vinaceous purple soluble pigments, with colony reverse either similarly colored or brown. Each of these species is easily distinguished from P. menonorum on these bases.
Raper & Thom (1949) regarded P. guttulosum to be a synonym of P. janthinellum, differing primarily by the production of copious amounts of exudate. Penicillium guttulosum as represented by Gilman & Abbott’s ex-type strain is distinct from P. janthinellum as well as the species studied here. Penicillium guttulosum cultures on CYA resemble the cultures of P. vinaceum, differing most noticeably in the production of dark purple exudate in large quantities, while P. vinaceum exudate is more red in color. Penicillium rubidurum colonies also resemble P. vinaceum and P. guttulosum but produce pale yellow exudate. Pitt (1980) treated P. papuanum as a synonym of P. parvum and they are in the same strongly supported clade (Fig. 2). Phenotypically, they are very similar to each other. Additional isolates of each species are needed to further assess the phylogenetic and phenotypic distinctions of these species.
Phylogenetic systematics (Hennig 1966) is based on the principle that species must be monophyletic. Taylor et al. (2000) presented the genealogical concordance phylogenetic species recognition (GCPSR) concept as a means of determining the boundaries of species in fungi. Dettman et al. (2006) showed experimentally that GCPSR is effective in recognizing species boundaries in the genus Neurospora. GCPSR can be applied to P. pimiteouiense and P. parvum in this study and the species are supported by the GCPSR principles. Penicillium vinaceum, P. guttulosum, P. rubidurum, and P. menonorum are each on distinct branches, but the boundaries of the species cannot be determined from the single isolates available here. Phenotypic distinctions make each of these species recognizable and the phylogenetic placement of the species is consistent with the phenotypic descriptions of the species.
Acknowledgments
Amy McGovern provided highly skilled technical support that is greatly appreciated. Patricia Eckel kindly translated the diagnosis into Latin. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
REFERENCES
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. (2006) Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia 98: 436–446 [DOI] [PubMed] [Google Scholar]
- Hennig W. (1966) Phylogenetic Systematics. (English Translation). Urbana: University of Illinois Press; [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Peterson SW, Corneli S, Hjelle JT, Miller-Hjelle MA, Nowak DM, Bonneau PA. (1999) Penicillium pimiteouiense: a new species isolated from polycystic kidney cell cultures. Mycologia 91: 269–277 [Google Scholar]
- Peterson SW. (2000) Phylogenetic analysis of Penicillium based on ITS and LSU-rDNA sequences. In: Samson RA, Pitt JI. (eds), Classification of Penicillium and Aspergillus: Integration of modern taxonomic methods: 163–178 Harwood Publishers, UK: [Google Scholar]
- Peterson SW, Horn BW. (2009) Penicillium parvulum and Penicillium georgiense, sp. nov. isolated from the conidial heads of Aspergillus species. Mycologia 101: 71–83 [DOI] [PubMed] [Google Scholar]
- Peterson SW, Jurjevic Z, Bills GF, Stchigel AM, Guarro J, Vega FE. (2010) The genus Hamigera, six new species and multilocus DNA sequence based phylogeny. Mycologia 102: 847–864 [DOI] [PubMed] [Google Scholar]
- Pitt JI. (1980) [‘1977’] The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press, UK: [Google Scholar]
- Raper KB, Thom C. (1949) The genus Penicillium. Williams and Wilkins, USA: [Google Scholar]
- Ridgway R. (1912) Color standards and color nomenclature. Published by the author, USA: [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, USA: [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32 [DOI] [PubMed] [Google Scholar]