Abstract
The morphology and phylogeny of anther smut specimens on Tractema verna collected in the United Kingdom were investigated using light microscopy, scanning electron microscopy and partial rDNA sequence analyses. The anther smut of Tractema verna shows similarity to Antherospora eucomis, A. scillae, A. tourneuxii, A. urgineae, A. vaillantii, and A. vindobonensis but differs in spore size range, spore wall thickness, host plant genera and considerable divergences of ITS and LSU sequences. Consequently, the smut is described here as a new species, Antherospora tractemae. The host plant was formerly included in the genus Scilla (S. verna), but recently moved to a distinct genus Tractema. Molecular phylogenetic analyses reveal that Antherospora tractemae is sister to the lineage of Muscari-parasitizing Antherospora and only distantly related to the Scilla-parasitizing Antherospora species. Thus, the phylogenetic placement of the smut fungus supports the systematic placement of its host plant.
Keywords: Molecular Analysis, Phylogeny, Plant Pathogens, Scilla verna, Smut Fungi, Tractema verna, Coevolution, Ustilaginomycotina
INTRODUCTION
The smut fungi sporulating in the anthers and on the surface of the inner floral organs of different Hyacinthaceae have recently been accommodated in a separate genus Antherospora (Bauer et al. 2008). Antherospora resides in the family Floromycetaceae (Urocystidales), together with the genus Floromyces, which produces sori in the inner floral organs of Anemarrhena asphodeloides (Agavaceae) (Vánky et al. 2008). Antherospora includes eight species, parasitic on hosts in seven different plant genera (Bauer et al. 2008, Vánky 2009). Despite phenotypic similarity, molecular phylogenetic analyses of Antherospora specimens parasitic on species of Muscari and Scilla have revealed significant genetic divergence between accessions from different host species (Bauer et al. 2008). For example, two closely related Central European Scilla species, S. bifolia and S. vindobonensis, harbour two morphologically similar but phylogenetically different Antherospora species. On the other hand, the phylogenetic results demonstrated that Antherospora vaillantii s. str. could infect two different hosts, Muscari comosum and M. neglectum (Bauer et al. 2008), indicating that some Antherospora spp. infect more than one host species. It is probable that host specificity is a widespread phenomenon and evolutionary driver in the genus Antherospora, similar, for example, to the anther smuts classified in the genus Microbotryum (Lutz et al. 2005, 2008, Le Gac et al. 2007, Refrégier et al. 2008, Denchev et al. 2009, Kemler et al. 2009, Piçtek et al. unpubl. data). Nevertheless, the DNA sequence data for Antherospora available in the NCBI’s GenBank nucleotide database is scant due, in part, to the inaccessibility of recently collected material. Much collecting and sequencing effort is necessary to understand the level of host specificity and the phylogenetic relationships within the genus.
Infected specimens of Tractema verna were collected recently in Wales and the Outer Hebrides (United Kingdom). The host plant is commonly known by its synonym, Scilla verna, while its anther smut has been referred to as Ustilago vaillantii (Vánky 1994, Legon et al. 2005). This study aimed to clarify whether the collected specimens could be assigned to any of the described Antherospora species, especially to A. vaillantii or one of the recognized species sporulating in anthers of Scilla, or whether it represented a distinct species. A further aim was to check the phylogenetic affinity within the genus Antherospora and to expand the sampling of Antherospora species for which DNA sequence data are available.
MATERIALS AND METHODS
Specimen sampling and documentation
The specimens examined during the present work are listed in Table 1. The voucher specimens are deposited in KR, KRAM F and H.U.V. The latter abbreviation refers to the personal collection of Kálmán Vánky named as Herbarium Ustilaginales Vánky (Gabriel-Biel-Str. 5, D-72076 Tübingen, Germany). The nomenclatural novelty was registered in MycoBank (www.MycoBank.org, Crous et al. 2004).
Table 1. List of specimens, with host plants, GenBank accession numbers, spore size range, mean spore sizes with standard deviation, and reference specimens, newly examined in the course of this study.
| Smut species | Host species | GenBank acc. no. (ITS/LSU) | Spore size range (μm) | Average spore size with standard deviation (μm) | Reference specimens1 |
|---|---|---|---|---|---|
| Antherospora tractemae | Tractema verna | JN104589/JN104590 | 7.0–12.0 × 6.5–9.5 | 9.3 ± 1.3 × 7.8 ± 0.7 | UK, Scotland, Outer Hebrides, Sgeir Ghlas Leac an Aiseig, Lewis, NA–993–215 [grid reference on UK national grid], 6 May 2010, P.A. Smith, KR 28182 |
| Antherospora tractemae | Tractema verna | JN204283/JN204279 | 7.5–14.5(–16.5) × 7.0–9.5(–10.5) | 10.5 ± 2.1 × 8.5 ± 0.8 | UK, Wales, Ceredigion, Llangranog Head, SN–312–551 [grid reference on UK national grid], 19 April 2011, A.O. Chater, KRAM F-48879 – holotype |
| Antherospora tractemae | Tractema verna | JN204284/JN204280 | 6.5–12.5(–16.0) × (5.5–)6.0–9.5(–10.0) | 9.3 ± 1.8 × 7.7 ± 0.9 | UK, Wales, Ceredigion, 100 m SW of mouth of Cwm Soden, SN–361–582 [grid reference on UK national grid], 29 April 2011, A.O. Chater, KRAM F-48878 |
| Antherospora tractemae | Tractema verna | JN204285/JN204281 | 7.0–11.5(–15.0) × 6.0–10.5 | 9.6 ± 1.7 × 8.2 ± 1.1 | UK, Wales, Ceredigion, 200 m NE of Mwnt church, SN–196–521 [grid reference on UK national grid], 6 May 2011, A.O. Chater, KRAM F-48877 |
| Antherospora tractemae | Tractema verna | JN204286/JN204282 | (7.0–)8.0–13.0(–14.0) × 6.0–10.5(–11.5) | 10.1 ± 1.6 × 8.6 ± 1.2 | UK, Wales, Ceredigion, 500 m E of Mwnt church, SN–200–521 [grid reference on UK national grid], 6 May 2011, A.O. Chater, KRAM F-48876 |
| Floromyces anemarrhenae | Anemarrhena asphodeloides | JN104591/- | not analysed | not analysed | China, Inner Mongolia, Chifeng city (Ulanhad), Hongshan Distr., Hongshan, 15 July 2007, T.Z. Liu, H.U.V. 21482 |
1H.U.V. – Herbarium Ustilaginales Vánky, Gabriel-Biel-Str. 5, D-72076 Tübingen, Germany; KR – Herbarium of the Staatliches Museum für Naturkunde, Karlsruhe, Germany; KRAM F – Mycological Herbarium of the W. Szafer Institute of Botany, Polish Academy of Sciences, Kraków, Poland.
Nomenclature of anther smuts on Hyacinthaceae
The nomenclature of anther smuts on Hyacinthaceae follows Bauer et al. (2008) and Vánky (2009). The collective name Ustilago vaillantii (syn. Vankya vaillantii) refers to all anther smuts on hyacinthaceous genera and species. The name Antherospora vaillantii refers to a species complex on Muscari spp., while Antherospora vaillantii s. str. refers to the species in its narrow sense (Bauer et al. 2008).
Morphological examination
Dried fungal teliospores of the investigated specimens were mounted in lactic acid, heated to boiling point and cooled, and then examined under a Nikon Eclipse 80i light microscope at a magnification of ×1000, using Nomarski optics (DIC). Spores were measured using NIS-Elements BR 3.0 imaging software. Spore size range, and the mean and standard deviation of the size of 50 measured spores were calculated for each investigated specimen (Table 1). The species description includes the combined values from all measured specimens. LM micrographs were taken with a Nikon DS-Fi1 camera.The ornamentation of the spore surface was studied using scanning electron microscopy (SEM). For this purpose, dry spores were mounted on carbon tabs and fixed to an aluminium stub with double-sided transparent tape. The tabs were sputter-coated with carbon using a Cressington sputter-coater and viewed with a Hitachi S-4700 scanning electron microscope, with a working distance of ca. 12−13 mm. SEM micrographs were taken in the Laboratory of Field Emission Scanning Electron Microscopy and Microanalysis at the Institute of Geological Sciences, Jagiellonian University, Kraków (Poland).
DNA extraction, PCR, and sequencing
Genomic DNA was isolated directly from the herbarium specimens. For methods of isolation and crushing of fungal material, DNA extraction, amplification, purification of PCR products, sequencing, and processing of the raw data see Lutz et al. (2004). ITS 1 and ITS 2 regions of the rDNA including the 5.8S rDNA (ITS) were amplified using the primer pair ITS1-F (Gardes & Bruns 1993) and ITS4 (White et al. 1990). The 5′-end of the nuclear large subunit ribosomal DNA (LSU) was amplified using the primer pairs LR0R and LR5 or NL1 and NL4, respectively (O’Donnell 1992, 1993, White et al. 1990). Primers were used for both PCR and cycle sequencing. For amplification the annealing temperature was adjusted to 45 °C. DNA sequences determined in this study were deposited in GenBank. GenBank accession numbers are given in Fig. 1 and Table 1.
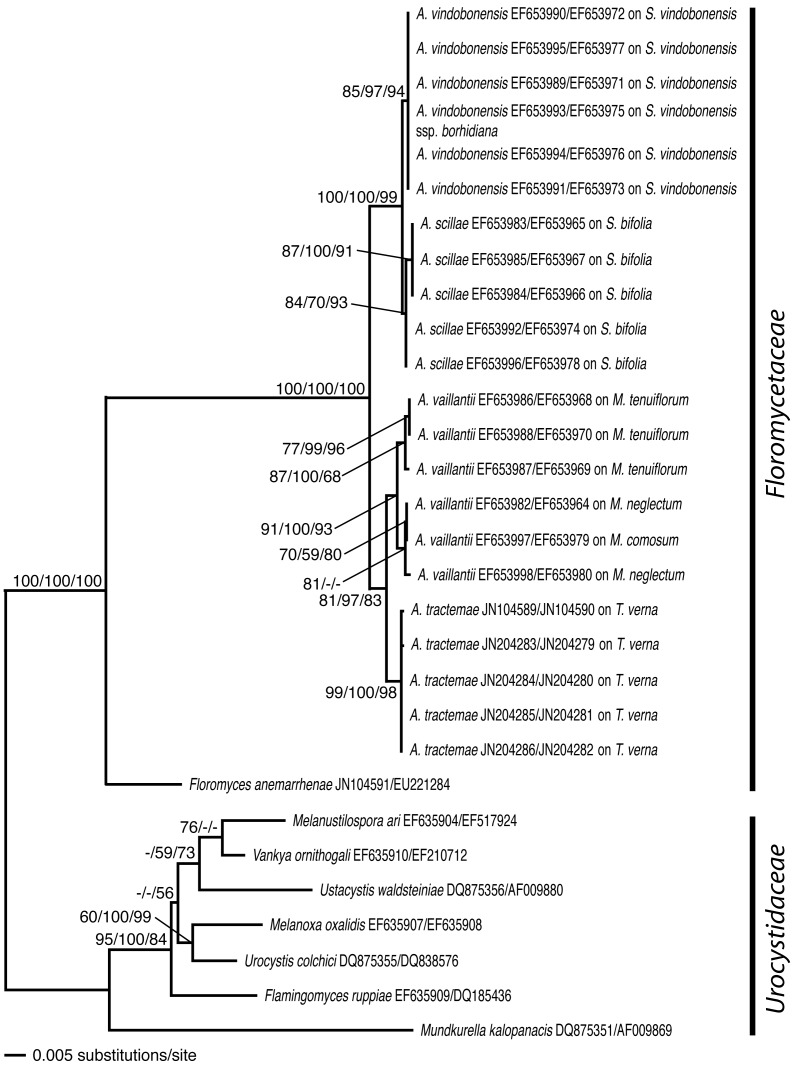
Fig. 1.
Hypothesis on phylogenetic relationships within the sampled Urocystidales based on neighbour-joining analysis of an alignment of concatenated ITS + LSU base sequences using the TrN + G model of DNA substitution. The topology was rooted with the urocystidacean species. NJ bootstrap values of 1000 replicates are indicated before slashes, numbers on branches between slashes are estimates for a posteriori probabilities, numbers on branches after slashes are ML bootstrap support values. A. = Antherospora, M. = Muscari, S. = Scilla, T. = Tractema.
Phylogenetic analyses
In addition to the sequences of Antherospora sp. on Tractema verna (ITS and LSU) and Floromyces anemarrhenae (ITS) newly obtained in this study, sequences from GenBank of the following species were used for molecular phylogenetic analyses (Begerow et al. 1997, 2006, Bauer et al. 2007, 2008, Vánky et al. 2008, Lutz et al. in press): Antherospora scillae, A. vaillantii, A. vindobonensis, Flamingomyces ruppiae, Floromyces anemarrhenae, Melanoxa oxalidis, Melanustilospora ari, Mundkurella kalopanacis, Urocystis colchici, Ustacystis waldsteiniae, and Vankya ornithogali (GenBank accession numbers included in Fig. 1).
To elucidate the phylogenetic position of the Antherospora specimens from Tractema verna their concatenated ITS + LSU sequences were analysed within a dataset covering all sequences of Floromycetaceae available in GenBank and representatives of all genera of Urocystidaceae. If present in GenBank, the respective type species were used.
Sequence alignment was obtained using MAFFT v. 6.853 (Katoh et al. 2002, 2005, Katoh & Toh 2008) using the L-INS-i option. To obtain reproducible results, manipulation of the alignment by hand as well as manual exclusion of ambiguous sites were avoided as suggested by Giribet & Wheeler (1999) and Gatesy et al. (1993), respectively. Highly divergent portions of the alignment were omitted using GBlocks 0.91b (Castresana 2000) with the following options: “Minimum Number of Sequences for a Conserved Position” to 16, “Minimum Number of Sequences for a Flank Position” to 16, “Maximum Number of Contiguous Non-conserved Positions” to 8, “Minimum Length of a Block” to 5 and “Allowed Gap Positions” to “With half”.
The resulting alignment [new number of positions: 1308 (65 % of the original 1990 positions) number of variable sites: 300] was used for phylogenetic analyses using Neighbour-Joining (NJ), a Bayesian Approach (BA) and Maximum Likelihood (ML). For NJ analysis the data were first analysed with Modeltest 3.7 (Posada & Crandall 1998) to find the most appropriate model of DNA substitution. The hierarchical likelihood ratio test proposed the TrN + G DNA substitution model. Bootstrap values were calculated from 1000 replicates. NJ analyses were carried out using PAUP v. 4.0b10 (Swofford 2001). For BA a Bayesian approach to phylogenetic inference using a Markov chain Monte Carlo technique was used as implemented in the computer program MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003). Four incrementally heated simultaneous Markov chains were run over 5 000 000 generations using the general time reversible model of DNA substitution with gamma distributed substitution rates and estimation of invariant sites, random starting trees and default starting parameters of the DNA substitution model as recommended by Huelsenbeck & Rannala (2004). Trees were sampled every 100th generation, resulting in an overall sampling of 50 001 trees. From these, the first 5 001 trees were discarded (burnin = 5 001). The trees sampled after the process had reached stationarity (45 000 trees) were used to compute a 50 % majority rule consensus tree to obtain estimates for the a posteriori probabilities of groups of species. This Bayesian approach to phylogenetic analysis was repeated five times to test the independence of the results from topological priors (Huelsenbeck et al. 2002).
ML analysis (Felsenstein 1981) was conducted with the RAxML v. 7.2.6 software (Stamatakis 2006), using raxmlGUI (Silvestro & Michalak 2010), invoking the GTRCAT and the rapid bootstrap option (Stamatakis et al. 2008) with 1000 replicates.
In line with Vánky et al. (2008), trees were rooted with the urocystidacean species Flamingomyces ruppiae, Melanoxa oxalidis, Melanustilospora ari, Mundkurella kalopanacis, Urocystis colchici, Ustacystis waldsteiniae, and Vankya ornithogali.
RESULTS
Morphological analyses
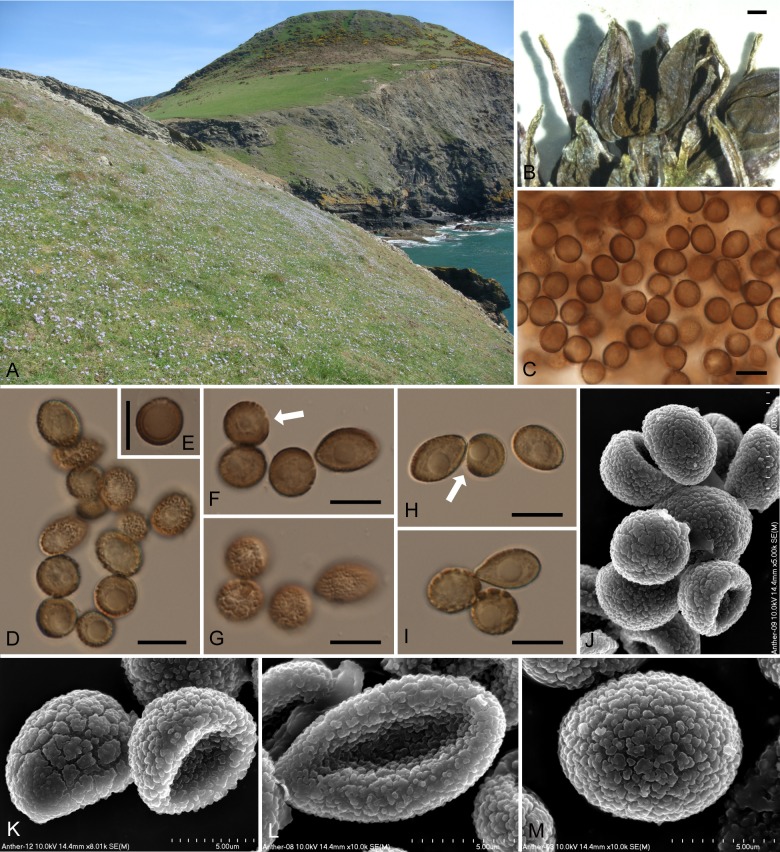
The examined specimens on Tractema verna produced olivaceous sori with teliospores in all anthers of the inflorescences. The spores in all specimens were verruculose, variable in shape and size within particular collections, and variable in spore size range and mean spore size between different collections (Table 1). The spore wall was two-layered, although the layers were not always clearly visible in some spores. The detailed morphological characteristics of anther smut on Tractema verna are included in the species description and depicted in Fig. 2.
Fig. 2.
Antherospora tractemae (KRAM F-48879 – holotype). A. Type locality on Llangranog Head, Wales, United Kingdom. B. Flower of Tractema verna with infected anthers. C–I. Spores seen by LM, median and superficial views. Note somewhat lighter coloured and thinner one side of spores indicated by arrows on pictures F and H, and two-layered spore wall visible on picture E. J–M. Ornamentation of spores seen by SEM. Bars: B = 1 mm; C–J = 10 μm; K–M = 5 μm.
Phylogenetic analyses
The ITS sequences of the five Tractema verna anther smut specimens analysed differed in one base pair (0.15 %) from each other or were identical, LSU sequences were identical. ITS and LSU sequence divergences of Antherospora species used in phylogenetic analyses are included in Table 2.
Table 2. ITS and LSU sequence divergences of Antherospora species used in phylogenetic analyses.
| Smut species | Antherospora scillae | Antherospora vaillantii | Antherospora vindobonensis | |||
| Host | Scilla bifolia | Muscari comosum | Muscari neglectum | Muscari tenuiflorum | Scilla vindobonensis | |
| Antherospora tractemae on Tractemaverna (no. of characters) | ITS (653a) | 3.4–3.5 % (22–23b) | 1.8–2.0 % (12–13) | 1.4–2.0 % (9–13) | 5.7–6.0 % (37–39) | 3.5–3.7 % (23–24) |
| LSU (658) | 1.5–1.7 % (10–11) | 0.9 % (6) | 0.9–1.2 % (6–8) | 1.2–1.4 % (8–9) | 1.7–1.8 % (11–12) | |
a A total number of nucleotide characters.
b The number of different nucleotide characters.
The different runs of BA that were performed and the ML analyses yielded consistent topologies which were congruent to the results of the NJ analysis in respect to well supported branchings (a posteriori probability greater than 54, ML bootstrap support values greater than 29). To illustrate the results, the phylogenetic hypothesis resulting from the NJ analysis is presented in Fig. 1. Bootstrap values from the NJ analysis are indicated on branches before slashes, estimates for a posteriori probabilities are indicated between slashes, numbers on branches after slashes are ML bootstrap support values.
In all analyses the Antherospora species included in previous work (Bauer et al. 2008) were inferred with high support values, and phylogenetic relationships between floromycetacean species were as in Bauer et al. (2008) and Vánky et al. (2008). The Antherospora specimens from Tractema verna formed a well supported clade that clustered as a sister group of Antherospora vaillantii with high (NJ, BA) to moderate (ML) support values. Thus, the Antherospora specimens from Tractema verna were well separated from the Antherospora species growing on Scilla species, A. scillae and A. vindobonensis.
TAXONOMY
Antherospora tractemae M. Pi¹tek & M. Lutz, sp. nov.
MycoBank MB563318
(Fig. 2)
Etymology: Named after the host plant genus.
Sori in antheris Tractemae vernae. Massa sporarum pulverulenta, olivaceo-brunnea. Sporae globosae, subglobosae, late ellipsoideae, late ovales, nonnumquam elongatae, pyriformae vel asymmetricae, 6.5–14.5(–16.5) × (5.5–)6.0–10.5(–11.5) μm, olivaceae vel flavido-brunneae, parietibus 0.5–1.3 μm crassis, dense verruculosis, a latere visae fere levigatae vel subtiliter sinuatae.
Typus: UK: Wales: Ceredigion, Llangranog Head, on Tractema verna (syn. Scilla verna), 19 Apr. 2011, A.O. Chater (KRAM F-48879 – holotypus; ITS/LSU sequences GenBank accession nos JN204283 and JN204279).
Parasitic on Tractema verna. Sori in the anthers, producing olive-brown, powdery mass of spores inside the pollen sacs. Infection systemic, all anthers of a plant infected. Spores globose, subglobose, broadly ellipsoidal, broadly ovoid, sometimes elongated, pyriform or asymmetrical, 6.5–14.5(–16.5) × (5.5–)6.0–10.5(–11.5) μm [av. ± SD, 9.8 ± 1.8 × 8.2 ± 1.0 μm, n = 250/5], olivaceous or yellowish-brown, sometimes lighter coloured on one side; wall two-layered, ca. 0.5–1.3 μm thick, thinner on the lighter side, finely, densely verruculose, spore profile almost smooth or finely wavy.
Additional specimens examined (paratypes): UK: Scotland: Outer Hebrides, Sgeir Ghlas Leac an Aiseig, Lewis, on Tractema verna, 6 May 2010, P.A. Smith (KR 28182); Wales: Ceredigion, 100 m SW of mouth of Cwm Soden, on Tractema verna, 29 Apr. 2011, A.O. Chater (KRAM F-48878); Ceredigion, 200 m NE of Mwnt church, on Tractema verna, 6 May 2011, A.O. Chater (KRAM F-48877); Ceredigion, 500 m E of Mwnt church, on Tractema verna, 6 May 2011, A.O. Chater (KRAM F-48876).
Ecology: The infected plants were found in April and May, peak flowering time for Tractema verna. The habitats for all the collections are broadly similar, consisting of short coastal turf on shallow soils. The Outer Hebrides locality is on pockets of maritime peat on a rocky substrate, referable to the MC10 Festuca rubra–Plantago spp. maritime grassland community (plant community nomenclature follows Rodwell et al. 1991–2000). The Welsh sites are in coastal heath vegetation referable to the H7 Calluna vulgaris–Scilla verna heath and H8d Scilla verna subcommunity of Calluna vulgaris–Ulex gallii heath, as well as in the Armeria subcommunity of the MC10 community. Tractema verna is locally common around the rocky western coasts of Britain, and occurs in a number of maritime communities. The smut often infects large numbers of individuals within a population, and its incidence varies greatly from year to year.
DISCUSSION
The anther smuts of the genus Antherospora offer few morphological characteristics for inter-specific differentiation. This is the reason why they were formerly identified as a single species, Ustilago vaillantii (syn. Vankya vaillantii)(e.g. Vánky 1994). It appears that only a few species could be differentiated based on differences in spore sizes, and to a lesser extent also the spore wall thickness and the localization of the sori which is limited to the anthers or to the anthers and the surface of the inner floral organs (Bauer et al. 2008, Vánky et al. 2008, Vánky 2009). The recognition of Antherospora species that lack morphological differences is difficult or impossible without the support of molecular data. The variation of spore sizes between different collections of the same species (Bauer et al. 2008) may additionally complicate the situation with species delimitation. The differences in spore size range and mean spore size between different collections on Tractema verna (Table 1) confirm the variability of this character in Antherospora species, and it seems that whenever possible the morphology should be characterized based on collections from different populations.
In spore size range (assessed from five specimens), the anther smut of Tractema verna is intermediate between Antherospora tourneuxii and A. urgineae on the one side and A. eucomis, A. scillae, A. vaillantii, and A. vindobonensis on the other side. Molecular data are not available for Antherospora eucomis, A. tourneuxii and A. urgineae. Other than infecting different host plant genera (Eucomis, Bellevalia and Charybdis respectively), A. eucomis can be separated by having smaller spores and a thinner spore wall, while the two remaining species have somewhat larger spores and thinner spore walls (Bauer et al. 2008, Vánky 2009, Vánky et al. 2010). The spores of Antherospora scillae, A. vindobonensis (on Scilla) and A. vaillantii (on Muscari) are smaller than those of the anther smut of Tractema verna, and the spore wall is additionally thinner in A. vaillantii. The spore wall thickness of Antherospora scillae and A. vindobonensis (0.8–1.5 μm, according to the key to Antherospora species by Vánky 2009) is comparable to those of the anther smut of Tractema verna, and different from all remaining Antherospora species that have spore walls of 0.5–0.8 μm thick. The molecular phylogenetic analyses separate the specimens on Tractema verna from these three Antherospora spp. (Fig. 1) and both ITS and LSU sequences differ significantly (Table 2). In conclusion, the morphology, the genetic difference, the results of the molecular phylogenetic analyses and different host plant genera support the recognition of the anther smut of Tractema verna as a new species, for which the name Antherospora tractemae is proposed in this study.
The host plant was at first assigned to the genus Scilla (S. verna) according to most taxonomic treatments (e.g. McNeill 1980), and thus it was initially assumed that its anther smut could be related to the two Antherospora species on Scilla already described. The molecular phylogenetic analyses revealed that the smut sporulating in anthers of Tractema verna occupies a position basal to the lineage of Muscari-parasitizing Antherospora. A subsequent survey of the botanical literature revealed that Scilla is a polyphyletic genus and that Scilla verna actually belongs to the distinct genus Tractema (Speta 1998). Tractema verna has not as yet been included in phylogenetic analyses, but the related species Tractema monophyllos, the type of the genus Tractema, clusters distantly from the lineage attributed to Scilla s. str. (Pfosser & Speta 1999). Thus, the phylogenetic position of Antherospora tractemae supports the disentanglement of Scilla verna from Scilla s. str. This emphasizes the importance of the assignment of host species to the correct genus in order to promote the understanding of the evolutionary relationships between smut fungi and their host plants. Furthermore, it supports the long known hypothesis that plant parasitic fungi, especially rusts and smuts, can indicate relationships between their host plants (e.g. Savile 1954, 1979, Nannfeldt 1968, Hijwegen 1979, 1988, Kukkonen & Timonen 1979).
The large number of host plants reported for Ustilago vaillantii (syn. Vankya vaillantii) (Zundel 1953, Vánky 1994) suggested that further species of Antherospora are likely to emerge. The descriptions of new species that lack morphological characteristics or have very subtle morphological differences needs to be supported by molecular data. The introduction of new species names based solely on supposed host specificity may be risky, because it is probable that within Antherospora there are species that have more than one host species as was evidenced for Antherospora vaillantii s. str. which is able to infect both Muscari comosum and M. neglectum (Bauer et al. 2008).
Acknowledgments
We thank Krzysztof Pawłowski (Kraków, Poland) and Michael Weiß (Tübingen, Germany) for translating the Latin description, Michael Weiß, Sigisfredo Garnica and Robert Bauer (Tübingen, Germany) for providing the molecular lab, Anna Łatkiewicz (Kraków, Poland) for help with the SEM pictures, and Roger G. Shivas (Indooroopilly, Australia) for helpful comments on the manuscript.
REFERENCES
- Bauer R, Lutz M, Begerow D, Piçtek M, Vánky K, Bacigálová K, Oberwinkler F. (2008) Anther smut fungi on monocots. Mycological Research 112: 1297–1306 [DOI] [PubMed] [Google Scholar]
- Bauer R, Lutz M, Piçtek M, Vánky K, Oberwinkler F. (2007) Flamingomyces and Parvulago, new genera of marine smut fungi (Ustilaginomycotina). Mycological Research 111: 1199–1206 [DOI] [PubMed] [Google Scholar]
- Begerow D, Bauer R, Oberwinkler F. (1997) Phylogenetic studies on nuclear LSU rDNA sequences of smut fungi and related taxa. Canadian Journal of Botany 75: 2045–2056 [Google Scholar]
- Begerow D, Stoll M, Bauer R. (2006) A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98: 906–916 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Denchev CM, Giraud T, Hood ME. (2009) Three new species of anthericolous smut fungi on Caryophyllaceae. Mycologia Balcanica 6: 79–84 [Google Scholar]
- Felsenstein J. (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution 17: 368–376 [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118 [DOI] [PubMed] [Google Scholar]
- Gatesy J, DeSalle R, Wheeler W. (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Molecular Phylogenetics and Evolution 2: 152–157 [DOI] [PubMed] [Google Scholar]
- Giribet G, Wheeler WC. (1999) On gaps. Molecular Phylogenetics and Evolution 13: 132–143 [DOI] [PubMed] [Google Scholar]
- Hijwegen T. (1979) Fungi as plant taxonomists. Symbolae Botanicae Upsalienses 22(4): 146–165 [Google Scholar]
- Hijwegen T. (1988) Coevolution of flowering plants with pathogenic fungi. In: Coevolution of Fungi with Plants and Animals (Pirozynski KA, Hawksworth DL, eds): 63–77 London: Academic Press; [Google Scholar]
- Huelsenbeck JP, Larget B, Miller RE, Ronquist F. (2002) Potential applications and pitfalls of Bayesian inference of phylogeny. Systematic Biology 51: 673–688 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Rannala B. (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Systematic Biology 53: 904–913 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics Applications Note 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2008) Recent developments in the MAFFT multiple sequence alignment program (outlines version 6). Briefings in Bioinformatics 9: 286–298 [DOI] [PubMed] [Google Scholar]
- Kemler M, Lutz M, Göker M, Oberwinkler F, Begerow D. (2009) Hidden diversity in the non-caryophyllaceous plant-parasitic members of Microbotryum (Pucciniomycotina: Microbotryales). Systematics and Biodiversity 7: 297–306 [Google Scholar]
- Kukkonen I, Timonen T. (1979) Species of Ustilaginales, especially of the genus Anthracoidea, as tools in plant taxonomy. Symbolae Botanicae Upsalienses 22(4): 166–176 [Google Scholar]
- Le Gac M, Hood ME, Fournier E, Giraud T. (2007) Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution 61: 15–26 [DOI] [PubMed] [Google Scholar]
- Legon NW, Henrici A, Roberts PJ, Spooner BM, Watling R. (2005) Checklist of the British and Irish Basidiomycota. Royal Botanic Gardens, Kew, Great Britain: [Google Scholar]
- Lutz M, Bauer R, Begerow D, Oberwinkler F, Triebel D. (2004) Tuberculina, rust relatives attack rusts. Mycologia 96: 614–626 [PubMed] [Google Scholar]
- Lutz M, Göker M, Piçtek M, Kemler M, Begerow D, Oberwinkler F. (2005) Anther smuts of Caryophyllaceae: molecular characters indicate host-dependent species delimitation. Mycological Progress 4: 225–238 [Google Scholar]
- Lutz M, Piçtek M, Kemler M, Chlebicki A, Oberwinkler F. (2008) Anther smuts of Caryophyllaceae: molecular analyses reveal further new species. Mycological Research 112: 1280–1296 [DOI] [PubMed] [Google Scholar]
- Lutz M, Vánky K, Bauer R. (In press) Melanoxa, a new genus in the Urocystidales (Ustilaginomycotina). Mycological Progress. DOI 10.1007/s11557-010-0737-7 [Google Scholar]
- McNeill J. (1980) Scilla L. In: Flora Europaea (Tutin TG, Heywood VH, Burges NA, Valentine DH, Walters SM, Webb DA, eds) 5: 41–43 Cambridge University Press; [Google Scholar]
- Nannfeldt JA. (1968) Fungi as plant taxonomists. Acta Universitatis Upsaliensis 17: 85–95 [Google Scholar]
- O’Donnell KL. (1992) Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris). Current Genetics 22: 213–220 [DOI] [PubMed] [Google Scholar]
- O’Donnell KL. (1993) Fusarium and its near relatives. In: The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. (Reynolds DR, Taylor JW, eds): 225–233 CAB International, Wallingford, United Kingdom: [Google Scholar]
- Pfosser M, Speta F. (1999) PhylogeneticsofHyacinthaceae based on plastid DNA sequences. Annals of the Missouri Botanical Garden 86: 852–875 [Google Scholar]
- Posada D, Crandall KA. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- Refrégier G, Le Gac M, Jabbour F, Widmer A, Shykoff JA, Yockteng R, Hood ME, Giraud T. (2008) Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation. BMC Evolutionary Biology 8: 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell JS, Pigott CD, Ratcliffe DA, Malloch AJC, Birks HJB, et al (1991–2000) British Plant Communities Vols 1–5. Cambridge University Press, Cambridge, UK: [Google Scholar]
- Ronquist FR, Huelsenbeck JP. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Savile DBO. (1954) The fungi as aids in the taxonomy of the flowering plants. Science 120: 583–585 [DOI] [PubMed] [Google Scholar]
- Savile DBO. (1979) Fungi as aids to plant taxonomy: methodology and principles. Symbolae Botanicae Upsalienses 22(4): 135–145 [Google Scholar]
- Silvestro D, Michalak I. (2010) raxmlGUI: a graphical front-end for RAxML. Available at http://sourceforge.net/projects/raxmlgui/ [Google Scholar]
- Speta F. (1998) Systematische Analyse der Gattung Scilla L. s.l. (Hyacinthaceae). Phyton 38: 1–141 [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2001) PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts, U.S.A: [Google Scholar]
- Vánky K. (1994) European Smut Fungi. Stuttgart: G. Fischer Verlag; [Google Scholar]
- Vánky K. (2009) Taxonomic studies on Ustilaginomycetes – 29. Mycotaxon 110: 289–324 [Google Scholar]
- Vánky K, Abbasi M, Samadi S. (2010) Additions to the knowledge of smut fungi (Ustilaginomycetes) of Iran. Rostaniha 11(2): 191–198 [Google Scholar]
- Vánky K, Lutz M, Bauer R. (2008) Floromyces, a new genus of Ustilaginomycotina. Mycotaxon 104: 171–184 [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols, a guide to methods and applications. (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds): 315–322 Academic Press, San Diego, U.S.A: [Google Scholar]
- Zundel GL. (1953) The Ustilaginales of the world. Pennsylvania State College School of Agriculture Department of Botany Contribution 176: xi + 1–410 [Google Scholar]